Abstract
Fibroblast growth factor 23 (FGF23) is a hormone required for normal renal phosphate reabsorption. FGF23 gain-of-function mutations result in autosomal dominant hypophosphatemic rickets (ADHR), and FGF23 loss-of-function mutations cause familial hyperphosphatemic tumoral calcinosis (TC). In this study, we identified a novel recessive FGF23 TC mutation, a lysine (K) substitution for glutamine (Q) (160 C > A) at residue 54 (Q54K). To understand the molecular consequences of all known FGF23-TC mutants (H41Q, S71G, M96T, S129F, and Q54K), these proteins were stably expressed in vitro. Western analyses revealed minimal amounts of secreted intact protein for all mutants, and ELISA analyses demonstrated high levels of secreted COOH-terminal FGF23 fragments but low amounts of intact protein, consistent with TC patients' FGF23 serum profiles. Mutant protein function was tested and showed residual, yet decreased, bioactivity compared with wild-type protein. In examining the role of the FGF23 COOH-terminal tail (residues 180–251) in protein processing and activity, truncated mutants revealed that the majority of the residues downstream from the known FGF23 SPC protease site (176RXXR179/S180) were not required for protein secretion. However, residues adjacent to the RXXR site (between residues 188 and 202) were required for full bioactivity. In summary, we report a novel TC mutation and demonstrate a common defect of reduced FGF23 stability for all known FGF23-TC mutants. Finally, the majority of the COOH-terminal tail of FGF23 is not required for protein secretion but is required for full bioactivity.
Keywords: fibroblast growth factor 23, phosphate, hyperphosphatemia, Klotho
proper control of serum phosphate concentrations is required for normal bone structure and function. Complex endocrine interactions maintain serum phosphate within a relatively narrow range (2.7–4.5 mg/dl in adults) (12, 28). The disruption of these regulatory pathways can lead to reciprocal human disorders of hyper- and hypophosphatemia, including familial tumoral calcinosis (TC; OMIM 211900) and autosomal dominant hypophosphatemic rickets (ADHR; OMIM 193100), respectively.
The autosomal recessive disorder, TC, is characterized by hyperphosphatemia secondary to increased renal phosphate reabsorption, normal or elevated 1,25(OH)2 vitamin D concentrations, and normocalcemia (16, 24, 25). Persistent hyperphosphatemia can result in the development of severe ectopic and vascular calcifications in these patients (16, 24, 25). Mutations responsible for familial TC were first reported in the GalNac transferase-3 (GALNT3) gene (30). GALNT3 is a UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyl transferase that initiates mucin-like O-linked glycosylation of nascent proteins within the trans-Golgi network (4). Subsequently, we (22) and others (2, 3, 6) have shown that homozygous missense mutations in the fibroblast growth factor 23 (FGF23) gene cause TC; however, whether a common disease mechanism underlies these mutants is unknown.
Of significance, patients with TC display clinical findings similar to those found in Fgf23- and Klotho (KL)-null mice (19, 26). α-KL is expressed as a type 1 membrane protein, as well as a secreted isoform, and shares homology with the β-glycosidase family of enzymes. KL is required for FGF23 intracellular signaling with interactions as a heteromeric complex with FGF23 and specifically FGFR1c in one report (31) or potentially with FGF23 and FGFR1c, -3c, or -4 (20). FGF23 binding to the FGFR-KL complex leads to activation of the MAPK pathway and phosphorylation of ERK1/2 as well the activation of the transcription factor early growth response-1 (EGR1) gene (11, 20, 31). Recessive inactivating KL mutations also result in TC, most likely due to an end-organ defect in renal responsiveness to FGF23 (14).
FGF23 controls the renal reabsorption of phosphate in concert with vitamin D metabolism, as exemplified by its causative role in ADHR, the phenotypic mirror-image disorder to TC (1). ADHR is characterized by hypophosphatemia with inappropriately normal 1,25(OH)2 vitamin D concentrations and osteomalacia or rickets (7). FGF23 mutations responsible for ADHR lead to amino acid substitutions at Arg 176 or Arg 179 (R176Q, R179Q, and R179W). These residues comprise a subtilisin-like proprotein convertase (SPC) proteolytic cleavage site (176RXXR179/S180), and the ADHR mutations stabilize full-length FGF23 by inhibiting the cleavage of the COOH-terminal tail from the NH2-terminal FGF-like domain (1, 32). However, the role of the COOH-terminal tail in the processing, secretion, and signaling activities of FGF23 is unknown.
Herein, we describe a novel FGF23 mutation in three patients with TC. Additionally, we have assessed the biochemical activity of FGF23 proteins containing the known TC mutations and determined the significance of the FGF23 COOH-terminal region. These studies expand our current understanding of the molecular genetic and metabolic etiologies of TC and reveal a common pathogenic mechanism for TC cases due to mutations in the FGF23 gene.
MATERIALS AND METHODS
TC patients.
All subjects provided written, informed consent, in accordance with the Institutional Review Board of Indiana University, which approved the study. The kindreds were of Iranian descent. Routine serum biochemistries were assessed using standard protocols.
FGF23 serum assays.
Intact FGF23 serum concentrations were determined using an ELISA according to the manufacturer's protocol (Kainos Laboratories International, Tokyo, Japan). This assay uses monoclonal antibodies and has been shown to recognize full-length human and rodent FGF23/Fgf23 (34). Serum FGF23 concentrations were also assessed using a COOH-terminal FGF23 serum assay kit (Immutopics, San Clemente, CA) (17) according to the manufacturer's protocol. This kit is a two-site sandwich ELISA that recognizes the COOH-terminal portion of human FGF23, thus reacting with both full-length and COOH-terminal fragments of FGF23.
FGF23 and GALNT3 mutational analyses.
Genomic DNA was extracted from blood samples using the Qiamp DNA Blood Extraction kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Gene exons, including the intron-exon splice junctions, were PCR-amplified with intronic primers (available upon request), using 20 ng of genomic DNA as template. PCR conditions for all experiments were 1 min at 95°C followed by 35 cycles of 1 min at 95°C, 1 min at 57°C, 1 min at 72°C, and a final extension of 7 min at 72°C. Amplified exons were analyzed by DNA sequencing with the appropriate forward primers for each exon using Big Dye Terminator Chemistry.
FGF23 exon 1 controls.
A primer pair (forward: 5′AATCTCAGCACCAGCCACTC 3′; reverse: 5′GATGGACAACAAGGGTGCTC 3′) was used to amplify a 270-bp region containing exon 1 of the FGF23 gene in the patients and in 200 control alleles. PCR conditions were the same as those described above for mutational analyses. The resulting PCR products were digested with PstI for 18 h at 37°C and analyzed on 2% ethidium bromide-stained agarose gels.
Site-directed mutagenesis.
Point mutations within the FGF23 cDNA were generated with the QuickChange II XL site-directed mutagenesis kit (Stratagene) using pCMV-FLAG containing wild-type (WT), full-length FGF23 (33) to create the FGF23-TC cDNAs: H41Q, Q54K, S71G, M96T, and S129F. Complimentary primer pairs (15–25 bases in length, available upon request) were designed to create the TC mutations using the PrimerX primer design program (http://bioinformatics.org/primerx/). PCR conditions for site-directed mutagenesis were 1 min at 95°C followed by 35 cycles of 50 s at 95°C, 50 s at 60°C, 7 min at 68°C, and a final extension of 7 min at 68°C. The mutagenesis reactions were digested with DpnI for 1 h at 37°C to remove template WT plasmid DNA. The plasmids containing mutant FGF23 cDNAs were sequenced to confirm replacement of the targeted residues.
TC mutant FGF23 in vitro expression and Western analyses.
The expression vectors containing the Q54K, H41Q, S71G, M96T, and S129F mutant FGF23 cDNAs were transiently expressed in human embryonic kidney (HEK)-293 cells as previously performed using the Fugene6 transfection reagent (Roche) (21, 32). The cellular lysates and media were collected in 1× lysis buffer (Cell Signaling) and assessed for the presence of FGF23 protein by Western analysis. The samples were electrophoresed on 15% SDS-PAGE minigels (Bio-Rad) and electrotransferred to PVDF membranes (Bio-Rad). The membranes were then probed with an anti-FLAG antibody (1:1,000; Sigma). Visualization was performed using the ECL Plus Western Blotting Detection Reagents (Amersham GE Healthcare).
Stable expression of FGF23 TC mutants.
Transfection of HEK-293 cells was performed using the Fugene6 Reagent (Roche) according to the manufacturer's protocol. Twenty-four hours after transfection, medium containing 750 ng/ml of G418 (Sigma) was added to the cells. Cells were cultured for 2 wk, with the media changed every 3–5 days until cell colonies were detectable. Single colonies were picked and expanded for conditioned media collection.
FGF23 activity assay for EGR1 mRNA.
The human KL expression vector was cloned (forward: 5′CCTGGCTCCCGCGCAG CATGCC 3′; reverse: 5′TTTGTAACTTCTTCTGCCTTTCTTCG 3′) from Human Placenta Marathon Ready cDNA (Clontech) into pcDNA3.1-V5-His-TOPO (Invitrogen) and verified by direct sequencing. To determine the relative activity of FGF23 WT, mutant, and truncated proteins, 5 × 105 HEK-293 cells stably expressing the membrane-bound form of KL were seeded into 12-well plates and cultured for 24 h in DMEM-F-12 medium. The cells were incubated for 24 h in serum-free medium and treated with WT, mutant, or truncated FGF23 for 1 h at 37°C. Total RNA was isolated from the cellular lysates using the RNeasy kit (Qiagen) according to the manufacturer's instructions.
Quantitative real-time PCR.
The TaqMan One-Step RT-PCR kit was used to perform quantitative real-time RT-PCR. Each 20-μl reaction consisted of 1× buffer (Applied Biosystems), 0.25 mM primer (forward and reverse), 0.125 mM tetramethyl-6-carboxyrhodamine probe, and 100 ng of total RNA and was performed in triplicate to measure the level of EGR1 gene expression for each treatment. The primers used for EGR1 were 5′-GGACACGGGCGAGCAG-3′ and 5′-CGTTGTTCAGAGAGATGTCAGGA-3′, and the EGR1 probe was 5′-CCTACGAGCACCTGACCGCAGAGTCT-3′; the primers for β-actin were 5′-GGCACCCAGCACAATGAAG-3′ and 5′-GCCGATCCACACGGAGTACT-3′, and the β-actin probe was 5′-TCAAGATCATTGCTCCTCCTGAGCGC-3′. The PCR conditions for all experiments were 30 min at 48°C and 10 min at 95°C followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Each RNA sample was analyzed in triplicate on an ABI-PRISM 7700 sequence detection system (Applied Biosystems), and relative mRNA levels were calculated using the comparative cycle threshold method. The results were compared with HEK-293 cells expressing the empty vector and treated with WT FGF23.
Statistical analysis.
Statistical analysis of the data was performed by Student's t-test, and significance for all tests was set at P < 0.05. Data are presented as means ± SD.
FGF23 activity assay for phospho-ERK1/2.
HEK-293 cells stably expressing the membrane form of KL were treated with conditioned medium containing 350 pg/ml of WT or Q54K FGF23 for 5 min following an overnight serum starve. The cells were washed three times with 1× PBS and then lysed with 0.125 ml of 1× lysis buffer (Cell Signaling Technologies, Danvers, MA) in the presence of the protease inhibitor 4-(2-aminoethyl)benzenesulfonyl fluoride (10 μg/ml). Cellular lysates were combined 1:1 with Laemmli Sample Buffer (Bio-Rad) and boiled for 5 min. Samples were electrophoresed and electrotransferred as described above. Membranes were incubated with 1:1,000 anti-total ERK1/2 or anti-phospho-ERK1/2 (Cell Signaling) for 1 h and then washed three times with 1% TBS with 0.05% Tween-20. Membranes were incubated with 1:3,000 of the secondary antibody anti-mouse horseradish peroxidase for 1 h and washed three times with 1% TBS with 0.05% Tween-20. Detection was performed using the ECL Plus Western Blotting Detection Reagents (Amersham GE Healthcare).
Truncated FGF23 in vitro expression and Western blot analyses.
Expression vectors containing WT FGF23 and FGF23 COOH-terminally truncated by 19, 48, 62, and 71 amino acid residues (FGF23Δ19, −Δ48, −Δ62, and −Δ71) were transiently transfected into HEK-293 cells. Due to the deletion of the epitope recognized by the anti-FGF23 antibody (22), both WT and truncated proteins contained an NH2-terminal FLAG tag. The cellular lysates and media were collected as described above and assessed for the presence of FGF23 protein by Western blot analysis using anti-FLAG (1:1,000; Sigma).
RESULTS
Clinical assessment of TC patients.
Three patients presented with a phenotype consistent with tumoral calcinosis. Patient 1 was an isolated case, whereas patients 2 and 3 were members of a large consanguineous family (Fig. 1). Patient 1, a 20-yr-old male, was referred for calcific lesions in the elbow, hip, and knee. Analyses of serum biochemistries revealed persistent hyperphosphatemia and normocalcemia as well as normal parathyroid hormone (PTH) and 1,25(OH)2 vitamin D concentrations (Table 1). The lesion in the patients’ elbows had been surgically removed due to restriction of movement, but it recurred.
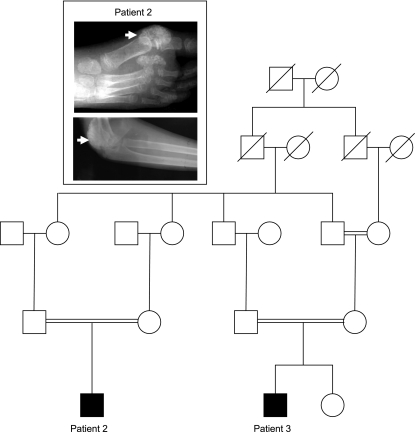
Fig. 1.
Family pedigree. Tumoral calcinosis (TC) patients 2 and 3 are members of a large consanguineous family (inset). Patient 2 had calcific lesion surrounding the large right toe and right elbow.
Table 1.
TC patient serum biochemistries
| Patient 1 (20 yr old) | Patient 2 (11 yr old) | Patient 3 (5 yr old) | |
|---|---|---|---|
| Phosphate, mg/dl | 7 (2.7–4.5) | 8.8 (4–6.5) | 6.5 (4–6.5) |
| Calcium, mg/dl | 9 (9–10.5) | 9.3 (8.5–10.4) | 9.2 (8.5–10.4) |
| PTH, pg/ml | 12 (9–55) | 17 (9–55) | 12 (4.4–15.6) |
| 1,25(OH)2D, pg/ml | 32 (15–60) | 48 (20–55) | Not available |
| Creatinine, mg/dl | 0.9 (0.6–1.2) | 1 (0.4–1.5) | 0.7 (0.5–0.8) |
TC, tumoral calcinosis; PTH, parathyroid hormone. Normal ranges are shown in parentheses.
Patient 2, an 11-yr-old male, was originally referred for the development of a painful lesion in the right big toe following physical trauma. The lesion in the toe was surgically removed, but it recurred (Fig. 1, inset). An additional lesion causing movement limitation developed in the patient's right elbow (Fig. 1, inset). Serum biochemistries revealed persistent hyperphosphatemia and normocalcemia as well as normal PTH and 1,25(OH)2 vitamin D (Table 1).
Patient 3, a 5-yr-old male, was referred following the development of a lesion in the left big toe. No physical trauma to the affected area was noted; however, the mass of the lesion increased over an 8-mo period. Serum biochemistries were significant for mild hyperphosphatemia with normocalcemia as well as normal PTH and 1,25(OH)2 vitamin D levels (Table 1).
FGF23 serum assays.
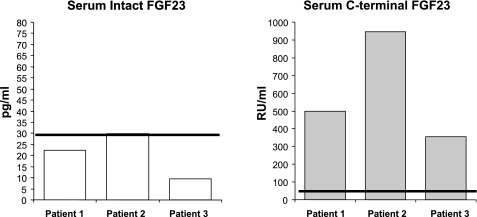
Serum FGF23 levels were assessed in these patients using two assays, the first being a “COOH-terminal” ELISA that detects both full-length and COOH-terminal fragments of FGF23 and the second being an “intact” ELISA that utilizes epitopes encompassing the 176RXXR179/S180 SPC protease site and therefore recognizes intact FGF23 protein. The COOH-terminal ELISA results showed that patients 1, 2, and 3 had FGF23 concentrations roughly nine, 17, and six times the normal mean, respectively (Fig. 2). In contrast, the intact FGF23 concentrations for the three patients were found to be inappropriately normal considering the marked degree of persistent hyperphosphatemia (Fig. 2). These values of elevated COOH-terminal FGF23 concentrations in parallel with normal levels of intact FGF23 are consistent with the aberrant circulating FGF23 profile detected in other TC cases (3, 22).
Fig. 2.
Fibroblast growth factor 23 (FGF23) serum levels in patients 1–3. Left: patients 1, 2, and 3 with the Q54K FGF23 mutations had low and normal serum concentrations of intact FGF23 compared with the normal mean. Right: the COOH-terminal ELISA values were 10-, 18-, and 6-fold above the normal mean for patients 1, 2, and 3, respectively. The normal means for both assays are shown as solid lines.
Mutational analyses.
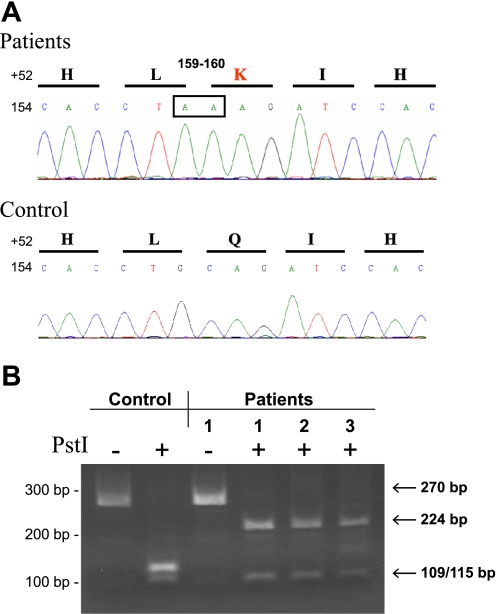
Genomic DNA from patients 1–3 was assessed for GALNT3 mutations as the etiology for the TC phenotype; however, no mutations were detected within the exons or intronic splice sites. In contrast, direct DNA sequence analyses of the patients' FGF23 exon 1 revealed a homozygous guanine-to-adenine (159 G > A) silent transition in concert with a cytosine-to-adenine transversion (160 C > A). The C > A change at position 160 resulted in a novel glutamine (Q)-to-lysine (K) amino acid substitution at position 54 (Q54K) (Fig. 3A). All other FGF23 exons and splice junction sites were negative for nucleotide substitutions.
Fig. 3.
A: cytosine to adenine base substitution at position 160 results in Q54K change. Sequence analyses of FGF23 exon 1 in patients 1, 2, and 3 revealed recessive guanine to adenine transition at position 159, affecting a wobble nucleotide, but a novel cytosine to adenine transversion at position 160 (nucleotides boxed at top) that creates a glutamine to lysine substitution at position 54 (Q54K) in the FGF23 protein. B: mutational analyses by restriction fragment length polymorphism. Amplification of FGF23 exon 1 results in a 270-bp DNA product (lanes 1 and 3) containing two PstI restriction sites. Digests of the 270-bp exon 1 product from normal individuals with PstI create 3 fragments that are 46 (not visible), 109, and 115 bp in size (lane 2). The presence of the guanine (G) to adenine (A) transition at position 159 (159 G > A) and cytosine (C) to A transversion at position 160 (160 C > A) mutations interrupt the PstI restriction site between the larger 109- and 115-bp fragments and results in the appearance of a 224-bp DNA fragment (lanes 4, 5, and 6). Position of molecular size markers are shown for reference at left. H, histidine; L, leucine; K, lysine; I, isoleucine; Q, glutamine.
FGF23 exon 1 controls.
The 159 G > A substitution and the 160 C > A substitution that results in the Q54K change were not found in dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/). Normal controls were then assessed by restriction fragment length polymorphism (RFLP) for the nucleotide changes, because either sequence alteration would interrupt a PstI site containing nucleotides 159 and 160. PCR amplification of FGF23 exon 1 results in a 270-bp DNA product containing two PstI restriction sites. Digestion of this product with PstI creates three fragments of 46, 109, and 115 bp in size in control individuals (Fig. 3B). The presence of the Q54K mutation in the three patients interrupts the restriction site between the 109- and 115-bp fragments and results instead in 46- and 224-bp DNA fragments (Fig. 3B). These RFLP analyses confirmed our sequencing findings described above by detecting the presence of a 224-bp band after the PstI digestion only in the patients' PCR products (Fig. 3B). In addition, similar analyses of 200 control alleles did not reveal this genetic alteration, confirming this substitution as a mutation in FGF23.
FGF23 mutant expression in vitro.
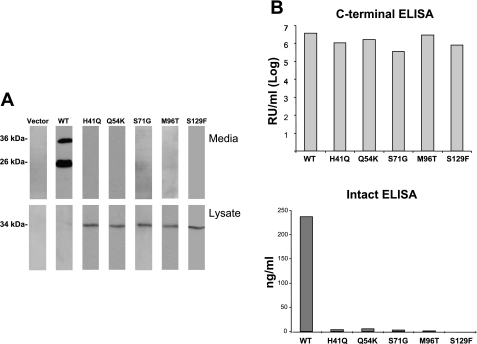
Previous studies indicated that FGF23 mutations resulting in TC caused destabilization of the intact FGF23 protein (21). The molecular consequences of the Q54K mutation on FGF23 protein stability, as well as other novel FGF23-TC mutants, including H41Q (23) and M96T (6), have not been evaluated; thus whether a common cellular mechanism exists for TC is currently unknown. Therefore, site-directed mutagenesis was used to produce FGF23 cDNAs harboring each of these three untested mutations (Q54K, H41Q, and M96T) as well as the S71G and S129F (Ser71Gly and Ser129Phe) FGF23-TC mutations that we have previously assessed for expression in vitro (22). These mutant FGF23 constructs were individually transfected into HEK-293 cells. As determined by Western analyses of the cellular lysates using an anti-FLAG antibody that recognizes an NH2-terminal FLAG epitope, all of the FGF23 TC mutants were retained within the cells, and intact FGF23 protein was not detectable in the unconcentrated medium from the same cells (Fig. 4A). In contrast, WT FGF23 displayed a known distribution pattern of low intracellular expression and high extracellular expression due to efficient processing of the native molecule (Fig. 4A). These results demonstrating similar aberrant protein production by the FGF23-TC mutants were then confirmed using the COOH-terminal and intact FGF23 ELISA on the cell medium. This assay showed markedly elevated levels of COOH-terminal fragments (Fig. 4B). Concomitant with these findings, low levels of intact FGF23 were detected by ELISA analysis in the same media samples (Fig. 4B), consistent with similar COOH-terminal and intact FGF23 ELISA patterns from the patients' serum samples (Fig. 2). Taken together, these results demonstrate that the known FGF23-TC mutants, including the novel Q54K mutation, have a common molecular property that results in the destabilization of intact FGF23 protein.
Fig. 4.
A: Western blot analyses of TC-mutant FGF23 proteins. Western blot analyses of conditioned media from human embryonic kidney (HEK)-293 cells transiently expressing wild-type (WT) FGF23 revealed that the protein is produced and secreted into the media (top) as 36- (full-length) and 26-kDa (NH2-terminal domain) proteins. These same cells were negative for the presence of the protein in the lysate (bottom). Media collected from cells expressing FGF23 cDNAs containing the TC mutations were negative for the presence of FGF23 (top). Lysates from the same cells were positive for a 34-kDa protein (bottom). B: intact and COOH-terminal FGF23 concentrations in conditioned media. ELISA analyses were performed to quantify intact and COOH-terminal FGF23 protein present in media samples from the mutant FGF23 species. The COOH-terminal ELISA results indicated mutant FGF23 protein levels similar to WT (Log10 concentration shown; top); however, the intact ELISA results showed that intact protein levels were >200-fold lower for the mutant proteins compared with WT FGF23 (bottom).
FGF23 mutant activity analyses.
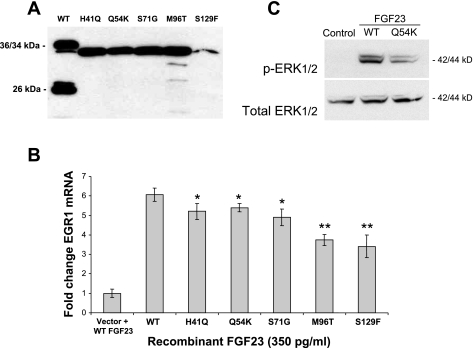
FGF23 stimulates the transcription of EGR1 in HEK-293 and other cell lines in the presence of the coreceptor KL (31). An assay based upon these findings has been developed for determining FGF23 bioactivity (11, 14). To produce mutant FGF23 protein at levels greater than that which could be derived from transiently transfected cells, we developed HEK-293 cell lines stably producing each of the FGF23 mutants Q54K, H41Q, S71G, M96T, and S129F (Fig. 5A). Due to the increased expression levels in the stable cell lines, Western blot analyses revealed that these lines produced the FGF23 mutants as a 34-kDa protein species, which represented a partially glycosylated full-length FGF23 protein (2). The FGF23 protein concentrations in the media samples were quantified using the intact FGF23 ELISA. FGF23 concentrations in the media were then standardized to 350 pg/ml (the concentration derived from the lowest-producing cell line, S129F). HEK-293 cells stably expressing KL were treated with FGF23-conditioned medium from each mutant as well as WT medium as positive control. Quantitative RT-PCR for EGR1 mRNA expression following treatment revealed that the FGF23-TC mutants retained biological activity but had statistically lower residual activity compared with the activity of WT FGF23 (H41Q, P = 0.031; Q54K, P = 0.027; S71G, P = 0.01; M96T, P = 0.0001; and S129F, P = 0.001; Fig. 5B). Native HEK-293 cells treated with WT FGF23 but not expressing KL showed only background EGR1 mRNA levels (Fig. 5B). To further test mutant FGF23 activity, we assessed the ability of the Q54K mutant (the mutant with the highest EGR1 expression; Fig. 5B) to stimulate phospho-ERK1/2 (pERK1/2) in vitro. Following treatment of the HEK-293-KL cells with control media or media containing 350 pg/ml WT or Q54K FGF23, pERK1/2 activity as assessed by Western blot analyses was reduced compared with WT FGF23. The control medium showed no ability to stimulate pERK1/2, and the cell total ERK1/2 content was not different for each treatment (Fig. 5C).
Fig. 5.
A: overexpression of TC-mutant FGF23 protein. Cell lines stably expressing the FGF23 mutants secreted high levels of intact FGF23 protein into the cellular media compared with cells transiently expressing the FGF23 proteins, as determined by Western blotting with the use of an anti-FLAG antibody. B: activity of TC-mutant FGF23. Quantitative PCR results of early growth response 1 (EGR1) expression in HEK-293 cells stably expressing the membrane form of Klotho (KL) and treated with WT and mutant FGF23 protein. Although significantly lower than WT FGF23 (H41Q, Q54K, and S71G P < 0.05; M96T and S129F P < 0.0005), the TC-causing FGF23 mutants retained biological activity. Error bars represent standard deviations, and each experiment was performed 3 times. Cells containing empty vector were treated with 350 pg/ml of WT FGF23 protein (“Vector + WT”). C: WT and Q54K-FGF23-mediated phosphorylated (p)ERK1/2 activity. Treatment of the HEK-293-KL stable cells with 350 pg/ml of WT or Q54K FGF23 resulted in the Q54K mutant showing less ability to stimulate pERK1/2, as assessed by Western blot analyses. Control media showed no pERK1/2 reactivity, and total ERK was similar for all treatments.
COOH-terminal FGF23 truncations.
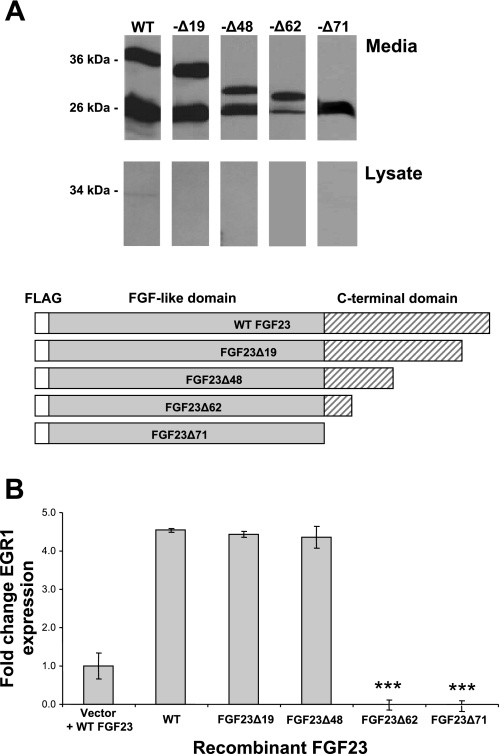
All of the known FGF23 mutations that result in TC occur NH2 terminal to the R176XXR179/S180 motif within the highly conserved FGF-like domain (residues 25–180); thus information on the function of the COOH-terminal tail is lacking. To test the importance of the COOH-terminal region in FGF23 processing, we performed nested PCR to serially delete 10–30 residue segments of the FGF23 COOH-terminus 3′ to the R176XXR179/S180 site. These truncated FGF23 analogs were designated as FGF23Δ19 residues, −Δ48, −Δ62, and −Δ71 (FGF23Δ71 has all of the COOH-terminal tail removed). Following expression of these constructs in HEK-293 cells, Western analyses revealed the presence of the FGF23Δ19, −Δ48, −Δ62, and −Δ71 proteins of the appropriate sizes, 33, 25, 23, and 20 kDa, respectively, in the cellular medium in amounts comparable to WT FGF23 (Fig. 6A). Only limited or no retention of these isoforms was detected in the cellular lysates (Fig. 6A). These results showed that in contrast to changes within the FGF23 NH2-terminal domain (Fig. 4), modification of the COOH terminus did not alter secretion of FGF23 protein compared with WT protein.
Fig. 6.
FGF23 COOH-terminal truncations. A: Western blot analyses. Conditioned media were assessed for expression of the truncated FGF23 analogs (diagrammed at bottom) by Western blot analyses using anti-FLAG. All species were secreted efficiently at the predicted molecular mass for each truncation. B: activity of COOH-terminally truncated FGF23. Quantitative PCR analyses of EGR1 expression determined that the activity of FGF23 protein lacking the 19 and 48 COOH-terminal residues was not significantly different from WT protein (P < 0.2 and 0.4, respectively). FGF23 protein lacking 62 and 71 COOH-terminal residues had no activity (compared with WT protein, P < 0.001). Control cells were transfected with empty vector and were treated with 350 pg/ml of WT FGF23 protein (Vector + WT).
Activity of truncated FGF23.
As determined above, our truncation analyses showed that the COOH-terminal region of FGF23 does not play a predominant role in regulating protein secretion; however, the functional consequences of this domain on FGF23 bioactivity remain unexplored. Therefore, we next examined the truncated FGF23 proteins for activity in the HEK-293-KL cell line. Due to the fact that the serial truncations resulted in removal of the epitopes required for the intact and COOH-terminal FGF23 ELISAs, confirmation of protein expression was assessed through examination of the intact FGF23 bands upon Western analyses of the truncated mutants (comparison of the 33-, 25-, 23-, and 20-kDa species). These analyses demonstrated that the FGF23Δ19 and FGF23Δ48 truncations possessed signaling capabilities and increased EGR1 mRNA five- to sixfold, which was not different from WT FGF23 (P < 0.167 and 0.427, respectively; Fig. 6B). However, FGF23 protein lacking the 62 and 71 COOH-terminal residues had no activity compared with WT FGF23 or the FGF23Δ19 and FGF23Δ48 truncations (P < 0.0001; Fig. 6B). Thus the residues between −62 and −48 (i.e., residues between +189 and +203 in mature 251-residue FGF23) are required for FGF23 activity, whereas the FGF23 amino acids 3′ to residue 203 are not necessary to initiate FGF23-dependent intracellular signaling.
DISCUSSION
In the present study, we identified a novel Q54K mutation in the FGF23 gene in three patients with TC and, through in vitro studies, tested the etiology of TC using biochemical analyses of FGF23 processing and activity. FGF23 is required for the control of serum phosphate and vitamin D metabolism, as highlighted by its role in ADHR (1, 33) and familial TC (2, 3, 9, 22). Stabilizing mutations in FGF23 cause ADHR through the interruption of the SPC 176RXXR179/S cleavage site (32), whereas destabilizing mutations in FGF23 lead to TC (3, 22). The phenotype of patients with TC varies considerably across and within kindreds. However, despite the clinical inconsistency in ectopic and vascular calcification severity, these patients share a common biochemical finding of persistent hyperphosphatemia. Furthermore, to date, all patients with TC due to GALNT3 or FGF23 mutations display markedly increased levels of COOH-terminal FGF23 fragments in tandem with low or low/normal levels of bioactive intact FGF23 protein (Fig. 2) (2, 9, 13, 15, 22). Importantly, all known FGF23-TC mutations occur in the portion of FGF23 conserved among the FGF family members NH2 terminal to the 176RXXR179/S SPC site.
The mutations in FGF23 that lead to TC have dramatic effects on the secretion of intact, active FGF23. Indeed, the ability of cells to secrete intact FGF23 protein containing the novel Q54K mutation and the untested H41Q and M96T alterations, as well as those mutations reported previously (2, 3, 6, 22, 23), was greatly reduced compared with wild-type FGF23, as shown by Western analyses of cellular lysates and conditioned media collected from cells transiently expressing FGF23 (Fig. 4). Consistent with our current findings, previous in vitro analyses of the FGF23 TC mutation S71G indicated that this amino acid alteration leads to the elevated secretion of FGF23 proteolytic fragments (3, 21). Disruption of the FGF23 176RXXR179/S180 SPC cleavage site using the ADHR R176Q and R179Q mutations in tandem with the S71G TC mutation resulted in a protein species less susceptible to cleavage (21), consistent with increased sensitivity of the FGF23-TC mutants to intracellular proteases.
In parallel with the FGF23 mutations, the known GALNT3 inactivating mutations that result in TC lead to decreased stability of full-length, biologically active FGF23 as the molecular pathogenesis of this disorder (5, 9, 13, 29). To date, all patients with FGF23 or GALNT3 mutations share similar COOH-terminal and intact FGF23 ELISA profiles (10). The FGF23 protein is O-glycosylated (27), and recent studies demonstrated improved secretion of FGF23 in the presence of GALNT3 in vitro (18). Thus it is likely that GALNT3 plays a posttranslational regulatory role in the production of bioactive FGF23. Consistent with this hypothesis, FGF23 peptides carrying mutations at the glycosylation site at T178 within the 176RHTR179 SPC motif are cleaved in vitro (8). Together with the GALNT3 findings, these results indicate that inappropriate exposure of the SPC site in FGF23 to proteases could occur through misfolding caused by the FGF23-TC mutations or by altered glycosylation caused by the lack of GALNT3 activity, resulting in the increased susceptibility of FGF23 protein to intracellular degradation and ultimately to insufficient amounts of circulating intact FGF23.
The bioactivity of FGF23 is dependent upon the expression of the KL coreceptor in the presence of a canonical FGFR (31). Of significance, the H41Q, Q54K, S71G, M96T, and S129F FGF23 mutant proteins retained the ability to activate KL-dependent signaling (Fig. 5); however, this activity was reduced over the dose and time tested compared with that of the wild-type FGF23 protein (Fig. 5). Determining that the FGF23-TC mutants possessed residual activity adds to our current understanding of the complexity of the disease etiology in TC patients. In this regard, our studies indicated that the decrease in circulating biologically active FGF23 in patients with TC is likely the primary cause for the persistent hyperphosphatemia in these patients. Taking into consideration the findings of the present study, as well as the normal or nearly normal intact serum levels of FGF23 in the current patients, as well as in other reported cases (2, 22), the decreased activity of the mutant protein may also have a role in TC. Decreased serum levels of biologically active FGF23 may be the initial step in disease pathology in patients with TC. As TC patients produce more FGF23 to attempt to compensate for their hyperphosphatemia, nearly normal serum levels of FGF23 are approached; however, these levels are insufficient to suppress the renal reabsorption of phosphate. Of note, Fgf23-null and KL-null mice display a severe hyperphosphatemic phenotype and survive an average of only 10–12 wk (26, 31). Therefore, the residual activity of the TC-mutant FGF23 isoforms is likely sufficient to sustain life in humans; however, these mutant isoforms are not produced in quantities high enough to adequately inhibit the renal reabsorption of phosphate.
Interestingly, the FGF23 mutations that are known to result in TC are located within the NH2-terminal region that shares a considerable degree of homology with all members of the FGF protein family (residues 25–180 in FGF23), and no disease-causing mutations have been found in the COOH-terminal variable region of FGF23. The consistent lack of COOH-terminal mutations in patients with disorders of phosphate homeostasis led to our investigation of this uncharacterized region. The deletion of 19 and 48 COOH-terminal residues had no effect on FGF23 processing or secretion (Fig. 6) or on FGF23 activity (Fig. 6). In contrast, the deletion of 62 COOH-terminal FGF23 residues, as well as the entire COOH-terminal variable region with the FGF23Δ71 truncation, did not affect protein production and secretion (Fig. 6) but abolished KL-dependent signaling (Fig. 6). These findings have two implications. First, FGF23 residues between 189 [the last residue in FGF23Δ62 (inactive)] and 203 [the last residue in FGF23Δ48 (active)] are critical for FGF23 bioactivity, and second, the NH2-terminal region produced by cleavage at the 176RXXR179/S180 SPC recognition site lacks signaling activity through EGR1, as shown by the FGF23Δ71 truncated species (Fig. 6). Since FGF23 is properly secreted and retains activity with large portions of the COOH-terminal tail truncated, this could also explain the lack of known disease-causing mutations within the nucleotides 3′ to the R176XXR179/S180 site in FGF23. In this regard, amino acid substitutions in this variable region are consistent with being less deleterious to FGF23 expression and function compared with the NH2-terminal conserved portions of FGF23 and thus may not lead to disease.
In summary, we report a novel recessive mutation in the FGF23 gene (Q54K) that results in familial TC. We have determined that all of the known FGF23 mutations that result in TC lead to destabilization of the intact molecule. We have also shown that the COOH-terminal variable region is not required for FGF23 processing and secretion; however, residues adjacent to the conserved NH2 terminus are necessary for full bioactivity. Collectively, these findings expand our molecular understanding of TC as well as define protein domains critical for FGF23 activity.
GRANTS
We acknowledge support by National Institutes of Health (NIH) Grants DK-063934 (K. E. White), AR-049698 (H. C. Dietz), and AR-042044 (H. C. Dietz), an NIH Musculoskeletal Training Grant to the Indiana University School of Medicine, AR-007581 (support of H. J. Garringer), the Howard Hughes Medical Institute (H. C. Dietz), and the Indiana Genomics Initiative of Indiana University, supported in part by the Lilly Endowment (K. E. White).
DISCLOSURES
H. J. Garringer, S. I. Davis, X. Yu, S. M. J. Mortazavi, F. Esteghamat, M. Malekpour, D. E. Arking, and H. C. Dietz have nothing to declare. K. E. White receives royalties from licensing FGF23 to Kirin Pharmaceuticals.
Acknowledgments
We acknowledge the participation of all patients.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.ADHR Consortium. Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat Genet 26: 345–348, 2000. [DOI] [PubMed] [Google Scholar]
- 2.Araya K, Fukumoto S, Backenroth R, Takeuchi Y, Nakayama K, Ito N, Yoshii N, Yamazaki Y, Yamashita T, Silver J, Igarashi T, Fujita T. A novel mutation in fibroblast growth factor 23 gene as a cause of tumoral calcinosis. J Clin Endocrinol Metab 90: 5523–5527, 2005. [DOI] [PubMed] [Google Scholar]
- 3.Benet-Pages A, Orlik P, Strom TM, Lorenz-Depiereux B. An FGF23 missense mutation causes familial tumoral calcinosis with hyperphosphatemia. Hum Mol Genet 14: 385–390, 2005. [DOI] [PubMed] [Google Scholar]
- 4.Bennett EP, Hassan H, Clausen H. cDNA cloning and expression of a novel human UDP-N-acetyl-alpha-d-galactosamine. Polypeptide N-acetylgalactosaminyltransferase, GalNAc-t3. J Biol Chem 271: 17006–17012, 1996. [DOI] [PubMed] [Google Scholar]
- 5.Campagnoli MF, Pucci A, Garelli E, Carando A, Defilippi C, Lala R, Ingrosso G, Dianzani I, Forni M, Ramenghi U. Familial tumoral calcinosis and testicular microlithiasis associated with a new mutation of GALNT3 in a white family. J Clin Pathol 59: 440–442, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chefetz I, Heller R, Galli-Tsinopoulou A, Richard G, Wollnik B, Indelman M, Koerber F, Topaz O, Bergman R, Sprecher E, Schoenau E. A novel homozygous missense mutation in FGF23 causes Familial Tumoral Calcinosis associated with disseminated visceral calcification. Hum Genet 118: 261–266, 2005. [DOI] [PubMed] [Google Scholar]
- 7.Econs MJ, McEnery PT. Autosomal dominant hypophosphatemic rickets/osteomalacia: clinical characterization of a novel renal phosphate-wasting disorder. J Clin Endocrinol Metab 82: 674–681, 1997. [DOI] [PubMed] [Google Scholar]
- 8.Frishberg Y, Ito N, Rinat C, Yamazaki Y, Feinstein S, Urakawa I, Navon-Elkan P, Becker-Cohen R, Yamashita T, Araya K, Igarashi T, Fujita T, Fukumoto S. Hyperostosis-hyperphosphatemia syndrome: a congenital disorder of O-glycosylation associated with augmented processing of fibroblast growth factor 23. J Bone Miner Res 22: 235–242, 2007. [DOI] [PubMed] [Google Scholar]
- 9.Garringer HJ, Fisher C, Larsson TE, Davis SI, Koller DL, Cullen MJ, Draman MS, Conlon N, Jain A, Fedarko NS, Dasgupta B, White KE. The role of mutant UDP-N-acetyl-alpha-d-galactosamine-polypeptide N-acetylgalactosaminyltransferase 3 in regulating serum intact fibroblast growth factor 23 and matrix extracellular phosphoglycoprotein in heritable tumoral calcinosis. J Clin Endocrinol Metab 91: 4037–4042, 2006. [DOI] [PubMed] [Google Scholar]
- 10.Garringer HJ, Mortazavi SM, Esteghamat F, Malekpour M, Boztepe H, Tanakol R, Davis SI, White KE. Two novel GALNT3 mutations in familial tumoral calcinosis. Am J Med Genet A 143: 2390–2396, 2007. [DOI] [PubMed] [Google Scholar]
- 11.Goetz R, Beenken A, Ibrahimi OA, Kalinina J, Olsen SK, Eliseenkova AV, Xu C, Neubert TA, Zhang F, Linhardt RJ, Yu X, White KE, Inagaki T, Kliewer SA, Yamamoto M, Kurosu H, Ogawa Y, Kuro-o M, Lanske B, Razzaque MS, Mohammadi M. molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Mol Cell Biol 27: 3417–3428, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg BG, Winters RW, Graham JB. The normal range of serum inorganic phosphorus and its utility as a discriminant in the diagnosis of congenital hypophosphatemia. J Clin Endocrinol Metab 20: 364–379, 1960. [DOI] [PubMed] [Google Scholar]
- 13.Ichikawa S, Guigonis V, Imel EA, Courouble M, Heissat S, Henley JD, Sorenson AH, Petit B, Lienhardt A, Econs MJ. Novel GALNT3 mutations causing hyperostosis-hyperphosphatemia syndrome result in low intact fibroblast growth factor 23 concentrations. J Clin Endocrinol Metab 92: 1943–1947, 2007. [DOI] [PubMed] [Google Scholar]
- 14.Ichikawa S, Imel EA, Kreiter ML, Yu X, Mackenzie DS, Sorenson AH, Goetz R, Mohammadi M, White KE, Econs MJ. A homozygous missense mutation in human KLOTHO causes severe tumoral calcinosis. J Clin Invest 117: 2684–2691, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ichikawa S, Lyles KW, Econs MJ. A novel GALNT3 mutation in a pseudoautosomal dominant form of tumoral calcinosis: evidence that the disorder is autosomal recessive. J Clin Endocrinol Metab 90: 2420–2423, 2005. [DOI] [PubMed] [Google Scholar]
- 16.Inclan A, Leon P, Camjeo MG. Tumoral calcinosis. JAMA 121: 490–495, 1943. [Google Scholar]
- 17.Jonsson KB, Zahradnik R, Larsson T, White KE, Sugimoto T, Imanishi Y, Yamamoto T, Hampson G, Koshiyama H, Ljunggren O, Oba K, Yang IM, Miyauchi A, Econs MJ, Lavigne J, Juppner H. Fibroblast growth factor 23 in oncogenic osteomalacia and X-linked hypophosphatemia. N Engl J Med 348: 1656–1663, 2003. [DOI] [PubMed] [Google Scholar]
- 18.Kato K, Jeanneau C, Tarp MA, Benet-Pagès A, Lorenz-Depiereux B, Bennett EP, Mandel U, Strom TM, Clausen H. Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. Secretion of fibroblast growth factor 23 requires O-glycosylation. J Biol Chem 281: 18370–18377, 2006. [DOI] [PubMed] [Google Scholar]
- 19.Kuroo M Introduction: aging research comes of age. Cell Mol Life Sci 57: 695–697, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurosu H, Ogawa Y, Miyoshi M, Yamamoto M, Nandi A, Rosenblatt KP, Baum MG, Schiavi S, Hu MC, Moe OW, Kuro-o M. Regulation of fibroblast growth factor-23 signaling by klotho. J Biol Chem 281: 6120–6123, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsson T, Davis SI, Garringer HJ, Mooney SD, Draman MS, Cullen MJ, White KE. Fibroblast growth factor-23 mutants causing familial tumoral calcinosis are differentially processed. Endocrinology 146: 3883–3891, 2005. [DOI] [PubMed] [Google Scholar]
- 22.Larsson T, Yu X, Davis SI, Draman MS, Mooney SD, Cullen MJ, White KE. A novel recessive mutation in fibroblast growth factor-23 causes familial tumoral calcinosis. J Clin Endocrinol Metab 90: 2424–2427, 2005. [DOI] [PubMed] [Google Scholar]
- 23.Masi L, Gozzini A, Carbonell S, Amedei A, Falchetti A, Capanna R, Tanini A, Brandi ML. A novel recessive mutation in fibroblast growth factor-23 (FGF23) causes a tumoral calcinosis (Abstract). J Bone Miner Res 20: S128, 2005. [Google Scholar]
- 24.Mitnick PD, Goldfarb S, Slatopolsky E, Lemann J Jr, Gray RW, Agus ZS. Calcium and phosphate metabolism in tumoral calcinosis. Ann Intern Med 92: 482–487, 1980. [DOI] [PubMed] [Google Scholar]
- 25.Prince MJ, Schaeffer PC, Goldsmith RS, Chausmer AB. Hyperphosphatemic tumoral calcinosis: association with elevation of serum 1,25-dihydroxycholecalciferol concentrations. Ann Intern Med 96: 586–591, 1982. [DOI] [PubMed] [Google Scholar]
- 26.Shimada T, Kakitani M, Yamazaki Y, Hasegawa H, Takeuchi Y, Fujita T, Fukumoto S, Tomizuka K, Yamashita T. Targeted ablation of Fgf23 demonstrates an essential physiological role of FGF23 in phosphate and vitamin D metabolism. J Clin Invest 113: 561–568, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimada T, Muto T, Urakawa I, Yoneya T, Yamazaki Y, Okawa K, Takeuchi Y, Fujita T, Fukumoto S, Yamashita T. Mutant FGF-23 responsible for autosomal dominant hypophosphatemic rickets is resistant to proteolytic cleavage and causes hypophosphatemia in vivo. Endocrinology 143: 3179–3182, 2002. [DOI] [PubMed] [Google Scholar]
- 28.Silve C, Friedlander G. Renal regulation of phosphate excretion. In: The Kidney: Physiology & Pathophysiology (3rd ed.), edited by Seldin DW and Giebisch G. Philadelphia, PA: Lippincott Williams & Wilkins, 2000, p. 1885–1904.
- 29.Specktor P, Cooper JG, Indelman M, Sprecher E. Hyperphosphatemic familial tumoral calcinosis caused by a mutation in GALNT3 in a European kindred. J Hum Genet 51: 487–490, 2006. [DOI] [PubMed] [Google Scholar]
- 30.Topaz O, Shurman DL, Bergman R, Indelman M, Ratajczak P, Mizrachi M, Khamaysi Z, Behar D, Petronius D, Friedman V, Zelikovic I, Raimer S, Metzker A, Richard G, Sprecher E. Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat Genet 36: 579–581, 2004. [DOI] [PubMed] [Google Scholar]
- 31.Urakawa I, Yamazaki Y, Shimada T, Iijima K, Hasegawa H, Okawa K, Fujita T, Fukumoto S, Yamashita T. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature 444: 770–774, 2006. [DOI] [PubMed] [Google Scholar]
- 32.White KE, Carn G, Lorenz-Depiereux B, Benet-Pages A, Strom TM, Econs MJ. Autosomal-dominant hypophosphatemic rickets (ADHR) mutations stabilize FGF-23. Kidney Int 60: 2079–2086, 2001. [DOI] [PubMed] [Google Scholar]
- 33.White KE, Jonsson KB, Carn G, Hampson G, Spector TD, Mannstadt M, Lorenz-Depiereux B, Miyauchi A, Yang IM, Ljunggren O, Meitinger T, Strom TM, Juppner H, Econs MJ. The autosomal dominant hypophosphatemic rickets (ADHR) gene is a secreted polypeptide overexpressed by tumors that cause phosphate wasting. J Clin Endocrinol Metab 86: 497–500, 2001. [DOI] [PubMed] [Google Scholar]
- 34.Yamazaki Y, Okazaki R, Shibata M, Hasegawa Y, Satoh K, Tajima T, Takeuchi Y, Fujita T, Nakahara K, Yamashita T, Fukumoto S. Increased circulatory level of biologically active full-length FGF-23 in patients with hypophosphatemic rickets/osteomalacia. J Clin Endocrinol Metab 87: 4957–4960, 2002. [DOI] [PubMed] [Google Scholar]