Abstract
Increased plasma and hepatic TNF-α expression is well documented in patients with alcoholic hepatitis and is implicated in the pathogenesis of alcoholic liver disease. We have previously shown that monocytes from patients with alcoholic hepatitis show increased constitutive and LPS-induced NF-κB activation and TNF-α production. Our recent studies showed that chronic ethanol exposure significantly decreased cellular cAMP levels in both LPS-stimulated and unstimulated monocytes and Kupffer cells, leading to an increase in LPS-inducible TNF-α production by affecting NF-κB activation and induction of TNF mRNA expression. Accordingly, the mechanisms underlying this ethanol-induced decrease in cellular cAMP leading to an increase in TNF expression were examined in monocytes/macrophages. In this study, chronic ethanol exposure was observed to significantly increase LPS-inducible expression of cAMP-specific phosphodiesterase (PDE)4B that degrades cellular cAMP. Increased PDE4B expression was associated with enhanced NF-κB activation and transcriptional activity and subsequent priming of monocytes/macrophages leading to enhanced LPS-inducible TNF-α production. Selective inhibition of PDE4 by rolipram abrogated LPS-mediated TNF-α expression at both protein and mRNA levels in control and ethanol-treated cells. Notably, PDE4 inhibition did not affect LPS-inducible NF-κB activation but significantly decreased NF-κB transcriptional activity. These findings strongly support the pathogenic role of PDE4B in the ethanol-mediated priming of monocytes/macrophages and increased LPS-inducible TNF production and the subsequent development of alcoholic liver disease (ALD). Since enhanced TNF expression plays a significant role in the evolution of clinical and experimental ALD, its downregulation via selective PDE4B inhibitors could constitute a novel therapeutic approach in the treatment of ALD.
dysregulated tnf metabolism in alcoholic hepatitis (AH) was first described by us almost two decades ago with the observation that cultured monocytes from patients with AH spontaneously produced TNF and produced significantly more TNF in response to an LPS stimulus than control monocytes (20). Furthermore, we and others have shown that there are increased serum levels of TNF, increased monocyte TNF production, and hepatic immunohistochemical staining for TNF in AH that frequently correlate with disease severity and mortality (2, 5, 12, 15, 19, 20, 22, 27). Although TNF plays an important role in the pathogenesis of liver injury and the clinical/biochemical abnormalities of AH (9, 20–22), the mechanism(s) by which ethanol enhances TNF expression, particularly in monocytes/macrophages, is only beginning to be understood (6, 14, 37, 38). We recently demonstrated that chronic ethanol exposure of monocytes/macrophages (including Kupffer cells) decreases both basal and LPS-stimulated cAMP levels and leads to the enhancement of LPS-inducible TNF production (6).
Increasing cellular cAMP levels with different types of cAMP enhancers has invariably resulted in the suppression of TNF production stimulated either in vitro or in vivo with LPS in monocytes/macrophages of both human and murine origin (39). Various studies have documented that, depending on the cell type, suppression of LPS-inducible TNF production by cAMP can occur in both NF-κB-dependent and -independent mechanisms (25, 28, 32, 34, 36). Our recent findings demonstrated that increasing the cellular cAMP levels with the use of a cell-permeable cAMP analog (dbcAMP) inhibits the synergistic enhancement in TNF-α expression caused by ethanol and LPS (6). To further extend our observations and clarify the mechanisms involved in the enhancement of LPS-inducible TNF expression in monocytes primed by ethanol exposure, we examined the role of phosphodiesterases (PDE), enzymes that degrade cAMP and play a critical role in cAMP-mediated signaling. Particularly, cAMP-specific PDE4, which is present in different tissues, is found to be the predominant PDE isoenzyme among 11 PDEs in human monocytes (4, 18). Numerous studies have shown that LPS-inducible TNF production by monocytes is markedly decreased when PDE activity is blocked by PDE4-specific inhibitors (3, 4, 16, 23, 29, 30, 31, 33, 35). PDE4 isozyme has four isoforms, namely A, B, C, and D (18). Specific to monocytes/macrophages, it has been established that PDE4B is involved in Toll-like receptor signaling and is essential for LPS-induced TNF-α expression (10, 11). Accordingly, the present work examined the putative role of PDE4B expression and activity in the ethanol-mediated enhancement of LPS-inducible TNF expression in monocytes.
The data obtained from these studies support our earlier findings and further extend them by showing that chronic ethanol exposure significantly increases LPS-inducible PDE4B expression and activity, resulting in decreased cellular cAMP in monocytes. Selective inhibition of PDE4 by rolipram abrogated LPS-mediated TNF expression at both protein and mRNA levels in control and ethanol-treated cells, strongly supporting a causal role for PDE4B in priming and augmentation of TNF expression. Notably, PDE4 inhibition did not affect LPS-inducible NF-κB activation but significantly decreased its transcriptional activity, thereby impacting TNF expression.
MATERIALS AND METHODS
Materials.
RAW 264.7 murine macrophage and THP-1 human monocytic cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). LPS (Escherichia coli 0111:B4) was purchased from Difco Laboratories (Detroit, MI). Before use, LPS was dissolved in sterile, pyrogen-free water, sonicated, and diluted with sterilized PBS. Penicillin, streptomycin, DMEM and RPMI media, and fetal bovine serum were purchased from Invitrogen (Grand Island, NY); murine and human TNF-α ELISA kits were from BioSource International (Camarillo, CA). Rolipram and p65 antibody was from Biomol Research Laboratories (Plymouth Meeting, PA). Histone 3 antibody was from Cell Signaling Technology (Beverly, MA). Supershift antibodies for p50 and p65 were from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell culture.
RAW 264.7 cells were cultured in DMEM, and undifferentiated THP-1 cells were cultured in RPMI media containing 10% (vol/vol) fetal bovine serum, 10 U/ml penicillin, and 10 μg/ml streptomycin at 37°C in a humidified 5% CO2 atmosphere. Cells were chronically exposed to 25 mM ethanol for a period of 6 wk.
TNF-α ELISA.
TNF-α in conditioned medium was quantified using ELISA kit in accordance with the manufacturer's instructions. All assays were run in triplicate.
RNA isolation and real-time PCR analysis.
RT-PCR assays were used to assess TNF-α and PDE4B mRNA levels in RAW 264.7 and THP-1 cells. Total RNA was isolated from cells using TRIzol Reagent (Invitrogen). For real-time PCR, the first-strand cDNA was synthesized using TaqMan Reverse transcription reagents (Applied Biosystems, Foster City, CA). The reverse transcription was carried out using 1× TaqMan RT buffer, 5.5 mM MgCl2, 500 mM of each dNTP, 2.5 mM random hexamer, 8 U of RNase inhibitor, and 25 U of Multiscribe Reverse Transcriptase with 200 ng of total RNA. The RT conditions were 10 min at 25°C, 30 min at 48°C, and 5 min at 95°C. Reactions in which the enzyme or RNA were omitted were used as negative controls. Real-time PCR was performed in triplicates with an ABI prism 7500 sequence detection system and SYBR green I dye reagents. The specific primers were designed for mouse GAPDH, TNF-α, and PDE4B with Primer3 software program. The following primers were used in real-time PCR: mGAPDH-FP, 5′TTGTGGAAGGGCTCATGACC3′; mGAPDH-RP, 5′TCTTCTGGGTGGCAGTGATG3′; mTNF-α-FP, 5′GAAGTTCCCAAATGGCCTCC3′; mTNF-α-RP, 5′GTGAGGGTCTGGGCCATAGA3′; mPDE4B-FP, 5′GCAGAAGGACAGGGAGAAGAA3′; and mPDE4B-RP, 5′ACCCAGCCACATTGAAGATG3′. Human 18S, TNF-α, and PDE4B-specific primers were purchased from SuperArray Bioscience (Frederick, MD).
The parameter threshold cycle (Ct) was defined as the fraction cycle number at which the fluorescence passed the threshold. The relative gene expression was analyzed using the 2−ΔΔCt method (17) by normalizing with GAPDH or 18S gene expression in all the experiments.
Nuclear protein extraction.
Cells were washed twice with PBS and lysed by hypotonic shock in cytoplasmic extraction buffer (10 mM HEPES, pH 7.8, 1.5 mM MgCl2, 0.5 mM DTT, 0.1% Nonidet P-40, and 0.5 μM PMSF) and 1 μg/ml each of the protease inhibitors aprotinin, leupeptin, and leucinethiol. The cells were collected by scraping, and the nuclei were separated from the cytosolic proteins by a low-speed centrifugation at 1,500 g for 5 min. The supernatant (cytoplasmic extract) was stored at −70°C. The nuclear pellet was resuspended in nuclear extraction buffer (20 mM HEPES, pH 7.8, 520 mM NaCl, 1.5 mM MgCl2, 0.1 mM EDTA, 25% glycerol, 0.5 mM DTT, 0.1% Nonidet P-40, and 0.5 μM PMSF) and 1 μg/ml each of the protease inhibitors aprotinin, leupeptin, and leucinethiol and incubated on ice for 60 min. The samples were centrifuged at 100,000 g for 15 min, and the supernatant containing nuclear proteins was stored in small aliquots at −70°C until further use. The protein concentration in nuclear extracts was measured with the use of the BioRad Dye Reagent (Bio-Rad, Hercules, CA) in accordance with the manufacturer's protocol.
EMSA.
Double-stranded oligonucleotide containing the binding site for NF-κB (5′-AGT-TGA-GGG-GAC-TTT-CCC-AGG-C-3′; Promega, Madison, WI) was end-labeled with (32P) dATP using T4 polynucleotide kinase (Gibco, Grand Island, NY). Nuclear proteins (5 μg) were incubated in binding buffer [50 mM Tris, pH 7.5, 500 mM NaCl, 5 mM DTT, 5 mM EDTA, 20% (vol/vol) glycerol, and 0.4 mg/ml sonicated salmon sperm DNA] with 0.3 ng of 32P-labeled probe for 20 min at room temperature. For supershift analysis, nuclear extracts were preincubated with 1 μg of p65 and/or p50 monoclonal antibody for 15 min before addition of the radioactive probe. The resultant DNA/protein complexes were separated by electrophoresis on a 5% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA (50 mM Tris, pH 8.0, 45 mM borate, and 5 mM EDTA). The gels were dried and analyzed by autoradiography for ∼18 h. The specificity of the reaction was evaluated by incubating the nuclear extract with a 100 molar excess of unlabeled wild-type (W) and mutant (M) NF-κB oligonucleotide.
Western blot analysis.
Nuclear proteins (30 μg) were analyzed by SDS-polyacrylamide gel electrophoresis using a Bio-Rad electrophoresis system, followed by immunoblotting according to the manufacturer's instructions. Immunoreactive bands were visualized using the enhanced chemiluminescence light detection reagents (Amersham, Arlington Heights, IL). Quantification (densitometry) of bands was performed using ImageQuantTLv2003.03. Histone 3 was used as a loading control.
Plasmids and transfections.
NF-κB-luciferase reporter construct was obtained from BD Biosciences/Clontech (Palo Alto, CA). Transfections were carried out using FuGENE according to instructions from the manufacturer (Roche Diagnostics, Indianapolis, IN). To control for differences in transfection efficiency, transfected cells were scraped and re-plated after 24 h at a density of 1 × 106 cells/well in 24-well plates and treated for 6 h.
Luciferase assay.
Luciferase activity was measured using Luciferase Assay Reagent (Promega). After treatments, total cell lysates were prepared in reporter lysis buffer (Promega). Luciferase enzymatic activity was measured in a TD 20/20 luminometer using a specific substrate provided by Promega.
PDE4 enzymatic assay.
PDE4 specific enzymatic activity was determined using PDE4 assay kit (FabGennix International, Frisco, TX) as described previously (13). Briefly, cells were lysed after treatment using SolObuffer (FabGennix International), and 25 μg protein was used per assay. Assay was carried in duplicate; the final concentration of cAMP was 2 μM. PDE4 activity was estimated from the difference between total and rolipram-resistant PDE activity.
Statistical analysis.
All experiments were repeated at least three times. Data were presented as means ± SD for the indicated number of independently performed experiments. Student's t-test was used for the determination of statistical significance. P < 0.05 was considered significant.
RESULTS
RAW 264.7 and THP-1 cells were continuously maintained in ethanol (25 mM)-supplemented medium, which was changed after every 12 h over a period of 6 wk. The concentration of ethanol chosen to treat the cells reflects the blood alcohol level that can be achieved during human alcohol consumption. Alcohol levels in media were routinely measured over the study duration using the Sigma Diagnostics Alcohol Reagent (Sigma Diagnostics, St. Louis, MO) (6). Typically, under the experimental conditions of ethanol treatment, a reduction in ethanol concentration by ∼12–15% occurred over the incubation period of 12 h. A minor reduction in the doubling time of both RAW 264.7 and THP-1 cells was observed in the first week only, with no further effect on either viability or proliferation (data not shown).
Ethanol exposure enhances PDE4 gene expression and activity in human THP-1 monocytes and murine RAW macrophages.
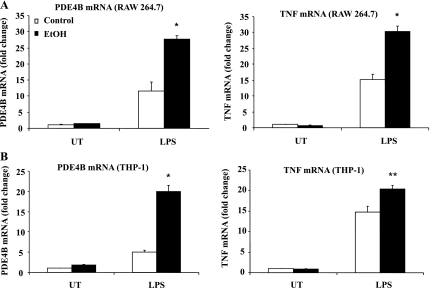
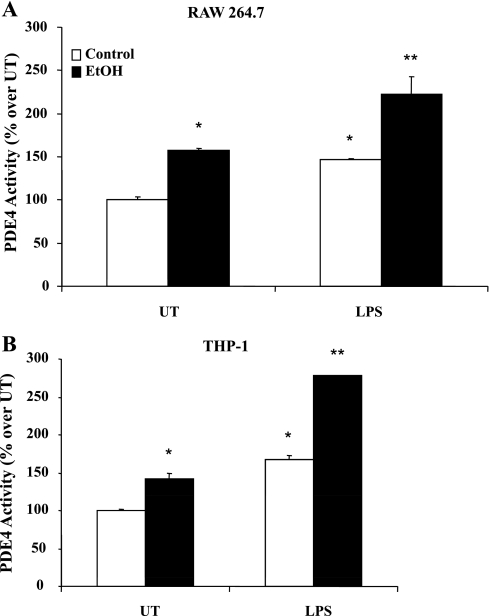
We have previously shown that chronic ethanol exposure significantly decreases intracellular cAMP levels in monocytes and Kupffer cells and is responsible for priming them to LPS-inducible TNF overproduction. PDE4B is the predominant subtype in monocytes/macrophages that degrades cAMP and is critical for LPS signaling and TNF expression (10, 11). Therefore, we examined the effect of chronic ethanol exposure on PDE4B expression and activity in monocytes/macrophages. After ethanol exposure, cells were plated into 60-mm culture dishes at 1 × 106 cells/ml density and stimulated with LPS (100 ng/ml) for 2 h for gene expression analysis and 4 h for PDE4 activity measurement. The time points for TNF and PDE4B mRNA expression and PDE4 activity were selected on the basis of previously established expression optima (6, 10, 11). Ethanol treatment significantly augmented LPS-inducible PDE4B mRNA with increased TNF expression in both RAW 264.7 (murine macrophage) and THP-1 (human monocytic) cells (Fig. 1, A and B, respectively). Furthermore, ethanol-treated cells showed increased basal and LPS-inducible PDE4 activity compared with control cells (Fig. 2, A and B).
Fig. 1.
Chronic alcohol exposure of monocytes/macrophages markedly upregulates LPS-inducible phosphodiesterase (PDE)4B and TNF-α mRNA expression. Cells were stimulated with LPS (100 ng/ml) and collected in 2 h for total RNA isolation. PDE4B and TNF-α mRNA were quantified using real-time PCR. Data are representative of 3 separate experiments. Data are presented as means ± SD (n = 3). A: RAW cells, *P < 0.05 compared with LPS-stimulated control. B: THP-1 cells, *P < 0.01 and **P < 0.05 compared with LPS-stimulated control. UT, untreated.
Fig. 2.
Chronic ethanol exposure significantly enhances PDE4-specific activity in monocytes/macrophages. Control and ethanol-treated cells were left untreated (UT) and/or stimulated with LPS (100 ng/ml) for 4 h. PDE4 activity was measured in 25-μg cell lysates and is represented as percent activity over control UT cells. Data are presented as means ± SD (n = 3). A: mouse macrophages (RAW), *P < 0.01 compared with control, UT, **P < 0.05 compared with control LPS and EtOH, UT. B: human monocytes (THP-1), *P < 0.05 compared with control, UT, **P < 0.01 compared with control, LPS.
PDE4 inhibition results in decreased LPS-inducible TNF production caused by ethanol.
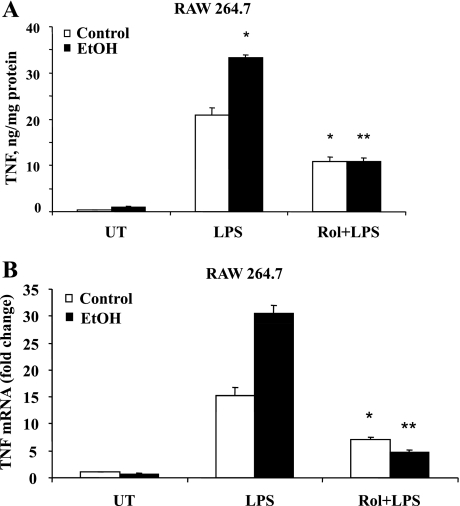
Our previous work has demonstrated that ethanol-mediated decrease in cAMP levels plays a significant role in priming monocytes, resulting in increased LPS-stimulatable TNF production. Also, since PDE4B specifically degrades cAMP in response to LPS, its role in modulating cAMP-dependent regulation of TNF expression in ethanol-treated monocytes was examined. We used a PDE4-specific inhibitor, rolipram (10 μM), to inhibit PDE4 activity in all the experiments. At this concentration, rolipram has a negligible inhibitory effect on other PDEs (11). RAW cells exposed to ethanol were treated with rolipram for 30 min before LPS (100 ng/ml) stimulation for 6 h. Supernatants were collected and analyzed for TNF production by ELISA. As observed previously, ethanol primed the monocytes, leading to a significant increase in the LPS-inducible TNF production. Notably, inhibition of PDE4B by rolipram effectively inhibited the enhancement in the TNF-α expression caused by ethanol and LPS (Fig. 3A).
Fig. 3.
PDE4 inhibition significantly decreases TNF production at both protein and mRNA level in RAW 264.7 cells. At 30 min before LPS (100 ng/ml) stimulation, cells were treated with PDE4-specific inhibitor rolipram (Rol) (10 μM). A: samples were collected in 6 h, and TNF was quantified using ELISA. Data are presented as means ± SD (n = 3). *P < 0.01 compared with LPS-stimulated control and EtOH; **P < 0.01 with LPS-stimulated EtOH. B: cells were collected in 2 h after LPS stimulation for RNA extraction. TNF-α mRNA was quantified using real-time PCR. Data are representative of 3 separated experiments. Data are presented as means ± SD (n = 3). *P < 0.05 compared with LPS-stimulated control and **P < 0.01 with LPS-stimulated EtOH.
Effect of chronic alcohol exposure and PDE4 inhibition on LPS-induced TNF gene transcription.
To address the mechanistic involvement of PDE4B in the effects of chronic ethanol exposure on TNF production, steady-state levels of TNF mRNA were quantified in cells pretreated with rolipram. Real-time PCR analysis of total RNA obtained from treated cells showed that the LPS-inducible steady-state TNF mRNA levels were significantly higher in ethanol-treated macrophages compared with control cells, corroborating our earlier findings and work done by others (Fig. 3B, open bars). Furthermore, this synergistic enhancement in the TNF mRNA levels caused by ethanol and LPS was significantly attenuated by PDE4 inhibition (Fig. 3B, solid bars).
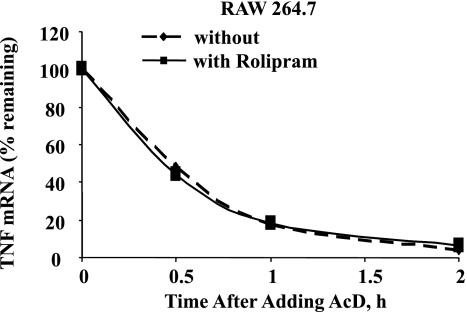
The effect of ethanol on the LPS-inducible steady-state TNF mRNA expression in monocytes/macrophages could be due to an increase in the synthesis of TNF mRNA or an increase in the stability of TNF mRNA. We did not observe the difference in the half-life (t1/2) of LPS-induced TNF mRNA between control and ethanol-treated cells (data not shown). We then examined the effect of PDE4 inhibition on stability of TNF mRNA. Ethanol-exposed cells with and without pretreatment with 10 μM rolipram for 30 min were stimulated with LPS (100 ng/ml) for 60 min followed by actinomycin D (1 μg/ml) up to 2 h. RNA was harvested at 0, 30, 60, and 120 min after actinomycin D addition and analyzed for TNF mRNA expression. The data obtained showed that PDE4 inhibition did not alter the stability of the LPS-induced TNF mRNA. The t1/2 of LPS-induced TNF mRNA was ∼25 min in both untreated as well as in rolipram-pretreated macrophages (Fig. 4). Thus pretreatment with PDE4 inhibitor, which abolished the enhancement in the LPS-inducible steady-state TNF mRNA expression in ethanol-treated macrophages, had no effect on the t1/2 of LPS-induced TNF mRNA.
Fig. 4.
PDE4 inhibition does not alter TNF mRNA half-life. RAW 264.7 cells chronically exposed to ethanol were pretreated with rolipram for 30 min and stimulated with LPS (100 ng/ml) for 1 h, followed by actinomycin D (AcD) (1 μg/ml), and total mRNA was isolated at times indicated above. The remainder TNF mRNA at each time point was quantified using real-time PCR.
PDE4 inhibition does not alter LPS-inducible NF-κB nuclear translocation and DNA binding but attenuates transcriptional activity of NF-κB.
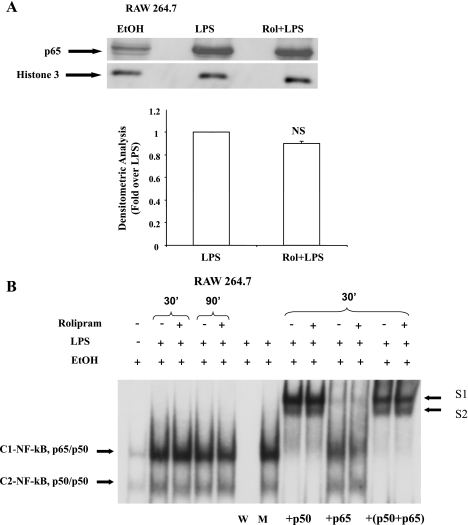
LPS stimulation of monocytes/ macrophages is known to induce TNF-α mRNA expression through a rapid and transient NF-κB activation leading to the induction of the TNF-α promoter. To assess whether the effect of PDE4 inhibition on LPS-stimulated TNF-α mRNA expression in ethanol-primed RAW cells was attributable to a decrease in NF-κB activation, nuclear levels of p65 and NF-κB DNA binding activity were examined in the nuclear proteins extracted from RAW cells pretreated with rolipram (10 μM for 30 min) followed by stimulation with LPS (100 ng/ml) for 30 min. Immunoblot analysis showed that pretreatment of cells with rolipram had no effect on the nuclear levels of LPS-inducible p65 (Fig. 5A); histone 3 was used as a loading control for the nuclear proteins.
Fig. 5.
LPS-inducible intranuclear level of p65 and NF-κB DNA binding is not affected by PDE4 inhibition in RAW 264.7 cells. Ethanol-exposed cells were pretreated with rolipram (10 μM) for 30 min and stimulated with LPS (100 ng/ml). A: p65 was analyzed by Western blotting from nuclear extracts obtained from cells stimulated with LPS for 30 min. Representative Western blot of p65 and histone 3 (as a loading control) is shown. Average density ratios of LPS and Rol+LPS from 3 separate experiments are shown in a bar graph. NS, not significant. Data are presented as fold change over LPS-inducible p65 levels normalized with histone 3. B: nuclear extracts obtained from LPS-stimulated cells for 30 and 90 min, respectively, were examined by EMSA to assess NF-κB DNA binding activity and supershift analysis to identify NF-κB DNA binding complexes using appropriate p50 and p65 antibodies. W, competition assay using unlabeled wild-type NF-κB; M, competition assay using mutant NF-κB oligonucleotides; +p50, supershift with p50; +p65, supershift with p65; +p50+p65, supershift with p50 and p65.
EMSA was performed to assess the effect of PDE4 inhibition on NF-κB DNA binding to the consensus κB sequence. Nuclear extracts were isolated from ethanol-treated monocytes without or with rolipram followed by LPS treatment for 30 and 90 min. Correspondent to the p65 nuclear levels, rolipram did not have any effect on the ethanol- and LPS-mediated NF-κB DNA binding (Fig. 5B). NF-κB/Rel transcription factors are homo- or heterodimeric complexes that consist of various combinations of Rel subunits. To identify the subunits present in the LPS-inducible NF-κB complexes, supershift experiments were performed using antibodies against p65 and p50 (Fig. 5B). In nuclear extracts from ethanol-treated cells stimulated with LPS for 30 min with and without rolipram, the p50-specific antibody completely shifted complex C2 and reduced the extent of binding of complex C1 and generating lower mobility complexes S1 and S2, showing that both C1 and C2 contain p50. The p65-specific antibody significantly shifted complex C1 to S1 with no effect on complex C2, demonstrating that C1 contains the p65 subunit of NF-κB. A combination of the two antibodies fully shifted both C1 and C2 to form S1 and S2. Thus supershift analysis indicates that the NF-κB/Rel complexes induced by LPS consist of p65/p50 heterodimer (C1) and p50/p50 homodimer (C2) and are not affected by rolipram.
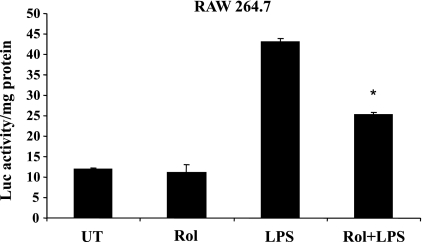
PDE4 inhibition downregulates LPS-stimulated NF-κB transcriptional activity.
We then examined the effect of PDE4 inhibition on the LPS-stimulated NF-κB transcriptional activity by performing transient transfection in RAW 264.7 cells with a luciferase reporter construct under the control of an NF-κB promoter containing three tandem repeats of the κB sequence (κB-luc). LPS stimulation for 6 h of ethanol-treated cells increased the NF-κB transcriptional activity approximately fourfold. Rolipram pretreatment significantly attenuated the increases in LPS-stimulated NF-κB activity (decreased by 42%) (Fig. 6). Taken together, these data indicate that PDE4 inhibition has minimal to no effect on the LPS-inducible signaling components leading to NF-κB activation; however, it can significantly decrease gene expression mediated by NF-κB-dependent transactivation.
Fig. 6.
PDE4 inhibition in alcohol-exposed cells significantly attenuates NF-κB activity in response to LPS in RAW 264.7 cells. Cells were transfected with a luciferase reporter construct containing the NF-κB-responsive IκB promoter as described in materials and methods. The transfected cells pretreated with rolipram (10 μM) for 30 min were further stimulated with LPS (100 ng/ml) for 6 h. Cytoplasmic extracts were prepared, and equal amounts were assayed for luciferase activity. Data are presented as means ± SD (n = 3). *P < 0.01 compared with LPS-stimulated cells.
DISCUSSION
Our earlier work documented that decreased cAMP levels played a causal role in ethanol-mediated priming of monocytes/macrophages leading to an increase in LPS-inducible TNF expression (6). PDEs are the only superfamily of enzymes that catalyze the hydrolysis of the cyclic nucleotides, cAMP and cGMP, and they play a critical role in regulating the intracellular levels of these important second messengers. Different functional pools of these cellular cyclic nucleotides are regulated by distinct PDEs in a cell-dependant manner. Particularly in the context of cAMP-regulating LPS-inducible TNF expression in monocytes, work done by Conti and colleagues (10, 11) has demonstrated the essential role of PDE4 (subtype B, PDE4B), a cAMP-specific PDE isozyme. Hence, effects of chronic ethanol exposure on PDE4 expression and activity and its functional role in upregulating LPS-inducible TNF expression was examined in the present work.
A major finding of this study was that chronic ethanol exposure led to a significant increase in the LPS-inducible PDE4B expression and activity in both monocytes and macrophages. Since PDE4B is required for TNF expression in response to LPS in monocytes/macrophages, these findings strongly suggest that its augmentation induced by ethanol plays a critical role in priming the monocytes/macrophages to mount an exaggerated inflammatory response to endotoxin. The role of PDE4B in the ethanol-mediated priming of macrophages was examined by using the prototypical PDE4-selective inhibitor rolipram. Cellular responses modulated by rolipram involve PDE4 regulation of cAMP levels. In this study, rolipram inhibited the enhanced TNF production caused by the synergism of ethanol and LPS, thus strongly supporting a causal role for a PDE4B-regulated pool of cAMP in controlling LPS-responsive TNF expression in macrophages chronically exposed to alcohol. These findings are consistent with earlier observations that PDE4 is the major cAMP-hydrolyzing PDE isozyme that regulates the cAMP-dependent mechanisms that play a central role in the LPS-inducible TNF expression in monocytes/macrophages (4, 23, 29, 35, 39). Furthermore, in accordance with our earlier work, which showed that the noncleavable cAMP analog (dbcAMP) inhibits TNF transcription in ethanol-treated macrophages, rolipram was observed to downregulate the steady-state levels of TNF mRNA expression without affecting TNF mRNA stability.
Numerous studies have demonstrated the critical role of NF-κB in the LPS-inducible transcriptional induction of the TNF gene (1, 7, 24). Our previous work (8) demonstrated that overproduction of TNF observed in peripheral blood mononuclear cells of alcoholic patients is associated with increased NF-κB activation and elevated TNF mRNA levels. In this regard, similar to our observations with dbcAMP, rolipram treatment had no effect on LPS-inducible NF-κB activation (nuclear translocation), as indicated by the nuclear p65 levels and DNA binding in ethanol-treated cells. However, functional studies in ethanol-treated monocytes demonstrated that rolipram significantly inhibited LPS-inducible transcriptional activation of the NF-κB reporter plasmid. Since the p50/p50 complex is not affected by rolipram pretreatment, it is unlikely that the observed repression of NF-κB activity caused by rolipram is occurring via the enhancement of p50/p50-mediated competition with the transcriptionally active p50/p65 heterodimer. These data and our previous observations suggest that chronic ethanol exposure may not only lead to an increase in LPS-inducible NF-κB activation but also further enhance the functional activity of nuclear NF-κB/Rel complexes because of decreased PDE4-regulated cellular pools of cAMP levels, resulting in overproduction of TNF by “ethanol-primed” monocytes/macrophages. This inference is also supported by several observations in different cell types, including monocytes, that show that cAMP represses NF-κB-mediated transcription and TNF mRNA expression without affecting NF-κB activation/DNA binding activity (25, 28, 32, 37). The exact mechanisms by which cAMP levels regulate NF-κB activity are unclear and are presently being investigated. In contrast to NF-κB binding sites, an activator protein-1 (AP-1)/cAMP response element (CRE)-like site in the proximal promoter region has been demonstrated to play an essential role in the cAMP-mediated downregulation of TNF gene transcription in murine macrophages (26). Hence, it is likely that the decreased levels of cAMP caused by enhanced PDE4B expression and activity in monocytes chronically exposed to ethanol could 1) increase NF-κB-dependent transactivation of the TNF promoter and 2) decrease AP-1/CRE-mediated suppression of TNF promoter activity, ultimately leading to overproduction of TNF.
Initial work by our laboratory showed increased priming of both human monocytes from patients with alcoholic hepatitis as well as Kupffer cells (macrophages) from rats chronically fed ethanol. Notably, our recent work has demonstrated that ethanol-mediated decrease in cAMP levels in cultured monocyte/macrophages and hepatic Kupffer cells plays a critical role in the priming of these cells leading to an exaggerated TNF response. Thus monocytes and macrophages, including hepatic Kupffer cells, play an important role in the dysregulated cytokine metabolism and pathogenesis in alcoholic liver disease (ALD). Our present data strongly suggest that decreased cellular cAMP levels in monocytes/macrophages chronically exposed to ethanol occur because of an increase in PDE4B expression. Importantly, the findings show that ethanol-mediated increase in PDE4B expression in monocytes/macrophages synergizes with LPS to upregulate the induction of TNF gene, leading to excessive TNF production. In terms of mechanisms, it is highly likely that chronic ethanol treatment decreases PDE4B-regulated pool of cAMP and influences cAMP signaling involved in the regulation of NF-κB-dependent and -independent LPS-inducible TNF expression. Overall, these data strongly support the pathogenic role of PDE4B in the ethanol-mediated priming of monocytes/Kupffer cells for increased LPS-inducible TNF production and the subsequent development of ALD. Importantly, since enhanced TNF expression plays a significant role in the evolution of clinical and experimental ALD, its downregulation via selective PDE4B inhibitors could constitute a novel therapeutic approach in attenuating or preventing the development/progression of ALD.
GRANTS
This work was supported by NIH grants R37AA010762 and 5R01AA010496 (to C. McClain) and R01AA014371 (to S. Barve).
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Barnes PJ, Karin M. Nuclear factor-kB: a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med 336: 1066–1071, 1997. [DOI] [PubMed] [Google Scholar]
- 2.Bird GL, Sheron N, Goka AK, Alexander GJ, Williams RS. Increased plasma tumor necrosis factor in severe alcoholic hepatitis. Ann Intern Med 112: 917–920, 1990. [DOI] [PubMed] [Google Scholar]
- 3.Draheim R, Egerland U, Rundfeldt C. Anti-inflammatory potential of the selective phosphodiesterase 4 inhibitor N-(3,5-dichloro-pyrid-4-yl)-[1-(4-fluorobenzyl)-5-hydroxy-indole-3-yl]-glyoxylic acid amide (AWD 12-281), in human cell preparations. J Pharmacol Exp Ther 308: 555–563, 2004. [DOI] [PubMed] [Google Scholar]
- 4.Essayan DM Cyclic nucleotide phosphodiesterases. J Allergy Clin Immunol 108: 671–680, 2001. [DOI] [PubMed] [Google Scholar]
- 5.Felver ME, Mezey E, McGuire M, Mitchell MC, Herlong HF, Veech GA, Veech RL. Plasma tumor necrosis factor alpha predicts decreased long-term survival in severe alcoholic hepatitis. Alcohol Clin Exp Res 14: 255–259, 1990. [DOI] [PubMed] [Google Scholar]
- 6.Gobejishvili L, Barve S, Joshi-Barve S, Uriarte S, Song Z, McClain CJ. Chronic ethanol mediated decrease in cellular cAMP leads to enhanced LPS-inducible NFkB activity and TNF expression in macrophages: Relevance to alcoholic liver disease. Am J Physiol Gastrointest Liver Physiol 291: G681–G688, 2006. [DOI] [PubMed] [Google Scholar]
- 7.Haas JG, Baeuerle PA, Riethmuller G, Ziegler-Heitbrock HW. Molecular mechanisms in down-regulation of tumor necrosis factor expression. Proc Natl Acad Sci USA 87: 9563–9567, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill DB, Barve S, Joshi-Barve S, McClain C. Increased monocyte nuclear factor-kappaB activation and tumor necrosis factor production in alcoholic hepatitis. J Lab Clin Med 135: 387–395, 2000. [DOI] [PubMed] [Google Scholar]
- 9.Hill DB, Shedlofsky SI, Schmidt J, Cohen D, McClain CJ. In vitro tumor necrosis factor cytotoxicity in HepG2 liver cells. Proceedings of the 1992 Conference on Alcohol, Drugs of Abuse, and Immunomodulators (AIDS). New York: Pergammon, 1993, p. 245–249.
- 10.Jin SL, Conti M. Induction of the cyclic nucleotide phosphodiesterase PDE4B is essential for LPS-activated TNF-alpha responses. Proc Natl Acad Sci USA 99: 7628–7633, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin SL, Lan L, Zoudilova M, Conti M. Specific role of phosphodiesterase 4B in lipopolysaccharide-induced signaling in mouse macrophages. J Immunol 175: 1523–1531, 2005. [DOI] [PubMed] [Google Scholar]
- 12.Khoruts A, Stahnke L, McClain CJ, Logan G, Allen JI. Circulating tumor necrosis factor, interleukin-1 and interleukin- 6 concentrations in chronic alcoholic patients. Hepatology 13: 267–276, 1991. [PubMed] [Google Scholar]
- 13.Kim JS, Bailey MJ, Ho AK, Møller M, Gaildrat P, Klein DC. Daily rhythm in pineal phosphodiesterase (PDE) activity reflects adrenergic/3′,5′-cyclic adenosine 5′-monophosphate induction of the PDE4B2 variant. Endocrinology 148: 1475–1485, 2007. [DOI] [PubMed] [Google Scholar]
- 14.Kishore R, McMullen MR, Nagy LE. Stabilization of tumor necrosis factor α mRNA by chronic ethanol: role of A + U-rich elements and p38 mitogen-activated protein kinase signaling pathway. J Biol Chem 276: 41930–41937, 2001. [DOI] [PubMed] [Google Scholar]
- 15.Lee E, Marsano L, McClain CJ. #275 Veterans Cooperative Study. Liver cytokines in alcoholic hepatitis as assessed by immunohistochemistry (Abstract). Hepatology 14: A141, 1991. [Google Scholar]
- 16.Kwak HJ, Song JS, No ZS, Song JH, Yang SD, Cheon HG. The inhibitory effects of roflumilast on lipopolysaccharide-induced nitric oxide production in RAW 264.7 cells are mediated by heme oxygenase-1 and its product carbon monoxide. Inflamm Res 54: 508–513, 2005. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and 2−ΔΔCt method. Methods 25: 402–408, 2001. [DOI] [PubMed] [Google Scholar]
- 18.Lugnier C Cyclic nucleotide phosphodiesterase (PDE) superfamily: a new target for the development of specific therapeutic agents. Pharmacol Ther 109: 366–398, 2006. [DOI] [PubMed] [Google Scholar]
- 19.McClain CJ, Barve S, Deaciuc I, Kugelmas M, Hill D. Cytokines in alcoholic liver disease. Semin Liver Dis 19: 205–219, 1999. [DOI] [PubMed] [Google Scholar]
- 20.McClain CJ, Cohen DA. Increased tumor necrosis factor production by monocytes in alcoholic hepatitis. Hepatology 9: 349–351, 1989. [DOI] [PubMed] [Google Scholar]
- 21.McClain CJ, Hill DB, Marsano L, Cohen D, Shedlofsky S. A role for cytokines in alcoholic hepatitis. Proceedings of the 1992 Conference on Alcohol, Drugs of Abuse, and Immunomodulators (AIDS). New York: Pergammon, 1993, p. 133–141.
- 22.McClain CJ, Hill DB, Schmidt J, Diehl AM. Cytokines and alcoholic liver disease. Semin Liver Dis 13: 170–182, 1993. [DOI] [PubMed] [Google Scholar]
- 23.Molnar-Kimber KL, Yonno Heaslip R, Weichman B. Modulation of TNF alpha and IL-1beta from endotoxin-stimulated monocytes by selective PDE enzyme inhibitors. Agents Actions 39: C77–C79, 1993. [DOI] [PubMed] [Google Scholar]
- 24.Muller JM, Ziegler-Heitbrock HW, Baeuerle PA. Nuclear factor kappa B, a mediator of lipopolysaccharide effects. Immunobiology 187: 233–256, 1993. [DOI] [PubMed] [Google Scholar]
- 25.Newman WH, Zhang LM, Lee DH, Dalton ML, Warejcka DJ, Castresana MR, Leeper-Woodford SK. Release of tumor necrosis factor-alpha from coronary smooth muscle: activation of NF-kappaB and inhibition by elevated cyclic AMP. J Surg Res 80: 129–135, 1998. [DOI] [PubMed] [Google Scholar]
- 26.O'Donnell PM, Taffet SM. The proximal promoter region is essential for lipopolysaccharide induction and cyclic AMP inhibition of mouse tumor necrosis factor-alpha. J Interferon Cytokine Res 22: 539–548, 2002. [DOI] [PubMed] [Google Scholar]
- 27.Ohlinger W, Dinges HP, Zatloukal K, Mair S, Gollowitsch F, Denk H. Immunohistochemical detection of tumor necrosis factor-alpha, other cytokines and adhesion molecules in human livers with alcoholic hepatitis. Virchows Arch A Pathol Anat Histopathol 423: 169–176, 1993. [DOI] [PubMed] [Google Scholar]
- 28.Ollivier V, Parry GCN, Cobb RR, Prost DD, Mackman N. Elevated cyclic AMP inhibits NF-kB-mediated transcription in human monocytic cells and endothelial cells. J Biol Chem 271: 20828–20835, 1996. [DOI] [PubMed] [Google Scholar]
- 29.Ouagued M, Martin-Chouly CA, Brinchault G, Leportier-Comoy C, Depincé A, Bertrand C, Lagente V, Belleguic C, Pruniaux MP. The novel phosphodiesterase 4 inhibitor, CI-1044, inhibits LPS-induced TNF-alpha production in whole blood from COPD patients. Pulm Pharmacol Ther 18: 49–54, 2005. [DOI] [PubMed] [Google Scholar]
- 30.Seldon PM, Meja KK, Giembycz MA. Rolipram, salbutamol and prostaglandin E2 suppress TNFalpha release from human monocytes by activating Type II cAMP-dependent protein kinase. Pulm Pharmacol Ther 18: 277–284, 2005. [DOI] [PubMed] [Google Scholar]
- 31.Semmler J, Wachtel H, Endres S. The specific type IV phosphodiesterase inhibitor rolipram suppress TNF alpha production by human mononuclear cells. Int J Immunopharmacol 15: 409–413, 1993. [DOI] [PubMed] [Google Scholar]
- 32.Shames BD, McIntyre RC, Bensard DD, Pulido EJ, Selzman CH, Reznikov LL, Harken AH, Meng X. Suppression of Tumor Necrosis Factor a Production by cAMP in Human Monocytes: Dissociation with mRNA Level and Independent of Interleukin-10. J Surg Res 99: 187–193, 2001. [DOI] [PubMed] [Google Scholar]
- 33.Torphy TJ Phosphodiesterase isozymes: molecular targets for novel anti-asthma agents. Am J Respir Crit Care Med 157: 351–370, 1998. [DOI] [PubMed] [Google Scholar]
- 34.Vincenti MP, Burrell TA, Taffet SM. Regulation of NF-kappa B activity in murine macrophages: effect of bacterial lipopolysaccharide and phorbol ester. J Cell Physiol 150: 204–213, 1992. [DOI] [PubMed] [Google Scholar]
- 35.Verghese MW, McConnell RT, Strickland AB, Gooding RC, Stimpson SA, Yarnall DP, Taylor JD, Furdon PJ. Differential regulation of human monocytes derived TNF alpha and IL-1 beta by type IV cAMP-phosphodiesterase (cAMP-PDE) inhibitors. J Pharmacol Exp Ther 272: 1313–1320, 1995. [PubMed] [Google Scholar]
- 36.Woo MS, Jang PG, Park JS, Kim WK, Joh TH, Kim HS. Selective modulation of lipopolysaccharide-stimulated cytokine expression and mitogen-activated protein kinase pathways by dibutyryl-cAMP in BV2 microglial cells. Brain Res Mol Brain Res 113: 86–96, 2003. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Z, Bagby GJ, Stoltz D, Oliver P, Schwarzenberger PO, Kolls JK. Prolonged ethanol treatment enhances lipopolysaccharide/phorbol myristate acetate-induced tumor necrosis factor-alpha production in human monocytic cells. Alcohol Clin Exp Res 25: 444–449, 2001. [PubMed] [Google Scholar]
- 38.Zhao XJ, Oliver P, Song K, Schurr J, Zhang Z, Kolls JK. Chronic ethanol enhances ectodomain shedding in T cells and monocytes. Alcohol Clin Exp Res 28: 1399–1407, 2004. [DOI] [PubMed] [Google Scholar]
- 39.Zidek Z Adenosine—cyclic AMP pathways and cytokine expression. Eur Cytokine Netw 10: 319–328, 1999. [PubMed] [Google Scholar]