Abstract
Protein aggregation cardiomyopathy is a life-threatening manifestation of a multisystem disorder caused by the exchange mutation in the gene encoding the human small heat shock protein αB-crystallin (hR120GCryAB). Genetic studies in mice have established cardiac hR120GCryAB expression causes increased activity of glucose 6-phosphate dehydrogenase (G6PD) and “reductive stress” (Rajasekaran et al., Cell 130: 427–439, 2007). However, the initiating molecular events in the pathogenesis of this novel toxic gain-of-function mechanism remain poorly defined. In an integrated systems approach using gene expression profiling, we identified a “biosignature,” whose features can be validated to predict the onset, rate of progression, and clinical outcome of R120GCryAB cardiomyopathy. At the 3 mo disease-related but compensated stage, we demonstrate that transcripts were only upregulated in three distinct pathways: stress response (e.g., Hsp70, Hsp90), glutathione metabolism (Gpx1, Gpx3, glutathione S-transferase), and complement and coagulation cascades in hR120GCryAB transgenic mouse hearts compared with either hCryAB WT transgenic mice or nontransgenic controls. In 6 mo old myopathic hearts, ribosomal synthesis and cellular remodeling associated with increased cardiac hypertrophy were additional upregulated pathways. In contrast, the predominant downregulated pathways were for oxidative phosphorylation, fatty acid metabolism, intermediate metabolism, and energetic balance, supporting their primary pathogenic roles by which G6PD-dependent reductive stress causes cardiac decompensation and overt heart failure in hR120GCryAB cardiomyopathy. This study extends and confirms our previous findings that reductive stress is a causal mechanism for hR120G CryAB cardiomyopathy and demonstrates that alteration in glutathione pathway gene expression is an early biosignature with utility for presymptomatic detection.
Keywords: reductive stress, microarray, transgenic mice, glutathione metabolism
protein aggregation cardiomyopathy (also termed desmin-related myopathy) is caused by missense arginine to glycine mutation at codon 120 in the gene encoding the human small heat shock protein (HSP) αB-crystallin (hR120GCryAB). We have previously described that increased recycling of glutathione (GSH), the major cellular nonprotein thiol, plays a major pathogenic role in the hR120GCryAB multisystem disorder that includes cardiomyopathy, heart failure and sudden cardiac death in mice (17). GSH is either synthesized de novo or recycled from oxidized glutathione (GSSG) and the enzymatic cofactor NADPH, which is generated from the pentose phosphate pathway enzyme, glucose 6-phosphate dehydrogenase (G6PD). Dysregulation of normal feedback mechanisms controlling glutathione homeostasis induces excessive pathogenic levels of reducing equivalents in the form of GSH and NAPDH, termed “reductive stress.” In contrast to well-established roles for oxidative stress in different cardiac diseases (3, 11), reductive stress has been demonstrated in lower eukaryotes but uncommonly in mammals and/or disease states. The distinction between reductive and oxidative stress is, therefore, neither arbitrary nor semantic but could be demonstrated in vivo using biochemical and genetic approaches in the intact animal (17). Specifically, the intercross of hR120GCryAB cardiomyopathic animals with G6PD-deficient (20% normal G6PD activity) mice was found to rescue hR120GCryAB/G6PDmut progeny from the pathogenic increase in G6PD activity, cardiac hypertrophy, and protein aggregation (17).
Such evidence, that dysregulation of G6PD activity is a causal mechanism for R120GCryAB cardiomyopathy, provides a conceptual advance for unraveling how perturbations of redox signaling and control initiate a toxic gain-of-function mechanism such as protein aggregation R120GCryAB cardiomyopathy. We further hypothesize that cardiotoxicity via similar mechanisms such as R120GCryAB are presently unrecognized among inherited and acquired cardiovascular conditions, thus paving the way for their future identification at the genetic, structural, biochemical, and clinical levels. There are, however, several fundamental issues that need to be resolved to test the validity of this idea. What changes in gene expression occur at the onset of the R120GCryAB cardiomyopathy? What changes in gene expression occur during the transition stage but preceding the onset of the R120CryAB cardiomyopathy? Can these early events be clustered into the discrete biochemical pathways linked causally to disease pathogenesis? Here we demonstrate the utility of gene expression profiling to identify a “biosignature,” which might be applied diagnostically to predict the onset, rate of progression, and clinical outcome of R120GCryAB cardiomyopathy with reproducibility and reliability.
MATERIALS AND METHODS
Transgenic constructs and mouse lines.
Generation of transgenic mice in our laboratory is described elsewhere (17). In brief, the full-length human αB-crystallin (CryAB; αBC) cDNA was kindly provided by Dr. James E. Goldman (Columbia University, New York, NY) (accession number S45360) (12). R120G was created from the human CryAB cDNA by PCR-based mutagenesis (Quick Change Site directed mutagenesis kit, Stratagene, La Jolla, CA) and confirmed by sequencing. Transgenic mice were generated by pronuclear injection according to standard procedures. Founders were identified by polymerase chain reaction and Southern blot and then crossed with wild-type (WT) C57BL/6 mice to establish the transgenic lines. In this study, we used a transgenic mouse line containing sixfold higher total cardiac-specific mutant CryAB expression under the control of the α-myosin heavy chain promoter, termed “hR120GCryAB High Tg” compared with nontransgenic (NTG) mice as recently described (17). Animal procedures were approved and monitored by the University of Utah Institutional Animal Care and Use Committee.
RNA isolation.
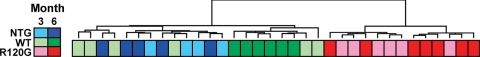
RNA was isolated from ventricles of 3- and 6-mo old NTG, human CryAB wild-type transgenic (hCryAB WT), and human R120GCryAB transgenic (hR120GCryAB) mice. Mice from each group were anesthetized and perfused in situ with 10 ml of sterile PBS followed by perfusion with 10 ml of RNALater (Ambion, Austin, TX). Hearts were immediately harvested, atria trimmed, and the ventricle immersed in RNALater solution for 45 min at room temperature before the samples were frozen at −80°C. Total RNA was extracted and purified from 25–30 mg heart tissue using the RNeasy mini kit (Qiagen, Valencia, CA) and the RNase-free DNase step according to the manufacturer's instructions. RNA quality was monitored by A260/A280 ratio, 1% agarose-formaldehyde gel electrophoresis and microfluidic electrophoresis (Agilent Bioanalyzer; Agilent, Foster City, CA). Originally, six mice were sampled for each of the six groups (three strains at two time points). Two of the 3-mo NTG samples were removed due to poor RNA quality. Genotype revalidation established that one of the 6 mo samples originally identified as an NTG mouse was actually a hR120G CryAB sample. Final numbers for 3 mo samples were NTG = 4, hCryAB WT = 6, hR120GCryAB = 6 and for 6 mo samples were NTG = 5, hCryAB WT = 6, hR120GCryAB = 7 (Fig. 1).
Fig. 1.
Hierarchical cluster analysis of experimental arrays. Log intensity ratios of all spots on each array were used to group the experimental samples into clusters. Each block represents individual samples with concatenated expression information from both mouse A and mouse B arrays. Sample groups are color coded as indicated by the legend at left. The dendrogram above the diagram indicates the similarity between the individual arrays. Arrays joined by shorter distances are more similar than arrays joined by longer distances. NTG, nontransgenic controls; WT, human wild-type CryAB transgene (hCryAB WT); R120G, human R120G mutant CryAB transgene (hR120GCryAB).
Microarray hybridization.
Microarray printing, labeling of RNA, microarray hybridization, scanning, and image processing were performed by the Huntsman Cancer Institute Microarray Resource using standard protocols (see Supplemental Material).1 Samples of between four and seven biological replicates for each condition (see Fig. 1) were each hybridized to two microarray slides (“mouse A” and “mouse B”) containing 9,278 and 9,047 different mouse cDNA clones respectively. Each PCR amplified clone was printed in duplicate on in-house fabricated arrays and consisted of the National Institute of Aging (NIA) 15K mouse cDNA clone set and a subset of the NIA 7.4K cDNA clone set (22) for a total of 18,325 unique features. This array design is available at the Gene Expression Omnibus (GEO) public database as accession number GPL6249 (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GPL6249). All experimental samples were labeled with Cy3 dye and hybridized vs. a standard reference sample (Universal Mouse Reference RNA; Stratagene, La Jolla, CA) labeled with Cy5 dye.
Microarray data analysis.
Microarray images where quantified using ImaGene software, version 6.0 (BioDiscovery, El Segundo, CA). The raw, nonnormalized data from the mouse A and mouse B spotted cDNA arrays were evaluated for overall quality with M vs. A and box plots (1) to check for any intensity-dependent or spatial artifacts. The plots revealed subtle intensity-dependent and spatial variations that were corrected using LOWESS normalization with print-tip scope. Normalization was performed in the AROMA software without background correction (2). Poor quality spots were removed, and log ratios were calculated in AROMA. Data for each sample from the two microarrays were concatenated together to form a single data set. Microarray experimental information and data have been deposited in the GEO public database under accession number GSE9924 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ltihbeayakaiuhc&acc=GSE9924).
For each of the ∼18,000 sequences (representing genes and expressed sequence tags) on the microarrays, we first calculated the experimental vs. reference standard intensity ratios to find the mean averaged intensity ratio over all replicate spots for that sequence. Although features represented by the same clone number (replicates) were averaged, features representing the same gene by different clone numbers were treated as different genes in the statistical analyses, leaving open the possibility that a single gene might be identified multiple times. Statistical analysis of differentially expressed genes and identification of potentially altered pathways were performed using GeneSifter software suite (VizX Labs, Seattle, WA; http://www.genesifter.net/web). Expression differences in NTG, hCryAB WT and hR120GCryAB mouse hearts were determined by ANOVA at 3 and 6 mo or by pairwise comparisons for each combination (NTG vs. hCryAB WT, NTG vs. hR120GCryAB, and hCryAB WT vs. hR120GCryAB) at both 3 and 6 mo. We used the method of Benjamini and Hochberg to adjust for multiple comparisons by controlling the false discovery rate (4). Sequences identified as having significantly altered expression and of known gene function were used to query the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg) annotated pathway database (13). Pathways represented in our dataset were filtered based on statistical z-scores of >2.5 or <−2.5.
Northern blot analysis.
For each condition (3 and 6 mo NTG, hCryAB WT, and hR120GCryAB), three RNA samples previously used for microarray analysis were randomly chosen for Northern blot analysis. In brief, 10 μg of total RNA was loaded and separated on 1.0% agarose gel with formaldehyde, capillary transferred (Turbo Blotter; Whatman, Florham Park, NJ) on supercharged nylon membrane (BrightStar-Plus, Ambion) and UV cross-linked. The agarose gels were imaged before transfer using an Image Station 2000R (Eastman Kodak, Rochester, NY) to monitor the 18S and 28S rRNA for loading normalization. cDNA probes were generated using their respective mouse clones by random priming in the presence of α-32P-ATP (Strip-EZ DNA, Ambion). Membranes were hybridized in Ultrahyb (Ambion) solution for 16–18 h and washed in low (2× 5 min, room temperature) and high (2× 15 min, 68°C) stringent solutions according to the manufacturer's instruction. Signals were detected using autoradiography and quantified using ImageJ software (National Institutes of Health, http://rsb.info.nih.gov/ij/).
Quantitative RT-PCR.
After quantifying RNA spectrometrically, we reverse transcribed 2–3 μg of total RNA using the Qiagen QuantiTect Reverse Transcription kit (catalog number 205311) for cDNA synthesis. The relative expression levels of nine genes were determined using quantitative real-time PCR assays performed and monitored in triplicate using an ABI PRISM 7000 Sequence detection system (Applied Biosystems, Foster City, CA). The PCR reactions contained 2.0 μl of cDNA (∼100 ng) in a reaction volume of 20 μl, 1× QuantiTect SYBR Green master-mix and the relevant mouse Qiagen QuantiTect Primer Assay. Thermocycling conditions were 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. The nine genes and corresponding QuantiTect Primer Assay catalog numbers were Gapdh (QT01658692), Arbp (QT00249375), G6pdx (QT00120750), Gpx1 (QT01195936), Gpx3 (QT01060465), Gsta4 (QT00098987), Gstm1 (QT00121191), Hsp90aa1 (QT00246967), and Hsp90ab1 (QT01039864). Relative gene expression levels were determined from observed CT data. The mean CT value for each gene was first subtracted from the mean CT value of Arbp1, which was used as the reference gene, from the same cDNA sample to normalize for any potential loading differences, and 2ΔCT calculated the linear expression value. Expression of Gapdh was included in the set of nine genes to verify the “housekeeping” status of Arbp1.
RESULTS
Mouse model of human R120GCryAB cardiomyopathy.
We investigated the disease-causing exchange mutation in human CryAB (R120G) by microarray analysis in our attempts to identify the early genetic events underlying its novel toxic gain-of-function mechanism involving dysregulation of glutathione homeostasis. In transgenic mice, dose-dependent expression of hR120GCryAB mimics the phenotype described in patients (7), including protein aggregation, cardiac hypertrophy, and sudden cardiac death (17). R120GCryAB mice exhibit no overt phenotype at 3 mo, but their survival declines precipitously after 6 mo. Therefore, our rationale for performing microarray analysis at these two intervals was to identify potential molecular markers corresponding to the transition between compensation and decompensation stages for this myopathic hR120GCryAB model.
Clusters for NTG and hCryAB WT segregate from hR120GCryAB expression.
To assess the overall reliability of the microarray intensity data, we first asked whether the expression profiles from the experimental samples would partition into meaningful groups (“array clustering”). For this analysis, we used mean log intensity ratio information for all sequences on each array and applied hierarchical clustering with no filtering for statistical differences in expression (Fig. 1). Besides the age-matched NTG strain, we analyzed hCryAB WT mice with WT human protein overexpression to independently assess for changes in gene expression likely attributed the nonmyopathic effects. The arrays partitioned into two main clusters: one consisting of the NTG and hCryAB WT samples and the other corresponding to hR120GCryAB samples. Such robust alterations in gene expression were attributable to the presence of hR120GCryAB when compared with either NTG or hCryAB WT. In the main cluster from hR120GCryAB hearts, the 3 mo and 6 mo groups segregated into subclusters indicating a profound shift in hR120GCryAB-induced gene expression, representing early (i.e., 3 mo) and late (i.e., 6 mo) phases of compensation (Fig. 1). The main cluster consisting of NTG and hCryAB WT was less structured than the hR120GCryAB main cluster, suggesting that such differences between the former groups were less robust. However, clustering of all six hCryAB WT 6 mo samples within one subcluster suggests that even WT transgene overexpression (50% increase vs. NTG) was sufficient to alter gene expression at 6 mo (Fig. 1).
hR120GCryAB triggers changes in overall gene expression at 3 mo.
We next used ANOVA modeling and filtered the 3 and 6 mo datasets for statistically significant changes in gene expression. So that only major changes in gene expression were identified, we arbitrarily restricted the search to those sequences exhibiting at least a twofold change in expression. For this analysis, levels of expression in hCryAB WT and hR120GCryAB hearts were compared with NTG hearts. For >50,000 comparisons, the correction for multiple comparisons resulted in only a modest decrease in the number of identified sequences. We identified 95 and 114 sequences as significantly altered at 3 mo and 6 mo with adjusted P values <0.005, respectively (Supplemental Table S1). Such stringent analysis predicts a false positive rate of <1 in 100 sequences.
Among the 95 sequences having significantly altered expression (68 upregulated, 27 downregulated) identified at 3 mo, three sequences, Rpl9, Rpl10, and Slc14a1, had almost identical changes in expression in both the hCryAB WT and hR120GCryAB strains. Eight had their major effects in the hCryAB WT strain in which they were all upregulated (Rbm12b, Lrrc50, and six expressed sequence tags). The remaining sequences identified showed only modest changes in the 3-mo-old hCryAB WT mouse line. Similarly, of the 114 sequences (47 upregulated, 67 downregulated) with significantly altered expression at 6 mo, only minor gene expression changes were evident in hearts of the hCryAB WT strain. Thus, whereas WT transgene overexpression per se had measurable but minimal effects, the R120G mutation induced significant changes in gene expression at both selected intervals.
Among 209 sequences identified by ANOVA exhibiting a twofold change in expression at 3 and/or 6 mo (P value <0.005), 142 had meaningful names representing 109 unique genes (several genes were identified two or more times). The relative levels of gene expression are shown in Fig. 2. Only 25 sequences (20 unique genes) were common to both the 3 and 6 mo analyses indicating that considerable cellular reprogramming occurred between 3 and 6 mo. Consistent with molecular events associated with protein aggregation cardiomyopathy, expression changes were noted for genes involved in stress response, cytoskeletal structure, protein synthesis, protein folding, and protein degradation (Fig. 2). Furthermore, the considerable gene expression changes evident at 3 mo provide suitable targets for potential molecular markers corresponding to the transition between compensation and decompensation stages for hR120GCryAB cardiomyopathy.
Fig. 2.
Major gene expression changes in transgenic hearts. All known genes identified with at least a 2-fold change in expression (P < 0.005) are shown. Green intensities denote decreased expression and red intensities denote increased expression according to the color bar (top right) normalized to the NTG control. Genes were identified as having significant expression changes at 3 mo, 6 mo, or both. Genes without significant change at either 3 or 6 mo are not represented on the heat map. Genes were assigned to arbitrarily selected functional groups but could participate in multiple processes.
hR120GCryAB altered specific cellular pathways.
Because mutant transgene overexpression likely imparts specific consequences on myocardial dysfunction through reprogramming of intracellular regulatory mechanisms, we next examined our dataset for significantly altered gene expression of cellular pathways related to cardiotoxicity. From the identified sequences with at least a 1.5-fold change and adjusted P values <0.05, we found 625 and 844 sequences at 3 and 6 mo, respectively, that were attributable to hR120GCryAB overexpression and whose chronic expression might produce dramatic phenotypic changes (Supplemental Fig. S1). Known genes from these sequences were then used to query an annotated pathway database to define interdependent networks among biological processes.
At 3 mo, several genes encoding major classes of heat-shock proteins and molecular chaperones (the majority of genes identified in the “antigen processing and presentation” pathway) (Supplemental Table S2) were significantly upregulated, consistent with dramatic cell adaptation during the compensation phase (i.e., normal cardiac function). We were quite intrigued to find among the “early response” pathways included the complement and coagulation cascades and the catabolic control of glutathione metabolism. Among the downregulated genes at 3 mo, no distinct pathways were identified at the chosen z-score threshold.
Accompanying the anticipated reprogramming and remodeling from disease progression and the transition towards heart failure, 13 pathways were identified at 6 mo (Table 1 and Supplemental Table S2). Both glutathione metabolism and antigen processing and presentation remained upregulated, but three additional pathways, engaged in cell communication, ribosomal biosynthesis, and aminoacyl-tRNA biosynthesis, were identified. Genes encoding cellular communication pathways were limited to cytoskeletal and extracellular matrix proteins such as collagen, β-actin, desmin, and lamin A, indicating cellular and tissue restructuring were characteristics of cardiomyopathy. Similarly, upregulation of ribosomal components, aminoacyl-tRNA biosynthetic pathways and downregulation of tRNA degradation enzymes indicate a concomitant demand for increased protein synthesis. Among downregulated pathways at 6 mo, the most dramatic responses were noted for the oxidative phosphorylation and fatty acid metabolism pathways as were pathways for multiple stages of intermediate metabolism, indicating a deficiency in metabolic precursor molecules and, perhaps, myocardial energetics.
Table 1.
Cellular pathways with altered expression in hR120GCryAB hearts relative to hCryAB WT hearts
| KEGG Pathway | Genes on Array |
Genes Identified |
z-Score
|
||
|---|---|---|---|---|---|
| Up | Down | Up | Down | ||
| 3 Month | |||||
| Upregulated | |||||
| Glutathione metabolism | 18 | 5 | 0 | 6.44 | −0.49 |
| Antigen processing and presentation | 24 | 5 | 2 | 5.38 | 3.02 |
| Complement and coagulation cascades | 25 | 4 | 0 | 4.02 | −0.58 |
| 6 Month | |||||
| Upregulated | |||||
| Antigen processing and presentation | 24 | 6 | 1 | 5.92 | −0.05 |
| Glutathione metabolism | 18 | 4 | 2 | 4.46 | 1.41 |
| Cell communication | 41 | 6 | 0 | 4.05 | −1.38 |
| Ribosome | 51 | 6 | 1 | 3.38 | −0.85 |
| Aminoacyl-tRNA biosynthesis | 23 | 3 | 0 | 2.59 | −1.03 |
| Downregulated | |||||
| Oxidative phosphorylation | 75 | 1 | 19 | −1.00 | 9.09 |
| Fatty acid metabolism | 26 | 1 | 9 | 0.14 | 7.61 |
| Valine, leucine and isoleucine degradation | 34 | 1 | 9 | −0.14 | 6.38 |
| Carbon fixation | 17 | 1 | 6 | 0.58 | 6.28 |
| Pyruvate metabolism | 22 | 0 | 5 | −0.88 | 4.25 |
| Citrate cycle (TCA cycle) | 24 | 0 | 5 | −0.92 | 3.98 |
| Glycolysis/Gluconeogenesis | 31 | 1 | 5 | −0.04 | 3.24 |
| Alanine and aspartate metabolism | 15 | 1 | 3 | 0.71 | 2.98 |
hR120GCryAB, mutant human small heat shock protein αB-crystallin; hCryAB, human αB-crystallin; WT, wild type.
Microarray validation.
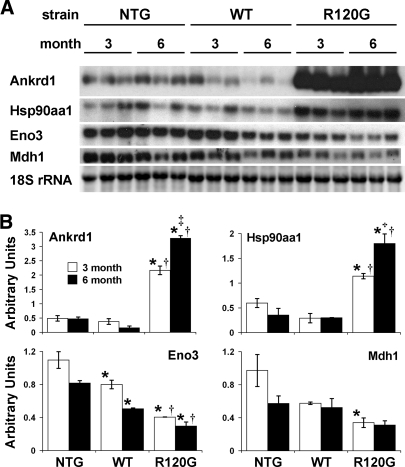
To validate the array expression, we performed Northern blot analysis of four representative genes identified as either upregulated [ankyrin repeat domain 1 (cardiac muscle), heat shock protein 90 kDa alpha (cytosolic), class A member 1] or downregulated [enolase 3, beta muscle, malate dehydrogenase 1, NAD (soluble)] in hR120GCryAB hearts (Fig. 3, Supplemental Table S3). All transcripts showed remarkable concordance for both magnitude and direction with the microarray data. Additionally, two other genes, catalase and glutathione peroxidase 3, were previously shown to be upregulated in hR120GCryAB hearts (17). By microarray analysis, Ankrd1, Cat, and Eno3 had significantly altered expression at both 3 and 6 mo, whereas Gpx3 was upregulated at 3 mo only and Hsp90aa1 was upregulated at 6 mo only. Northern blot analysis indicated that Gpx3 and Hsp90aa1 were upregulated at both 3 and 6 mo. The citric acid cycle enzyme malate dehydrogenase 1 (Mdh1) was identified as significantly downregulated by the less stringent criterion used for the pathway analysis (>1.5-fold change and adjusted P values <0.05), but not by the more stringent analysis used to identify the major changes (>2-fold change and adjusted P values <0.005) depicted in Fig. 2. Although the Northern blot analysis confirmed the significant downregulation of Mdh1, it also showed high variability in control samples, which probably explains why Mdh1 was not identified by the stringent analysis.
Fig. 3.
Expression validation for specific genes by Northern blot analysis. A: representative Northern blots and corresponding 18S rRNA from ethidium bromide stained gel. B: analysis of densitometry data presented as mean gene density/18S rRNA density ± SE. For each gene, significant differences between conditions were determined by ANOVA and Fisher's PLSD post hoc test. *P < 0.001 vs. NTG at corresponding time point, †P < 0.002 vs. hCryAB WT at corresponding time point, ‡P < 0.002 vs. hR120GCryAB at 3 mo; n = 3 RNA samples per group. Ankrd1, ankyrin repeat domain 1 (cardiac muscle); Hsp90aa1, heat shock protein 90 kDa alpha (cytosolic) class A member 1; Eno3, enolase 3 beta muscle; Mdh1, malate dehydrogenase 1 NAD (soluble).
For independent verification of the cDNA-based microarray data, we performed hybridizations for a subset of the samples using a different microarray platform. We examined expression in two samples each from 6 mo hR120GCryAB and hCryAB WT hearts on an oligonucleotide-based microarray platform (Affymetrix). Similar expression changes in the 109 unique genes identified by ANOVA were seen for both platforms (Supplemental Table S3 and data not shown). This concordant expression from an independent microarray platform provides further validation of the original cDNA-based data set.
Expression of G6PD.
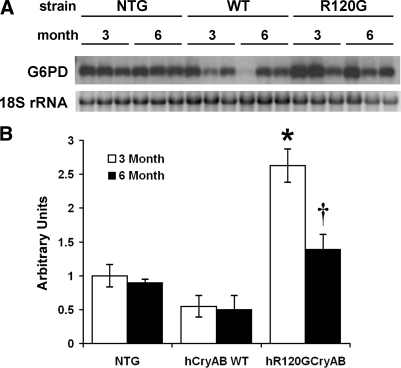
We were surprised that the microarray analysis did not identify G6PD mRNA as being upregulated in R120G hearts since our previous analysis demonstrated that G6PD protein was elevated about fourfold relative to control tissue (17). G6PD was represented on the microarray by a single cDNA clone. Northern blot analysis using a probe generated from this cDNA clone was unsuccessful, indicating that the microarray may not have accurately assessed G6PD mRNA levels. However, using a second probe, representing exon 13 of the G6PD gene, Northern blot analysis showed a 4.8- and 2.8-fold increase in G6PD mRNA expression at 3 mo and 6 mo, respectively (Fig. 4).
Fig. 4.
Glucose 6-phosphate dehydrogenase (G6PD) expression by Northern blot analysis. Northern blot band intensities were assessed by densitometry and normalized to 18S rRNA levels. Significant differences between conditions were determined by ANOVA and Fisher's PLSD post hoc test. A: representative Northern blot and corresponding 18S rRNA from ethidium bromide stained gel. B: analysis of densitometry data presented as mean G6PD density/18S rRNA density ± SE of 3 replicates. *P < 0.0005 vs. all other groups, †P = 0.005 vs. hCryAB WT at 6 mo.
Biosignature validation.
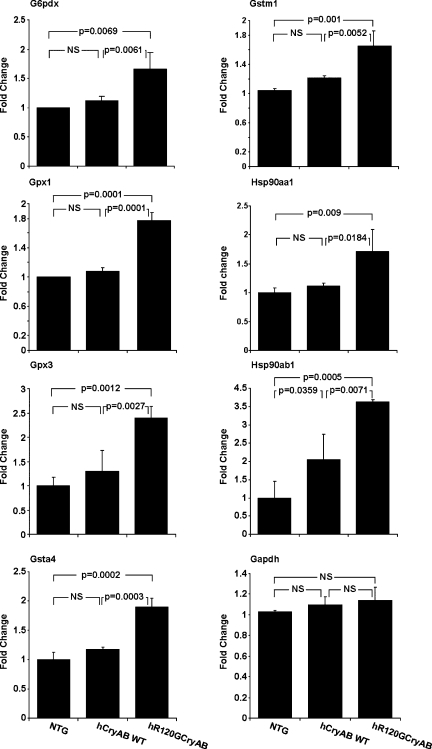
The compendium of gene expression changes identified in this analysis gives us the opportunity to identify a biosignature. In this context, a biosignature reflects specific changes in gene expression with sensitivity and specificity for the biological pathway(s) relevant to disease pathogenesis. To identify those signatures present before overt presentation of symptoms (early response) we asked if differential expression of individual genes could effectively discriminate between the normal and presymptomatic phase of hR120GCryAB cardiomyopathy. At 3 mo (see Fig. 2 and Supplemental Fig. S1), we found upregulation of the glutathione pathway genes (Gpx1, Gpx3, Gstm1, Gsta4) in hR120GCryAB hearts, but not in hCryAB WT hearts, consistent with activation of glutathione metabolism during pathogenesis in this presymptomatic phase. For each gene, a threshold microarray gene expression value could be identified that gave 100% accuracy in discriminating these two experimental groups. To validate these results from the microarray gene expression data, we next performed RT-PCR for glutathione pathway and other stress response genes (Fig. 5). In this analysis, three RNA samples that been previously been analyzed by microarray were randomly chosen from each group (3 mo NTG, hCryAB WT, and hCryAB R120G) and subjected to quantitative RT-PCR analysis for the genes Gpx1, Gpx3, Gstm1, Gsta4, G6pdx, Hsp90aa1, and Hsp90ab1. Six of the seven selected genes showed no difference in expression between the NTG and hCryAB WT hearts, indicating that expression of the human WT gene was nonpathogenic. However, expression of each gene was significantly higher in hR120GCryAB, indicating that the gene expression effects in hR120GCryAB were highly sensitive and specific for identification of presymptomatic disease. Because the above analysis relied on the RNA samples originally used to identify the biosignature, our strategy potentially risked overfitting the data. To circumvent this limitation we repeated and confirmed our analysis using quantitative RT-PCR on new samples isolated from 3 mo hearts (6 NTG and 6 hCryAB R120G), indicating the reproducibility of the biosignature with high specificity and sensitivity (data not shown). We conclude that expression of glutathione and stress-response pathway genes are appropriate molecular markers that predict development of overt symptoms in hR120GCryAB cardiomyopathy, although we recognize that other genes may also be appropriate targets for inclusion in such a biosignature.
Fig. 5.
Genes encoding stress and glutathione pathway proteins, causally linked to reductive stress, were validated as an early biosignature for hR120GCryAB cardiomyopathy. Gene expression was evaluated by quantitative RT-PCR in 3 mo hearts. Data were first normalized to Arbp1 expression and then to the corresponding gene expression in the nontransgenic group (ΔΔCT method). No significant differences between NTG and hCryAB WT hearts for the glutathione metabolic pathway genes (Gpx1, Gpx3, Gsta4, Gstm1), G6pdx, and Hsp90aa1 were evident, but all were significantly elevated in age-matched hR120GCryAB hearts establishing an early diagnostic marker for this pathogenic process before development of overt disease. NS, nonsignificant; G6pdx, glucose 6-phosphate dehydrogenase X-linked; Gstm1, glutathione S-transferase μ1; Gpx1, glutathione peroxidase 1; Hsp90aa1, heat shock protein 90-α (cytosolic) class A member 1; Gpx3, glutathione peroxidase 3; Hsp90ab1, heat shock protein 90 kDa-α (cytosolic) class B member 1; Gsta4, glutathione S-transferase-α 4; Gapdh, glyceraldehyde-3-phosphate dehydrogenase.
DISCUSSION
The most significant finding of this study was that identification of the glutathione metabolic pathway, at the onset and preceding overt heart failure, provides predictive value for profound changes in gene expression of, perhaps, prognostic significance for R120GCryAB cardiomyopathy. During the preclinical asymptomatic phase (i.e., <3 mo), we found among the earliest and only upregulated pathways belonged to the “classical” stress (HSP) response, complement/coagulation cascades, and glutathione metabolic pathways. Misfolded proteins such as hR120GCryAB serve as bona fide stress stimulus of stress response pathways (23), in which specialized HSPs, termed molecular chaperones, exhibit ATP-dependent and -independent properties to prevent protein aggregation, improve quality control and/or promote protein degradation (9, 21). However, our genetic studies in mice have established that hR120GCryAB expression causes increased G6PD activity and reductive stress from increased equivalents (i.e., GSH, NADPH) resulting in cardiac decompensation (i.e., >6 mo) and premature death. As the major nonprotein thiol in the cell, the metabolite glutathione (GSH), a tripeptide γ-glutamylcysteinylglycine, serves essential roles as a redox buffer in protection against oxidative stress (15, 19), but such reductive stress would be predicted to alter redox-sensitive pathways including gene expression (8), mitochondrial energetics (14), cardiac metabolism, cellular processes (e.g., oxidative protein folding) (10), and the disulfide proteome (24).
Alterations in specific redox signaling pathways by GSH overproduction might impart pleiotropic consequences ranging from cardiac remodeling to mitochondrial dysfunction and myocardial energetics in experimental cardiomyopathy in mice. To date, aberrant glutathione metabolism and overproduction have been elegantly addressed using the Saccharomyces cerevisiae genome-wide deletion library at the genetic and biochemical levels (16, 18). Such genetic screens have implicated diverse cellular processes such as reprogramming of ribosomal biosynthesis, mitochondrial electron chain, vacuolar function, ion homeostasis, cell communication, and aminoacyl-tRNA biosynthesis (16). Consistent with such findings, we have identified similar pathways by our genomic profiling during the decompensation stage of hR120GCryAB cardiomyopathy, thereby extending the relevance of GSH-dependent redox regulation to a clinically relevant model.
Sanbe and colleagues (20) have recently shown markedly increased protein oligomers in human myocardial tissue associated with idiopathic dilated cardiomyopathy and ischemic heart disease, suggesting that the full repertoire of protein degradation pathways, besides polyubiquitination, might be activated in these pathologic states. Of interest, the gene encoding WD repeat and FYVE domain containing 3 (Wdfy3), which targets cytosolic protein aggregates for autophagic degradation (32), was downregulated by hR120GCryAB during early compensation at 3 mo. Sequestosome 1 protein (Sqstm1) (5), which is involved in linking polyubiquitinated protein aggregates to the autophagy machinery, was induced 2.11-fold by hR120GCryAB expression at 6 mo. While the precise role(s) of GSH overproduction on major components of multivesicular body formation remains poorly defined, the discovery of such novel pathways underscores the hypothesis-generating potential of an integrated systems approach.
Several genes (e.g., Rps5, Rpl3, Rpl10A, and Rpl15) encoding protein biosynthesis were induced by hR120GCryAB expression at both early and late compensation stages (Supplemental Table S1). Increased ribosome and aminoacyl-tRNA biosynthesis induced by hR120GCryAB have been reported in other models of cardiac hypertrophy including pressure overload, spontaneously hypertensive rats, and the Dahl salt-sensitive rats concomitant with the demand for increased protein synthesis (6). Among several downregulated mRNA transcripts at the later stage of hR120GCryAB expression were genes encoding the mitochondrial electron transport chain, supporting a role for decreased ATP production on myocardial energetics (Fig. 2 and Supplemental Table S2). Downregulation of both oxidative phosphorylation and fatty acid metabolic pathways at 6 mo is consistent with evolutionarily conserved responses for shifts in substrate utilization after environmental stress. The concordance between yeast deletion mutants and the biological processes implicated by gene expression profiling of hR120GCryAB is striking and also supports the hypothesis that reductive stress has relevance in pathologic human biology. Taken together, our results suggest that the molecular signatures related to anabolic and catabolic control of glutathione metabolism feature prominently during progression of R120GCryAB cardiomyopathy in mice.
In conclusion, we have confirmed our previous findings that alterations in glutathione pathway are a causal mechanism for hR120G CryAB cardiomyopathy and demonstrated that gene expression profiling associated with reductive stress is a suitable diagnostic marker, termed a biosignature, for presymptomatic detection. We have further identified the compendium of gene expression events associated with the development of overt disease, which should prove useful for dissecting the mechanisms associated with the pathogenesis of protein aggregation hR120GCryAB cardiomyopathy. Interventions to reverse or prevent cardiac dysfunction in at-risk individuals will require future studies that address validation of relevant redox-dependent transcriptional events and, perhaps, redox proteomic assessment, in humans.
NOTE ADDED IN PROOF
During manuscript review, it was noticed that some information shown in Fig. 3 largely duplicated previously published results. The duplicated data have been removed from the final version of the manuscript.
GRANTS
Awards from National Heart, Lung, and Blood Institute (5RO1 HL-63874), the Christi T. Smith Foundation, and the University of Utah Health Sciences Center Catalyst Research Grant Program provided support for this work.
Supplementary Material
Acknowledgments
We appreciate the helpful suggestions and comments from our colleagues Joseph Prchal and Shannon Odelberg. Tandy Bales provided expert editorial assistance during preparation of this manuscript. Expert technical assistance was provided by Patti Larrabee for mouse breeding and Diane Dunn in performing the Affymetrix microarrays.
Address for reprint requests and other correspondence: I. J. Benjamin, Center for Cardiovascular Translational Biomedicine, Cardiology Div., Dept. of Medicine, Univ. of Utah School of Medicine, 50 No. Medical Dr., Salt Lake City, Utah 84132 (e-mail: ivor.benjamin@hsc.utah.edu).
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Amaratunga D, Cabrera J. Exploration and Analysis of DNA Microarray and Protein Array Data. Hoboken, NJ: John Wiley, 2004, p. xiv.
- 2.Bengtsson H, Jonsson G, Vallon-Christersson J. Calibration and assessment of channel-specific biases in microarray data with extended dynamical range. BMC Bioinformatics 5: 177, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamin IJ, Schneider MD. Learning from failure: congestive heart failure in the postgenomic age. J Clin Invest 115: 495–499, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B 57: 289–300, 1995. [Google Scholar]
- 5.Bjorkoy G, Lamark T, Johansen T. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy 2: 138–139, 2006. [DOI] [PubMed] [Google Scholar]
- 6.Blaxall BC, Spang R, Rockman HA, Koch WJ. Differential myocardial gene expression in the development and rescue of murine heart failure. Physiol Genomics 15: 105–114, 2003. [DOI] [PubMed] [Google Scholar]
- 7.Fardeau M, Godet-Guillain J, Tome FM, Collin H, Gaudeau S, Boffety C, Vernant P. [A new familial muscular disorder demonstrated by the intra-sarcoplasmic accumulation of a granulo-filamentous material which is dense on electron microscopy] (author's transl.). Rev Neurol (Paris) 134: 411–425, 1978. [PubMed] [Google Scholar]
- 8.Fratelli M, Goodwin LO, Orom UA, Lombardi S, Tonelli R, Mengozzi M, Ghezzi P. Gene expression profiling reveals a signaling role of glutathione in redox regulation. Proc Natl Acad Sci USA 102: 13998–14003, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gething MJ, Sambrook J. Protein folding in the cell. Nature 355: 33–45, 1992. [DOI] [PubMed] [Google Scholar]
- 10.Ghyczy M, Torday C, Boros M. Simultaneous generation of methane, carbon dioxide, and carbon monoxide from choline and ascorbic acid: a defensive mechanism against reductive stress? FASEB J 17: 1124–1126, 2003. [DOI] [PubMed] [Google Scholar]
- 11.Giordano FJ Oxygen, oxidative stress, hypoxia, and heart failure. J Clin Invest 115: 500–508, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwaki A, Iwaki T, Goldman JE, Ogomori K, Tateishi J, Sakaki Y. Accumulation of alpha B-crystallin in brains of patients with Alexander's disease is not due to an abnormality of the 5′-flanking and coding sequence of the genomic DNA. Neurosci Lett 140: 89–92, 1992. [DOI] [PubMed] [Google Scholar]
- 13.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucl Acids Res 34: D354–D357, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lash LH Mitochondrial glutathione transport: physiological, pathological and toxicological implications. Chemico-biological Interactions 163: 54–67, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meister A, Anderson ME. Glutathione. Annu Rev Biochem 52: 711–760, 1983. [DOI] [PubMed] [Google Scholar]
- 16.Perrone GG, Grant CM, Dawes IW. Genetic and environmental factors influencing glutathione homeostasis in Saccharomyces cerevisiae. Mol Biol Cell 16: 218–230, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajasekaran NS, Connell P, Christians ES, Yan LJ, Taylor RP, Orosz A, Zhang XQ, Stevenson TJ, Peshock RM, Leopold JA, Barry WH, Loscalzo J, Odelberg SJ, Benjamin IJ. Human alpha B-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell 130: 427–439, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rand JD, Grant CM. The thioredoxin system protects ribosomes against stress-induced aggregation. Mol Biol Cell 17: 387–401, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rebrin I, Kamzalov S, Sohal RS. Effects of age and caloric restriction on glutathione redox state in mice. Free Radic Biol Med 35: 626–635, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanbe A, Osinska H, Saffitz JE, Glabe CG, Kayed R, Maloyan A, Robbins J. Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc Natl Acad Sci USA 101: 10132–10136, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med 81: 678–699, 2003. [DOI] [PubMed] [Google Scholar]
- 22.VanBuren V, Piao Y, Dudekula DB, Qian Y, Carter MG, Martin PR, Stagg CA, Bassey UC, Aiba K, Hamatani T, Kargul GJ, Luo AG, Kelso J, Hide W, Ko MS. Assembly, verification, and initial annotation of the NIA mouse 7.4K cDNA clone set. Genome Res 12: 1999–2003, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao X, Benjamin IJ. Stress-response proteins in cardiovascular disease. Am J Hum Genet 64: 685–690, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Song Y, Loscalzo J. Regulation of the protein disulfide proteome by mitochondria in mammalian cells. Proc Natl Acad Sci USA 104: 10813–10817, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.