Abstract
Rhizobia are nitrogen-fixing soil bacteria that establish endosymbiosis with some leguminous plants. The completion of several rhizobial genome sequences provides opportunities for genome-wide functional studies of the physiological roles of many rhizobial genes. In order to carry out genome-wide phenotypic screenings, we have constructed a large mutant library of the nitrogen-fixing symbiotic bacterium, Mesorhizobium loti, by transposon mutagenesis. Transposon insertion mutants were generated using the signature-tagged mutagenesis (STM) technique and a total of 29 330 independent mutants were obtained. Along with the collection of transposon mutants, we have determined the transposon insertion sites for 7892 clones, and confirmed insertions in 3680 non-redundant M. loti genes (50.5% of the total number of M. loti genes). Transposon insertions were randomly distributed throughout the M. loti genome without any bias toward G+C contents of insertion target sites and transposon plasmids used for the mutagenesis. We also show the utility of STM mutants by examining the specificity of signature tags and test screenings for growth- and nodulation-deficient mutants. This defined mutant library allows for genome-wide forward- and reverse-genetic functional studies of M. loti and will serve as an invaluable resource for researchers to further our understanding of rhizobial biology.
Key words: Mesorhizobium loti, signature-tagged mutagenesis, mutant library, reverse genetics
1. Introduction
Nitrogen-fixing rhizobia are of great agronomic benefit, allowing many leguminous crops to be grown without nitrogen fertilizer by forming endosymbiotic relationships with the plants. In the course of the symbiosis, rhizobia elicit formation of specialized organs, ‘root nodules’, on the roots of compatible host legumes. Inside the nodules, the bacteria convert inert atmospheric dinitrogen into biologically usable ammonia. Since this symbiotic relationship is established by the results of highly regulated molecular dialogues between host plant and rhizobia, the system of symbiotic nitrogen fixation is therefore a very suitable model for studies of plant–microbe interactions.
The agronomic and biological importance of the rhizobia has accelerated the determination of the full genome sequences of several rhizobial species.1–6 The availability of complete genome sequences has drastically changed the strategy for studying rhizobial genetics. Gene identification and functional assignment have been accelerated by utilizing the genome sequences. Simultaneously with the completions of rhizobial genome sequences, post-genomic researches on several rhizobia have rapidly progressed. Comprehenzsive transcriptome or proteome analyses have been conducted to examine physiological states of rhizobia under a variety of conditions, including symbiosis with the host legume or nutrient-depleted conditions.7–12 In addition, we previously conducted a large-scale interactome analysis in order to propose functional relationships between known and unknown rhizobial genes.13 These advances in rhizobial genetics have generated a vast amount of information about gene function and regulation; however, the advances have simultaneously raised the need for development of novel molecular tools which make use of the results of these functional analyses.
One of the most utilized genetic approaches to reveal gene function is the disruption of particular genes and subsequent phenotypic characterization of the mutants. Comprehensive collections of single gene disruptants have been systematically constructed in several model organisms by transposon random mutagenesis or targeted gene disruption approaches.14–18 These analyses have contributed to the discovery of essential genes of the organisms and have provided fundamental tools for functional studies of many known and unknown genes. In rhizobia, recent genome-wide analyses have provided various intriguing and useful starting points for more detailed functional studies;7–13 however, there are still large gaps between such systematic analyses and the availability of the corresponding gene mutants. Such gaps have resulted primarily because targeted gene disruption achieved by homologous recombination is not suitable for systematic generation of mutants in rhizobia because of its low efficiency. In addition, screening of mutants in some test conditions is problematic because phenotypes of individual mutants have to be evaluated one-by-one. Therefore, more effective approaches to generate large mutant collections and allow simultaneous screening of many mutants are needed.
In this study, we conducted the first large-scale random mutagenesis of Mesorhizobium loti, an endosymbiont of the model legume Lotus japonicus, using a signature-tagged mutagenesis (STM) technique. STM is based on transposon insertional mutagenesis that allows large numbers of mutants to be analyzed simultaneously. This is accomplished by tagging each mutant with a unique short DNA sequence so that it can subsequently be distinguished from other mutants by detecting unique signature tags. This system has been used with a variety of bacteria, and, in particular, has been applied to discovery of virulence genes in several microbial pathogens.19 Recently, a large STM library was constructed in Sinorhizobium meliloti strain 1021,20 a symbiont of Medicago truncatula and contributed to the discovery of many genes relevant to symbiosis and competitiveness.21 However, genome-wide generation of disruption mutants in M. loti, in addition to the set of S. meliloti, is also very valuable for several reasons. First, M. loti and S. meliloti exhibit different aspects as symbiont for legumes. They naturally exhibit distinct host-specificity and form different types of nodules (determinate and indeterminate nodules) on their respective model legume hosts, L. japonicus and M. truncatula. With respect to this symbiosis, previous studies have discovered several rhizobial factors and metabolic cycles required for nodulation and nitrogen-fixation.22–24 However, comparative verifications of the significance of these factors on respective symbioses have not been fully explored. Second, comparative genomics analyses have revealed the existence of genes that are conserved, as well as many that are specific to each species,2,25 but the molecular tools available for the functional comparisons of these genes are extremely limited. Therefore, large collections of disruption mutants on the individual species are necessary to reveal common and unique biological aspects of the two species.
In this report, we developed a large signature-tagged mutant library of M. loti in order to provide fundamental tools for functional studies of M. loti genes and characterized it with the gene coverage and transposon insertion distribution of the library. Sequencing of transposon insertion sites enabled us to collect defined sets of transposon mutants. Furthermore, pilot experiments showed the utility of the mutants and provide basic experimental conditions that can be used for future mutant screenings. This large signature-tagged mutant library will be an important and powerful resource for future functional genomics in M. loti.
2. Materials and methods
2.1. Bacterial strains and growth conditions
Escherichia coli strains DH5α and MT616 were used as hosts for the transposon plasmids and pRK600 helper plasmid,26 respectively. Transposon insertion mutants were generated in the wild-type M. loti strain MAFF303099. Mesorhizobium loti was grown at 30°C in tryptone yeast (TY) extract medium27 supplemented with phosphomycin (100 µg/ml). Escherichia coli was grown at 37°C in Luria-Bertani (LB) medium. LB medium was supplemented with streptomycin (100 µg/ml) and spectinomycin (100 µg/ml) for DH5α donor cells harboring transposon plasmids and with chloramphenicol (20 µg/ml) for MT616 helper cells. Transposon insertion mutants were selected on TY medium supplemented with streptomycin, spectinomycin and phosphomycin at the same concentration as above.
2.2. Construction of signature-tagged transposon plasmids
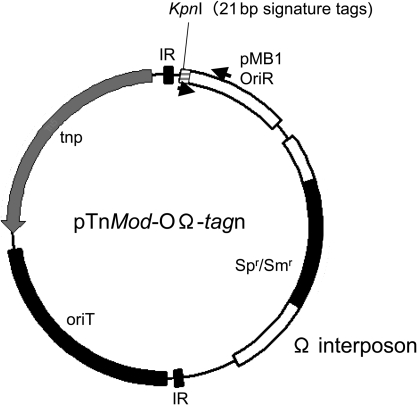
The backbone of the transposon plasmid was pTnMod-OGm.28 The Ω interposon cassette was excised from pHP45Ω29 and inserted by replacing a SacI fragment containing the gentamicin resistance gene cassette of pTnMod-OGm. The resultant construct was designated pTnMod-OΩ. Twenty-seven different 21-base oligonucleotides (Supplementary Table S1) were annealed to complementary molecules to make double-strand tags. Each oligonucleotide tag was inserted individually into the blunted KpnI site of pTnMod-OΩ (Fig. 1). These 27 signature-tagged constructs were designated pTnMod-OΩ-tag1 to pTnMod-OΩ-tag41.
Figure 1.
Vector construct used for STM of M. loti. The tagged transposon plasmids were constructed from the backbone of pTnMod-OGm.28 The omega-interposon cassette (Spr/Smr) was excised from pHP45Ω29 and inserted into the SacI site of pTnMod-OGm. Oligonucleotide tags (21 bp) were incorporated into the KpnI site. Arrows indicate the locations of the STM common primer and the tag-specific primer (Supplementary Table S1). OriT is an RP4 origin of transfer. IR and tnp represent the inverted repeat and Tn5 transposase, respectively.
2.3. Transposon mutagenesis
Introduction of tagged transposon plasmids into M. loti was carried out by tri-parental mating.30 Escherichia coli strains DH5α and MT616 (containing the pRK600 helper plasmid) were used as the conjugation donor and helper, respectively. An overnight culture of donor and helper (0.5 ml each) was mixed with 1 ml of a 2-day culture of M. loti. After washing several times with sterilized water, the cell mixture was suspended in 50 µl of sterilized water and spotted onto a sterilized membrane filter (MILLIPORE, Billerica, MA, USA, pore size 0.45 µm, Cat. No. HAWG047S3) that was placed on TY agar medium without antibiotics. After an 8 h incubation on the membrane filter at 30°C, all the cells were collected and suspended in 1 ml of sterilized water. Suspended cells were plated on TY selection medium supplemented with phosphomycin, spectinomycin and streptomycin, and grown further until positive colonies appeared. Finally, individual positive colonies were picked up randomly in 96-well plates and stored at −80°C.
2.4. Determination of transposon insertion sites
Transposon insertion sites were determined by direct sequencing of transposon-genome flanking region. Cell culture and genomic DNA isolation were carried out in 96-well plate format. Five microlitter of stocked mutant clones were inoculated into 1 ml of TY medium containing phosphomycin (100 µg/ml), streptomycin (100 µg/ml) and spectinomycin (100 µg/ml), and the cultures were grown in 96-well deep-well plate (Axygen Bioscience, Union City, CA, USA) at 30°C for 2 days. After centrifugation of cultured cells at 4000 rpm for 15 min, genomic DNA of mutants was prepared using AquaPure Genomic DNA isolation kit (Bio-Rad Laboratories, Tokyo, Japan). Collected cells in each well were suspended in 150 µl of Genomic DNA Lysis Solution and were incubated at 80°C for 15 min. RNaseA solution (0.75 µl) was added to cell lysate, and the mixture was incubated at 37°C for 60 min. After the incubation on ice for 15 min, 50 µl of protein precipitation solution was added to each well and the samples were mixed well by vortex. Samples were then transferred individually to 1.5 ml microcentrifuge tube and were centrifuged at 15 000 rpm for 15 min. Supernatants (100–125 µl) were transferred to new 96-well PCR plate containing 100 µl of isopropanol and were mixed well by inverting plate. Genomic DNA precipitated by centrifugation (4000 rpm, 30 min) was rinsed three times with 200 µl of 70% ethanol. Genomic DNA was then dried and dissolved in 20 µl of DNA hydration solution. After heating the samples at 65°C for 60 min, concentration of genomic DNA was measured and ∼2 µg of genomic DNA was used for direct sequence. Direct sequencing was performed with the STM-seq primer (5′-TTTGCTGGCCTTTTGCTCACATGTTCTTTC-3′) using the following PCR conditions: 98°C for 3 min, followed by 70 cycles of 97°C for 15 s and 60°C for 4 min. The composition of reaction mixture for direct sequence was as follows. Two micrograms of genomic DNA, 1 µl of STM-seq primer (3.2 µM), 1 µl of BigDye Terminator premix (Applied Biosystem, Foster City, CA, USA), 3.5 µl of 5× sequencing buffer (Applied Biosystems) and sterilized water were added up to 20 µl.
For some clones which were not successfully identified by direct sequencing, we determined the transposon integration sites by two-round inverse PCR. For inverse PCR, genomic DNA (∼1 µg) isolated from each mutant was digested with SalI and XhoI, and then self-ligated using a DNA ligation kit (TAKARA Bio, Shiga, Japan). Using a self-ligated DNA template and a primer pair (5′-TTCGCCACCTCTGACTTGAGCGTCG-3′ and 5′-GAATTGATCCGGTGGATGAC-3′), first round PCR was performed using the following cycling conditions: 98°C for 2 min, followed by 35 cycles of 97°C for 15 s, 55°C for 45 s, 72°C for 3 min and a final extension at 72°C for 10 min. A 1 µl aliquot from the first round PCR product was added to a secondary PCR reaction mixture containing a nested primer pair (5′-TTTGCTGGCCTTTTGCTCACATGTTCTTTC-3′ and 5′-ACGGTTTACAAGCATAAAGC-3′). Second round PCR was performed using the following PCR conditions: 98°C for 2 min, followed by 35 cycles of 97°C for 15 s, 57°C for 45 s, 72°C for 3 min and a final extension at 72°C for 10 min. After the secondary PCR, products were purified by polyethylene glycol precipitation and suspended in 10 µl distilled water. The composition of sequence reaction mixture with amplified PCR product was as follows. Two microliters of PCR product, 1 µl of sequencing primer (5′-ACGGTTTACAAGCATAAAGC-3′) (3.2 µM), 1 µl of BigDye Terminator premix (Applied Biosystems), 3.5 µl of 5× sequencing buffer (Applied Biosystems) and 12.5 µl of sterilized water. Cycle sequencing was performed at 96°C for 1 min and then at 96°C for 10 s, 50°C for 5 s and 60°C for 4 min for 25 cycles. All sequencings were analyzed by ABI 3730 autosequencer. The resulting sequences were subsequently subjected to BlastN search against the M. loti genome sequence (http://www.kazusa.or.jp/rhizobase/Mesorhizobium/index.html) to determine the transposon insertion sites.
2.5. PCR and real-time PCR
The specificity of oligonucleotide tags was examined by PCR. Tag-containing DNA fragments (160 bp) were amplified from genomic DNA using the Herculase II Fusion DNA Polymerase (STRATAGENE, La Jolla, CA, USA), and forward (Supplementary Table S1) and reverse (STM-common, 5′-TTCGCCACCTCTGACTTGAGCGTCG-3′) primers were used for amplification. PCR reactions were conducted in GeneAmp PCR system 9700 (Applied Biosystems) using the following conditions: initial denaturation at 98°C for 2 min, followed by 25 cycles of 97°C for 15 s, 68°C for 1 min and a final extension at 72°C for 10 min. Amplified products were analyzed on 2% agarose gels.
The population of growth-deficient mutant within a mutant pool was examined by real-time PCR. Genomic DNA isolated from input and output pools was used as the DNA template. Real-time PCR was performed using a DyNAmo HS SYBR Green qPCR kit (Finnzyme, Espoo, Finland) and reactions were detected by a DNA Engine Opticon system (Bio-Rad). The conditions used for real-time PCR are as follows: 15 min at 95°C, followed by 35 cycles of 94°C for 30 s, 58°C for 30 s and 72°C for 30 s and a final extension at 72°C for 10 min. Quantitative and specificity (melting curve) analyses of amplified PCR products were carried out using associated software in the Opticon system (Opticon monitor3) by following the manufacture's protocol.
2.6. Detection of signature tags from plant samples
Seeds of L. japonicus (MG-20 Miyakojima) were sterilized and germinated on half-strength B&D agar medium. Six-day-old seedlings were transferred to B&D agar medium or inoculation pots filled with vermiculite and were then inoculated with 1 × 107 cell/ml of mutant mixture.
For tag detection from nodules, detached nodules were washed twice with a solution of 0.5%-SDS-100 mM NaCl and then soaked in 70% ethanol for 5 min. After five times washes with sterilized water, total DNA was isolated using a DNeasy plant mini kit (Qiagen, Hilden, Germany). For amplification of a tag-containing fragment from a single nodule, nodule was washed as above and crushed in 700 µl sterilized water. After boiling the samples at 95°C for 5 min, 5 µl of supernatant was used for PCR. PCR reactions with nodule samples used the following conditions: initial denaturation at 98°Cfor 2 min, followed by 25 cycles of 97°C for 15 s, 68°C for 1 min and a final extension at 72°C for 10 min. Amplified products were analyzed on 2% agarose gels.
3. Results and discussion
3.1. Construction of a signature-tagged mutant library
In this study, we adapted transposon-based STM techniques to genome-wide mutagenesis of the symbiotic bacterium, M. loti. As a transposon construct, the plasposon plasmid pTnMod-OGm which carries a Tn5-based mini-transposon and conditional origin of replication28 was used for mutagenesis (Fig. 1). In order to screen for genes which were inactivated by transposon insertion and those affected polarity effect of the inserted transposon, we introduced the Ω interposon cassette by replacing the gentamycin resistance gene (aacC1) of pTnMod-OGm (Fig. 1). Since pTnMod is a broad host range suicide vector and the insertion of the Ω fragment confers spectinomycin and streptomycin resistance, only mutants in which the transposon fragment is integrated into the M. loti genome can be selected effectively by their antibiotic resistance.
In order to distinguish individual transposon mutants, we introduced 21 bp short oligonucleotide tags that have been used in a previous study.31 Since these oligonucleotide tags were designed to have similar G+C content and melting temperature, they are feasible for PCR-based mutant screening. Considering the isolation of symbiotic mutants from root nodules in which reduction of variation was expected, number of tag variation need to be restricted to obtain reliable results of screening from a reasonable number of plant materials (root nodules). Therefore, we further selected tags from the original 41 tags31 by analyzing their specificity of PCR amplification. We selected a total of 29 unique tags based on the reproducibility of PCR amplification and the plasposon plasmids tagged with each of 29 tags (pTnMod-OΩ-tagn) were then individually mobilized into wild-type M. loti by tri-parental mating. Among the 29 tags, two tag plasmids which yielded significantly small numbers of transposon mutant colonies were eliminated, and the remaining 27 tags (Supplementary Table S1) were used for further analysis (name of tags are designated according to Hunt et al.31). In order to construct independent mutant libraries, tri-parental mating and collection of transposon mutants were carried out separately for each oligonucleotide tag. As the result of 63 independent tri-parental mating experiments, 29 330 independent transposon mutants were obtained (Table 1). Individual mutants were stored in 96-well plates and mutant names (ID) were assigned according to their position in the plate with prefix indicating tag number followed by ‘T’ (i.e. 07T02d09 is a mutant integrated with tag7 plasmid).
Table 1.
Summary of experimental results
| Mutants collected | 29 330 |
| Mutants sequenced for mapping of transposon insertion site | 9344 |
| Mapped insertion locations | 7892 |
| Unique insertion locations | 7586 |
| Insertion between ORFs (intergenic region) | 1156 |
| Chromosome | 1018 |
| pMLa | 80 |
| pMLb | 58 |
| Insertion inside ORFs | 6430 |
| Chromosome | 6000 |
| pMLa | 318 |
| pMLb | 112 |
| Insertion inside ORFs (non-redundant) | 3680 |
| Genes supported by more than two mutant alleles | 1592 |
The coverage of the collected mutants is approximately fourfold with respect to the 7281 ORFs of M. loti.1 If the transposon insertions are evenly distributed throughout the M. loti genome, the mean distance between independent insertions is estimated to be 259 bp, and thus 91% of M. loti genes are expected to carry at least one insertion since M. loti genes shorter than 259 bp account for 9% of all genes.
3.2. Utility of STM mutants
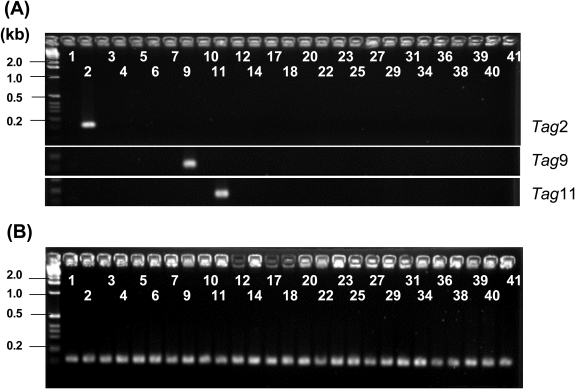
The STM technique allows for the simultaneous screening of multiple mutants in mixed populations by detecting unique DNA tags. However, previous studies have reported that non-specific amplification or failure in tag amplification from mutant chromosomal DNA is a major inherent problem that affects the efficiency and reproducibility of the screening procedure.31,32 Therefore, the specificity and efficiency of tag amplification after integration into the genome needs to be tested in the respective targeted organisms. To investigate the possibility of cross-detection among the 27 signature tags used in this study, we conducted test amplifications by PCR. Fig. 2A shows representative results in which PCR was performed with genomic DNA from three individual tagged mutants (Tag2, Tag9 and Tag11). When a tag-specific primer (Supplementary Table S1) and the STM common primer (5′-TTCGCCACCTCTGACTTGAGCGTCG-3′) that anneals 120 bp downstream of the integrated oligonucleotide tag were used for PCR, specific amplification was observed only in reactions containing the appropriate tag-specific primer. A series of PCR reactions performed on the other oligonucleotide tags produced the same results as Fig. 2A, indicating that there are no cross-amplifications among the designed oligonucleotide tags. In addition, all 27 oligonucleotide tags were amplified specifically when genomic DNA from a mixture of mutants was used as the template (Fig. 2B). These test amplifications demonstrated that the applied oligonucleotide tags can be recovered specifically from the M. loti genome and that the 27 individually tagged mutants can be distinguished from one another by detecting these signature tags.
Figure 2.
Representative gels of control experiments testing the specificity of signature oligonucleotide tags. PCR was performed with genomic DNA isolated from mutants with pTnMod-OΩ-Tag2 (upper), pTnMod-OΩ-Tag9 (middle) or pTnMod-OΩ-Tag11 (bottom) alone (A), and using genomic DNA isolated from cultures containing a mixture of 27 mutants (B). The STM-common primer was used with one of the 27 different tag-specific primers (Supplementary Table S1) in each reaction. The numbers indicate the combination of primers.
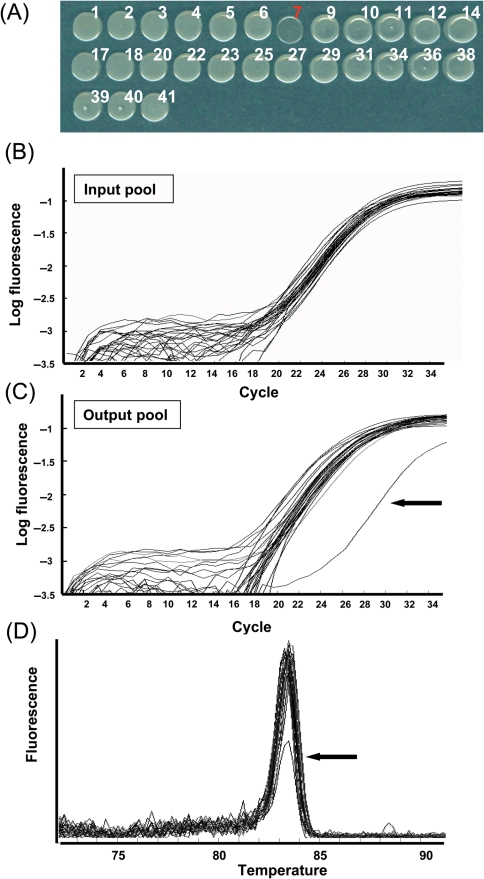
In order to test the utility of our mutant sets, we carried out additional pilot experiments using both in vitro and in vivo conditions. In vitro experiments were conducted with a pool of mutants that contains a mutant with an attenuated growth on rich medium. To make a mutant set for the test experiment, one mutant was selected randomly from each 27-tag library and cultured individually. One growth-deficient clone (07T02d09) was included in the pool (Fig. 3A). Independently cultured clones were then mixed at same concentration to make an input pool. As for an output pool, the input pool was diluted 500-fold and cultured further for 2 days. After isolation of genomic DNA from both pools, the tag population was examined by real-time PCR. Real-time PCR was performed using the SYBR Green detection system with the same primer set used to test the cross-reactivity (Fig. 2). Fig. 3 illustrates the results of real-time PCR of the input (Fig. 3B) and output (Fig. 3C) pool samples. Consistent with the growth on agar medium (Fig. 3A), the quantity of PCR product derived from the growth-deficient mutant (07T02d09) was significantly lower than the other tags which were all amplified to similar levels (Fig. 3B and C). In addition, after each round of PCR amplification, a melting curve was generated for each run of 27 samples (Fig. 3D). The melting curve of all reactions exhibited a sharp peak at ∼83.5°C and a low melting peak was observed in the reactions with the tag7 primer (Fig 3D arrow), indicating that all tag-containing fragments were amplified specifically and that the quantity of tag7-containing fragment was lower than the other tags. This test experiment demonstrated that we can distinguish differences in populations of each tagged mutant within pools by quantifying the signature tags.
Figure 3.
Test experiments for identifying a growth-deficient mutant within a pool of mutants. (A) Growth of 27-tagged mutants on rich TY medium. Same concentration of individual mutants was spotted on TY broth medium. Mutant 07T2d09, integrated with pTnMod-OΩ-Tag7, exhibited reduced growth compared with other mutants. Quantitation of PCR amplifications of the input pool (B) and output pool (C). X and Y axes show the number of PCR cycles and log fluorescence of PCR products, respectively. Arrow in C indicates the quantitative curve from PCR reaction with Tag7 primer. The amounts of amplified products in all reactions of input pool were similar (ranging from 0.37- to 1.7-fold compare with average). In output pool, the amount of amplified products of all but tag7 were similar to that of input pool (ranging from 0.25- to 1.6-fold compared with that of input pool), whereas the amount in reaction of tag7 primer was 90-fold lower than that of input pool. (D) Melting curve of the output pool showing the change in fluorescence from 72°C to 91°C. All melting curves are shown as relative intensities among all reactions. Arrow in D indicates the melting curve of PCR reaction with Tag7 primer.
In consideration of future screening experiments that would be explored in planta, we next examined whether tagged mutants can also be detected from plant samples (root nodules). For these test experiments, one mutant from each of the 27 tag libraries was mixed in roughly equal quantities and inoculated onto L. japonicus roots to form nodules. Four weeks after inoculation, the nodules were detached and used for mutant detection. In order to determine how many mutants exist in a single nodule, we examined 22 nodules formed by the inoculation of two different pools of mutants. This confirmation enables us to estimate how many nodules should be analyzed for mutant screenings. When the presence of mutants in a single nodule was examined by the procedures described in the Materials and methods section, most nodules (18 of 22 tested nodules) were occupied by a mutant with a single tag (Supplementary Fig. S1). This result was consistent with a previous study in S. meliloti,21 indicating that the colonization or infection process may have similar features for both determinate and indeterminate nodules, and competition of infection may already occur at the colonization stage on host root hairs. And, as far as we tested, all tags were recovered in a screen of 70 nodules, suggesting that at least 70 nodules should be analyzed for mutant screenings in nodules.
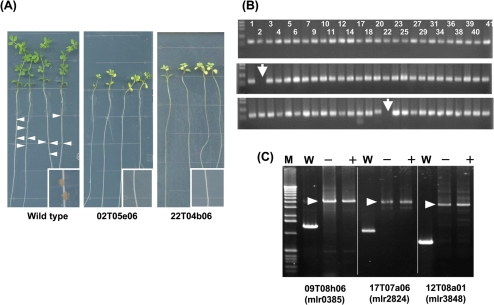
Additionally, we tested whether nodulation mutants can be screened properly for in nodules formed by pools of mutants. Two mutants (02T05e06 and 22T04b06) that exhibit defective in nodulation (Fig. 4A) were included in two distinct test sets (input samples) and inoculated onto L. japonicus roots. When we examined the presence of mutants in the total DNA isolated from 120 nodules (output samples), tag2 and tag22 containing fragments were not recovered, whereas all the other tags were detected (Fig. 4B). These results are consistent with the defective nodulation phenotype of the mutants (02T05e06 and 22T04b06), as tested individually (Fig. 4A). During the infection process, integrated transposon elements were stable since identical amplification patterns were obtained before and after infection (Fig. 4C, three representative results are shown).
Figure 4.
Pilot experiments identifying mutants with a nodulation defective phenotype. (A) Phenotype of L. japonicus after inoculation with wild-type M. loti or with two nodulation-deficient mutants (02T05e06 and 22T04b06). Arrowheads indicate the nodules formed on the roots of L. japonicus and the inset show the enlarged image of root and nodules. (B) Tag amplification from genomic DNA isolated from input pool (upper) and output pool (middle and bottom). The numbers indicate the combination of primers used. The arrows indicate the absence of PCR products in reactions with the Tag2 or Tag22 primer. (C) Representative PCR amplification showing the stability of the integrated transposon fragment in three different mutants before (−) and after (+) inoculation. Arrowheads indicate the fragment containing the integrated transposon element. W, PCR amplification with genomic DNA from wild-type M. loti.
From the results of these pilot experiments, we concluded that the signature-tagged mutants can be used for future STM screenings both in vitro and in vivo using our experimental conditions.
3.3. Mapping of transposon insertion sites
Along with the collection of transposon mutants, we determined the transposon insertion sites of individual mutants. Transposon insertion sites were determined by sequencing of transposon-flanking genomic regions, followed by comparison with the reference genome sequence. Determination of transposon insertions revealed that the inverted repeat of both the left and right ends of the integrated transposon were followed by the individual 9 bp direct repeat of the target site, indicating that the transposition were occurred properly by Tn5-transposon integration.33
The distribution of transposon insertion sites in the M. loti genome is illustrated in Fig. 5. Among 9344 mutants examined, insertion sites of 7892 were determined (success rate, 84.5%) that were mapped to 7586 unique insertion sites (Table 1). This proportion indicates that ∼96% of mutants carry a unique transposon insertion in their genome and there is no significant redundancy of a clone arising from identical parent clones in the same tri-parental mating experiment. Among the unique insertions, 7018 (92.5%), 398 (5.2%) and 170 (2.3%) were mapped onto the chromosome and two plasmids of M. loti (pMLa and pMLb), respectively (all insertion sites are listed in Supplementary Table S2 and S3). The proportion of transposon distribution was consistent with the proportion of total size of each M. loti replicon (Chr, 92.6%, pMLa 4.6% and pMLb 2.7%). Furthermore, among the unique insertion sites, 6430 (84.8%) were within ORFs, corresponding to the overall proportion (85.6%) of coding sequences within the entire M. loti genome.1
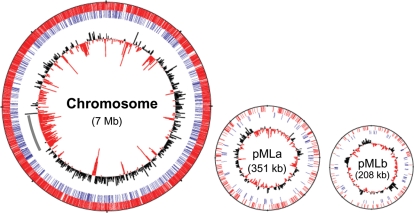
Figure 5.
Distribution of transposon insertions in the M. loti genome. The bars in the first and second circles show the position of transposon insertion sites within ORFs (red bars) and within intergenic regions (blue bars), respectively. The innermost circles represent the average G+C content calculated in each 10 kb. A positive deviation in G+C content from the average (62.7%) is shown by black bars pointing outward and a negative deviation by red bars pointing inward. Grey bar in chromosome indicates the region of symbiosis island.
Previous studies have demonstrated that the distribution patterns of transposon insertion into genomes vary depending on the types of transposon and organisms used for mutagenesis.34–36 To examine whether the transposon insertions were distributed randomly throughout the M. loti genome, we first investigated potential target site preferences for transposon insertion. Transposon insertions occurred in both intragenic and intergenic regions and the orientations of integrated transposons, with respect to the ORF directions of transcription, are present in nearly equal proportions (3274 clones in the same direction as transcription and 3397 clones in the opposite direction) (Supplementary Table S2). In addition, according to a previous report,37 G+C content was calculated from the 9 nucleotides of the insertion-target sites and 10 nucleotides up- and down-stream of target site of 50 randomly selected clones. It showed a high variance ranging from 30% to 88% (Supplementary Table S4), and the average G+C content of the target site (62.7%) was consistent with that of M. loti chromosome (62.7%).1 Furthermore, 50 randomly selected target sites showed no obvious sequence pattern, including palindromic or symmetrical sequences, when these were analyzed by TEIRESIAS, the sequence pattern search algorithm (http://cbcsrv.watson.ibm.com/Tspd.html).38 These features indicate that there are no clear preferences in insertion with regard to G+C content or structural features of the target sites.
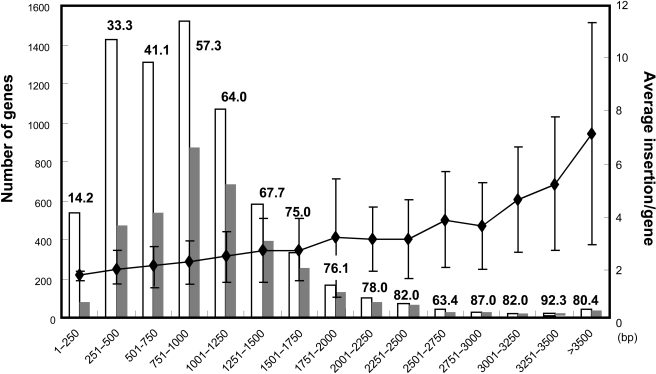
In order to verify the random distribution of transposon insertion, we additionally investigated the relationship between the number of genes carrying an inserted transposon and the size of the corresponding genes. Fig. 6 shows the distribution of transposon insertion within ORFs. As expected, more transposon insertions occurred in larger ORFs and the number of disrupted genes clearly correlate with the number of M. loti ORFs in each size range. In addition, as shown in Fig. 5, there is no obvious correlation between the distribution of transposon insertion and local G+C content of the M. loti genome. The symbiosis island, which exhibits features of horizontally transferred genomic element of low G+C content, also contained transposon insertions at rates similar to other chromosomal region (Fig. 5 grey line). Furthermore, this random distribution was observed for all tagged plasmid constructs.
Figure 6.
Distribution of transposon insertion within M. loti ORFs. The white bars represent the number of M. loti genes in each size range and the grey bars represent the number of genes with transposon insertions. The line chart represents the average number of insertion in genes of each size range with standard deviation. Percentages in each bar indicate the coverage of disrupted genes in each size range.
Several previous studies have provided sets of essential genes of organisms predicted from comprehensive random mutagenesis or targeted gene disruption.36,39,40 Although here we collected a large number of transposon mutants of M. loti, it is currently impossible to estimate the precise number of M. loti essential genes because the number of mutants for which transposon insertion sites have been determined is not sufficient to cover all non-essential genes. However, we can predict essential gene candidates among the relatively large size M. loti genes since large size M. loti genes (>3000 bp) were disrupted at high coverage (83.1%). Among 77 genes whose size is larger than 3000 bp, transposon insertions were not detected in 13 genes, whereas an average of 4.7 insertions occurred in other genes in this size range (Supplementary Table S5). Considering the random distribution of transposon insertion, these 13 genes are candidate M. loti essential genes required for growth on rich media. Among the genes, 11 genes (excluding mlr9704 and mll1090) are highly conserved in the genome of completely sequenced rhizobia and four genes, mlr8250 (isoleucyl-tRNA synthetase), mll0870 (DNA polymerase III alpha subunit), mlr0276 (RNA polymerase beta subunit) and mlr0277 (RNA polymerase, beta prime subunit), showed high similarity to essential genes of E. coli (ileS, dnaE, rpoB and rpoC, respectively). Furthermore, mlr2517 is orthologous to the essential gene of Acinetobacter baylyi ADP1 (carB, carbamoyl-phosphate synthase, large subunit).41 Other essential gene candidates could be those with insertions only near 3′-terminus of the ORF. Mutants for mll1426, mlr0796 and mlr0585 could be the candidate of essential genes since relatively small number of insertions, all of which were in their 3′-terminus, were detected as listed in Supplementary Table S5.
In bacteria, essential genes are more evolutionarily conserved than non-essential genes.42 We compared E. coli essential genes and M. loti genes [BlastP search was carried out against E. coli essential genes listed in the PEC data base (www.shigen.nig.ac.jp/ecoli/pec/index.jsp)] and revealed that a total of 196 M. loti ORFs showed high similarity to E. coli essential genes. Transposon mutants were obtained for only 21 of these 196 genes (10.7%). This ratio is significantly low since the result of sequence analysis transposon insertion sites identified mutants for ∼50% of M. loti genes. Among the 21 genes with transposon insertion, eight genes have putative palarogous genes which are considered to complement the function of the disrupted genes. These results indicate that significant overlap exists among sets of essential genes in E. coli and M. loti. However, saturating collections of transposon mutants under different selective conditions are, of course, needed to confirm these genes cannot actually be disrupted.
3.4. Mutant database
To provide resources for depositing and retrieving information on M. loti mutants, we developed a relational database, RhizoGenes (http://www.kazusa.or.jp/rhizobase/Mesorhizobium/genes2/index.html). This database operates as a public web database that provides general information about mutants, e.g. mutant IDs, transposon insertion site, list of mutant alleles and experimental results (which can be deposited by users). In this database, individual mutants can be retrieved through keyword search by clone ID, name and functional annotation of disrupted genes. Therefore, when genes have attracted interest on the basis of bioinformatic or functional genomic analyses, appropriate mutants can be searched immediately. The transposon mutants collected in this study will be available from the National Bioresource project of Japan (http://www.legumebase.agr.miyazaki-u.ac.jp/index.jsp).
Furthermore, we have integrated this database into the genome information database, RhizoBase (http://bacteria.kazusa.or.jp/rhizobase/Mesorhizobium/index.html), so as to link directly to more detailed information about M. loti ORFs, such as operon structure, predicted protein domains and orthologous (paralogous) protein groups.
3.5. Concluding remarks
In the present study, we have described a large-scale transposon mutagenesis in the nitrogen-fixing symbiotic bacterium, M. loti. The STM technique was adapted to barcode individual transposon mutants and a large mutant library containing >29 000 independent clones was created using a set of 27 signature tags. Sequencing of the transposon insertion sites of one-third of the mutants revealed a random distribution of transposon insertion and ∼50% of M. loti genes have been confirmed to be disrupted. Since such a large number of disruption mutants have not previously been available for M. loti and the low efficiency of gene-disruption mediated by homologous recombination has been a serious technical bottleneck for functional studies of this symbiont, our mutant collections should be a powerful tool for future functional studies of M. loti genes.
Applied STM technique and PCR-based screening allow for the simultaneous discovery of mutants with subtle phenotypic differences, such as reduced competitiveness or survival under various conditions. Additionally, random distribution of transposon insertion ensures the screenings of mutants over the entire M. loti genome and reduces the number of clones to be screened. Determination of transposon insertion sites also enables users to select defined, non-redundant sets of mutants depending on individual needs and facilitates rapid identification of causal genes of the mutant phenotypes. Furthermore, individual transposon mutants can be used as single gene disruptants. The availability of single gene disruptants allows functional verifications of individual or paralogous M. loti genes as well as comparative functional studies by examining with the orthologous genes of other rhizobium species. Additionally, our resource will be useful for in planta studies in the host legume. Actually, our STM mutants have recently contributed to reveal the importance of purine metabolisms in Lotus-M. loti symbiosis.24 Use of the mutant collections in Lotus-M. loti symbiosis will contribute to discovery of novel rhizobial factors or host plant recognition systems required for the symbiosis. By providing this mutant resource to the research communities, we hope to contribute to various functional analyses that lead to novel findings and understandings of rhizobial biology.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by KAKENHI (Grant-in-Aid for Scientific Research) on Priority Areas “Comparative Genomics” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and the Kazusa DNA Research Institute Foundation.
Supplementary Material
References
- 1.Kaneko T., Nakamura Y., Sato S.,, et al. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 2000;7:331–338. doi: 10.1093/dnares/7.6.331. [DOI] [PubMed] [Google Scholar]
- 2.Kaneko T., Nakamura Y., Sato S.,, et al. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 2002;9:189–197. doi: 10.1093/dnares/9.6.189. [DOI] [PubMed] [Google Scholar]
- 3.Galibert F., Finan T. M., Long S. R.,, et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 4.González V., Santamaría R. I., Bustos P.,, et al. The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc. Natl. Acad. Sci. USA. 2006;103:3834–3839. doi: 10.1073/pnas.0508502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young J. P., Crossman L. C., Johnston A. W.,, et al. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 2006;7:R34. doi: 10.1186/gb-2006-7-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giraud E., Moulin L., Vallenet D., Barbe V.,, et al. Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science. 2007;316:1307–1312. doi: 10.1126/science.1139548. [DOI] [PubMed] [Google Scholar]
- 7.Bergès H., Lauber E., Liebe C.,, et al. Development of Sinorhizobium meliloti pilot macroarrays for transcriptome analysis. Appl. Environ. Microbiol. 2003;69:1214–1219. doi: 10.1128/AEM.69.2.1214-1219.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Djordjevic M. A., Chen H. C., Natera S.,, et al. A global analysis of protein expression profiles in Sinorhizobium meliloti: discovery of new genes for nodule occupancy and stress adaptation. Mol. Plant Microbe. Interact. 2003;16:508–524. doi: 10.1094/MPMI.2003.16.6.508. [DOI] [PubMed] [Google Scholar]
- 9.Barnett M. J., Toman C. J., Fisher R. F., Long S. R. A dual-genome Symbiosis Chip for coordinate study of signal exchange and development in a prokaryote–host interaction. Proc. Natl. Acad. Sci. USA. 2004;101:16636–16641. doi: 10.1073/pnas.0407269101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoa L. T. P., Nomura M., Tajima S. Characterization of bacteroid proteins in soybean nodules formed with Bradyrhizobium japonicum USDA110. Microbiol. Environ. 2004;19:71–75. [Google Scholar]
- 11.Uchiumi T., Ohwada T., Itakura M.,, et al. Expression islands clustered on symbiosis island of Mesorhizobium loti genome. J. Bacteriol. 2004;45:2439–2448. doi: 10.1128/JB.186.8.2439-2448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarma A. D., Emerich D. W. Global protein expression pattern of Bradyrhizobium japonicum bacteroids: a prelude to functional proteomics. Proteomics. 2005;5:4170–4184. doi: 10.1002/pmic.200401296. [DOI] [PubMed] [Google Scholar]
- 13.Shimoda Y., Shinpo S., Kohara M., Nakamura Y., Tabata S., Sato S. A large scale analysis of protein–protein interactions in the nitrogen-fixing bacterium Mesorhizobium loti. DNA Res. 2008;15:13–23. doi: 10.1093/dnares/dsm028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchison C. A., Peterson S. N., Gill S. R.,, et al. Global transposon mutagenesis and a minimal mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi K., Ehrlich S. D., Albertini A.,, et al. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang Y., Durfee T., Glasner J. D.,, et al. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 2004;186:4921–4930. doi: 10.1128/JB.186.15.4921-4930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baba T., Ara T., Hasegawa M.,, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glass J. I., Assad-Garcia N., Alperovich N.,, et al. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA. 2006;103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saenz H. L., Dehio C. Signature-tagged mutagenesis: technical advances in a negative selection method for virulence gene identification. Curr. Opin. Microbiol. 2005;8:612–619. doi: 10.1016/j.mib.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Pobigaylo N., Wetter D., Szymczak S.,, et al. Construction of a large signature-tagged Mini-Tn5 transposon library and its application to mutagenesis of Sinorhizobium meliloti. Appl. Environ. Microbiol. 2006;72:4329–4337. doi: 10.1128/AEM.03072-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pobigaylo N., Szymczak S., Nattkemper T. M., Becker A. Identification of genes relevant to symbiosis and competitiveness in Sinorhizobium meliloti using signature-tagged mutants. Mol. Plant Microbe. Interact. 2008;21:219–231. doi: 10.1094/MPMI-21-2-0219. [DOI] [PubMed] [Google Scholar]
- 22.Fraysse N., François C., Véréna P. Surface polysaccharide involvement in establishing the rhizobium-legume symbiosis. FEBS J. 2003;270:1365–1380. doi: 10.1046/j.1432-1033.2003.03492.x. [DOI] [PubMed] [Google Scholar]
- 23.D'Antuono A. L., Casabuono A., Couto A., Ugalde R. A., Lepek V. C. Nodule development induced by Mesorhizobium loti mutant strains affected in polysaccharide synthesis. Mol. Plant Microbe. Interact. 2005;18:446–457. doi: 10.1094/MPMI-18-0446. [DOI] [PubMed] [Google Scholar]
- 24.Okazaki S., Hattori Y., Saeki K. The Mesorhizobium loti purB gene is involved in infection thread formation and nodule development in Lotus japonicus. J. Bacteriol. 2007;189:8347–8352. doi: 10.1128/JB.00788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerrero G., Peralta H., Aguilar A.,, et al. Evolutionary, structural and functional relationships revealed by comparative analysis of syntenic genes in Rhizobiales. BMC Evol. Biol. 2005;5:55. doi: 10.1186/1471-2148-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finan T. M., Kunkel B., De Vos G. F., Signer E. R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beringer J. E. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 28.Dennis J. J., Zylstra G. J. Plasposon: modular self-clonig minitransposon derivatives for rapid genetic analysis of Gram-negative bacterial genomes. Appl. Environ. Microbiol. 1998;64:2710–2715. doi: 10.1128/aem.64.7.2710-2715.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prentki P., Kirsch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 30.Simon R., Priefer U., Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 31.Hunt T. A., Kooi C., Sokol P. A., Valvano M. A. Identification of Burkholderia cenocepacia genes required for bacterial survival in vivo. Infect. Immun. 2004;72:4010–4022. doi: 10.1128/IAI.72.7.4010-4022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Autret A., Charbit A. Lessons from signature-tagged mutagenesis on the infectious mechanisms of pathogenic bacteria. FEMS Microbiol. Rev. 2005;29:703–717. doi: 10.1016/j.femsre.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Reznikoff W. S. Tn5 as a model for understanding DNA transposition. Mol. Microbiol. 2003;47:1199–1206. doi: 10.1046/j.1365-2958.2003.03382.x. [DOI] [PubMed] [Google Scholar]
- 34.Groh J. L., Luo Q., Ballard J. D., Krumholz L. R. A method adapting microarray technology for signature-tagged mutagenesis of Desulfovibrio desulfuricans G20 and Shewanella oneidensis MR-1 in anaerobic sediment survival experiments. Appl. Environ. Microbiol. 2005;71:7064–7074. doi: 10.1128/AEM.71.11.7064-7074.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki N., Okai N., Nonaka H., Tsuge Y., Inui M., Yukawa H. High-throughput transposon mutagenesis of Corynebacterium glutamicum and construction of a single-gene disruptant mutant library. Appl. Environ. Microbiol. 2006;72:3750–3755. doi: 10.1128/AEM.72.5.3750-3755.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallagher L. A., Ramage E., Jacobs M. A., Kaul R., Brittnacher M., Manoil A. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc. Natl. Acad. Sci. USA. 2007;104:1009–1014. doi: 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mormann S., Lömker A., Rückert C.,, et al. Random mutagenesis in Corynebacterium glutamicum ATCC 13032 using an IS6100-based transposon vector identified the last unknown gene in the histidine biosynthesis pathway. BMC Genomics. 2006;7:205. doi: 10.1186/1471-2164-7-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigoutsos I., Floratos A. Combinatorial pattern discovery in biological sequences: the TEIRESIAS algorithm. Bioinformatics. 1998;14:55–67. doi: 10.1093/bioinformatics/14.1.55. [DOI] [PubMed] [Google Scholar]
- 39.Jacobs M. A., Alwood A., Thaipisuttikul I.,, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liberati N. T., Urbach J. M., Miyata S.,, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Berardinis V., Vallenet D., Castelli V.,, et al. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol. Syst. Biol. 2008;4:174. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerdes S. Y., Scholle M. D., Campbell J. W.,, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.