Abstract
Low bone mineral density (BMD) is a phenotype associated with osteoporosis and increased risk of fracture. Since 60–80% of variation in BMD is associated with genetic factors, we used the novel approach of chromosome substitution strains (CSS) to identify chromosomes that harbor genes that regulate BMD. Specifically, we evaluated 24 wk old C57BL/6J-Chr #A/J/NaJ CSS (n = 7 to 18) in which each chromosome in the host C57BL/6J strain is replaced by a corresponding chromosome from the donor A/J strain (19 autosomes, X, Y). We determined several A/J chromosomes contribute to body weight (BW), percent body fat (BF), whole body areal BMD (aBMD), and serum insulin-like growth factor (IGF)-I in both a positive and negative manner when compared with C57BL/6J. Specifically, C57BL/6J-Chr 5A/J/NaJ (B.A-5) (males) and B.A-13 (females) contributed to increased BW, whereas B.A-3, 4, 8, 9, 12, and 18 (males) and B.A-3, 4, and 11 (females) contributed to reduced BW. B.A-5 (males) and B.A-13 (females) contributed to increased BF, whereas B.A-12 (males) and B.A-3, 4, 10, and 11 (females) contributed to reduced BF. B.A-14 (females) contributed to increased aBMD and B.A-1, 2, 6, 9, 10, and 18 (males) and B.A-8, 9, and 10 (females) contributed to reduced aBMD. To determine if similar chromosomes regulate aBMD and IGF-I, we determined serum concentrations of IGF-I. B.A-14 and Y (males) and B.A-6 (females) contributed to increased IGF-I and male B.A-3 and female B.A-8 contributed to reduced IGF-I. Overall, we identified several sex-dependent and -independent chromosomes that regulate aBMD and IGF-I in adult mice.
Keywords: A/J, C57BL/6J, quantitative trait loci, osteoporosis
osteoporosis is a major health concern in humans, and this disease is characterized by a low bone mineral density (BMD). BMD is an important determinant of bone strength and low BMD is the result of increased bone resorption and decreased bone formation. Osteoporosis leads to increased bone fragility and therefore an increase in fractures with age. Many factors contribute to regulation of BMD in adult life including sex, nutrition, exercise, environment, and genetics. Of the many factors that contribute to attainment and maintenance of bone mass, genetics accounts for up to 80% of the variation in BMD. Through identification of genes that regulate BMD, we can identify potential therapeutic targets to prevent and/or treat osteoporosis.
Several methods have been utilized to determine the role of genetics in regulating BMD. A well-accepted method is the use of quantitative trait locus (QTL) analysis. We and others have identified many chromosomal loci involved in regulating BMD using this method (2–4, 8, 11, 15, 21, 25). Although this method has successfully identified several chromosomal regions involved in regulating BMD, it requires a large number of mice that take several years to raise, phenotype, and genotype. Recently, a new resource tool, chromosome substitution strains (CSS), has been developed to more efficiently evaluate specific chromosomes that contribute to phenotypic differences between two strains of mice. For this study, we have used the C57BL/6J-Chr #A/J/NaJ consomic set where each chromosome in the host strain, C57BL/6J, was systematically replaced by the A/J donor chromosome (18, 23). The B.A CSS consist of 21 strains with one strain for each autosome and the two sex chromosomes. These strains of mice have been a useful tool for studying the genetic influence on several phenotypes in mice (23). Using CSS has several advantages including: 1) linkage crosses between two inbred strains of mice are not necessary; 2) genotyping of the different lines is not necessary; 3) all the mice are produced within the same genetic background; and 4) the CSS can be backcrossed to the host strain (B6) to break-up the donor chromosome for fine mapping. One disadvantage to this method is that it is only available for a limited number of strain combinations, the A/J and B6 strains being one (a set is also available for B6 and PWD - C57BL/6J-Chr#PWD/Ph/ForeJ); however, the advantages far outweigh this restriction.
Because CSS have been successfully used to identify chromosomes involved in regulating many phenotypes (23), we hypothesized that by comparing body composition and bone-related phenotypes between the CSS strains and the host B6 strain, we could identify A/J chromosomes that harbor genes that regulate the selected phenotypes in adult mice.
MATERIALS AND METHODS
Mice.
The C57BL/6J-Chr #A/J/NaJ chromosome substitution strains (B.A CSS) used in our study were originally developed by Nadeau et al. (18). Briefly, these CSS are C57BL/6J (B6) host mice in which a single B6 chromosome has been replaced with an A/J chromosome. Therefore, there are 21 B.A CSS strains (B.A-1 to 19, -X, and -Y). For our studies, male mice were compared with the host B6 strain. However, for the female mice, the B6 mice were collected at a later date than the other strains. Based on the knowledge that seasonal variation can affect skeletal phenotype, we chose to use B.A-Y strain females as controls for comparison with other female CSS for all phenotypes evaluated. The B.A-Y female strain was derived by breeding B.A-Y males with unrelated B6 females, which resulted in female offspring that are wild type for the B6 chromosome X. Their littermates are the male B.A-Y mice.
Husbandry.
Mice were housed in the Genetic Resource Repository at the Jackson Laboratory in groups of three or four of the same sex in polycarbonate cages (324 cm2) on sterilized White Pine shavings until ∼6 mo of age. Mice were provided free access to acidified water (pH 2.5 with HCl to retard bacterial growth) and NIH31 diet containing 6% fat, 17% protein, Ca:P of 1.5:0.85, plus vitamin and mineral fortification (Purina Mills International, Brentwood, MO). The full composition of this open formula diet is available at www.labdiet.com/indexlabdiethome.htm. All procedures involving mice were reviewed and approved by the Institutional Animal Care and Use Committee of The Jackson Laboratory.
Dual energy X-ray densitometric analyses of areal bone mineral density and percent body fat.
At 6 mo of age, mice (n = 6 to 18/strain) were necropsied for trunk blood samples, followed by determination of whole body composition data (lean, fat, bone mineral) using the PIXImus dual energy X-ray densitometer (DEXA) as described below. Serum was extracted from the blood samples and stored at −70°C until analysis was performed. Data for body fat and areal bone mineral density (aBMD) were collected using the PIXImus small animal DEXA system (software version 1.43.036.008; PIXImus, Fitchburg, WI). The PIXImus has been reconfigured with lower X-ray energy than in human DEXA machines to achieve contrast in small specimens. The resolution of the PIXImus is 0.18 × 0.18 mm pixels with a usable scanning area of 80 × 65 mm, allowing for measurement of single whole mice and collections of isolated specimens. The PIXImus has been calibrated with a phantom utilizing known values, and a quality assurance is performed daily with this same phantom. Assessment of accuracy for the PIXImus was done with a set of hydroxyapatite standards (0–2,000 mg), yielding a correlation of 0.999 between standards and PIXImus measurement of mineral. The precision for aBMD is excellent, <1% for whole body, ∼1.5% for specialized regions. Correlation with peripheral quantitative computerized tomography (pQCT) values for 614 isolated spinal vertebrae is significant (P < 0.001; r = 0.704). These data were acquired from euthanized mice.
Serum insulin-like growth factor-I RIA.
Insulin-like growth factor-I (IGF-I) was measured by RIA using rabbit polyclonal antiserum and recombinant IGF-I as standard and tracer, respectively. IGFBP were removed from serum prior to RIA by acid gel filtration protocol (16).
Statistical analysis.
Male and female data were analyzed separately by ANOVA, and each CSS was compared with the control host strain (females vs. B.A-Y and males vs. B6) using the Dunnett test. ANOVA and correlation analysis were performed using Statistica 6 software (StatSoft, Tulsa, OK). Data are expressed as a percentage of the control strain and presented as means ± SE. A significant difference was determined at P ≤ 0.05.
RESULTS
Body weight.
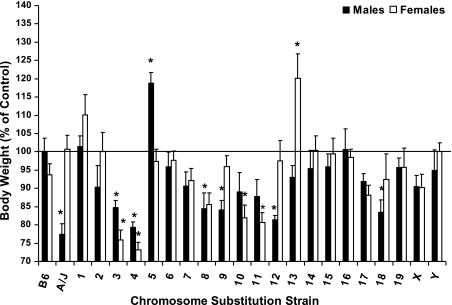
Several A/J chromosomes contributed to variations in body weight (BW) in both male and female mice (Fig. 1). Specifically, male A/J mice were smaller (77%) than B6 mice (P = 0.001). Similarly, in male B.A-3, 4, 8, 9, 12, and 18, BW was 79–85% of male B6 (P < 0.05). In contrast, BW was 119% of B6 in male B.A-5 (P = 0.01). Although female A/J mice have a similar BW compared with B.A-Y (P = 0.99), female B.A-3, 4, and 11 were 73–81% (P < 0.01), and B.A-13 were 120%, respectively (P < 0.05), of B.A-Y females.
Fig. 1.
Body weights of male and female C57BL/6J, A/J, and CSS mice at 6 mo of age. Data are presented as % of control mice [males vs. C57BL/6J and females vs. B.A-Y (B.A-Y females were derived by breeding B.A-Y males with unrelated B6 females, which resulted in female offspring that are wild type for the B6 Chr X.)] and means ± SE. Body weights for B6 males and B.A-Y females are 30.47 ± 1.15 and 22.84 ± 0.55 g, respectively. Data were analyzed by ANOVA and Dunnett's Test. *Significant difference from control at P ≤ 0.05.
Total body fat.
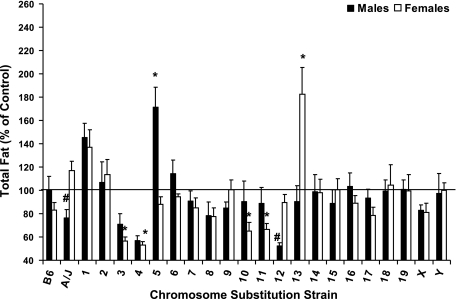
Although a significant difference in body fat was not observed between male A/J and B6 mice (P = 0.87), B.A-5 was 172% compared with B6 males (P < 0.001, Fig. 2). In females, B.A-13 were 182% of female B.A-Y mice (P < 0.01) for body fat, and B.A-3, 4, 10, and 11 were reduced 34–47% compared with B.A-Y females (P < 0.05).
Fig. 2.
Total body fat of male and female C57BL/6J, A/J, and CSS mice at 6 mo of age. Data are presented as % of control mice [males vs. C57BL/6J and females vs. B.A-Y (B.A-Y females were derived by breeding B.A-Y males with unrelated B6 females, which resulted in female offspring that are wild type for the B6 Chr X.)] and means ± SE. Total body fat for B6 males and B.A-Y females are 6.63 ± 0.79 and 5.28 ± 0.34 g, respectively. Data were analyzed by ANOVA and Dunnett's Test. *Significant difference from control at P ≤ 0.05; #tendency for a difference from control at P ≤ 0.10.
aBMD.
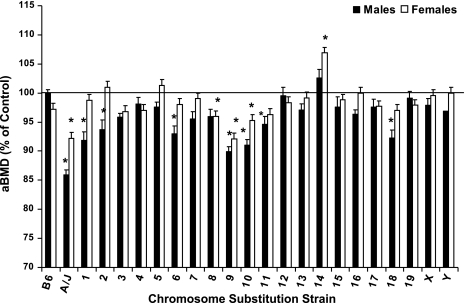
Total body aBMD was less in both male (86%) and female (92%) A/J mice compared with controls (P < 0.05, Fig. 3). Consistent with these data, aBMD was reduced in male B.A-1, 2, 6, 9, 10, 11, and 18 (90–95%, P < 0.05) and reduced in female B.A-8, 9, and 10 (92–96%, P = 0.05). Interestingly, aBMD was greater in female B.A-14 (107%, P < 0.001) compared with B6 females.
Fig. 3.
Total body areal bone mineral density (aBMD) of male and female C57BL/6J, A/J, and CSS mice at 6 mo of age. Data are presented as % of control mice [males vs. C57BL/6J and females vs. B.A-Y (B.A-Y females were derived by breeding B.A-Y males with unrelated B6 females, which resulted in female offspring that are wild type for the B6 Chr X.)] and mean ± SE. Total body aBMD for B6 males and B.A-Y females are 0.0515 ± 0.0006 and 0.0489 ± 0.0003 g/cm2, respectively. Data were analyzed by ANOVA and Dunnett's Test. *Significant difference from control at P ≤ 0.05.
Serum IGF-I.
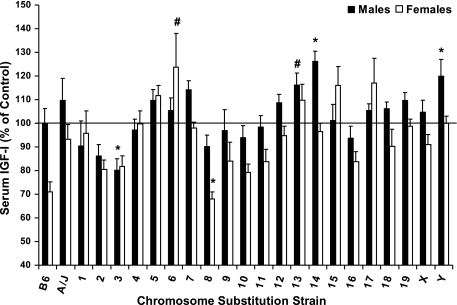
To determine if circulating IGF-I is regulated by specific chromosomes in A/J and B6 mice, we determined serum concentrations of IGF-I in all CSS (Fig. 4). Although we did not observe a significant difference between male A/J and B6 mice (P = 0.91), B.A-14 and -Y had greater IGF-I (136 and 120%, respectively) and B.A-3 had reduced IGF-I (80%) compared with male B6 (P < 0.05). Surprisingly, female B6 mice exhibited 30% lower serum IGF-I compared with female B.A-Y although both of these lines are genetically identical. Because female B6 mice were generated at a later time compared with all other strains, we chose to compare female CSS with female B.A-Y rather than and B6 female mice. Female B.A-6 had greater serum IGF-I (124%, P < 0.01) and B.A-8 had reduced serum IGF-I (68%, P = 0.06) compared with female B.A-Y mice.
Fig. 4.
Serum IGF-I of male and female C57BL/6J, A/J, and CSS mice at 6 mo of age. Data are presented as % of control mice [males vs. C57BL/6J and females vs. B.A-Y (B.A-Y females were derived by breeding B.A-Y males with unrelated B6 females, which resulted in female offspring that are wild type for the B6 Chr X.)] and mean ± SE. Serum IGF-I levels for B6 males and B.A-Y females are 444 ± 28 and 537 ± 16 ng/ml. Data were analyzed by ANOVA and Dunnett's Test. *Significant difference from control at P ≤ 0.05. #Tendency for a difference from control at P ≤ 0.10.
Correlations.
To determine if certain phenotypes are significantly correlated with each other, we performed correlation analysis. As expected, aBMD (R = 0.26 and 0.41), serum IGF-I (R = 0.21 and 0.26), and body fat (R=0.89 and 0.90) were positively correlated with BW in both male and female mice, respectively (P < 0.01 for all). Serum IGF-I was positively correlated with body fat in both males and females (R = 0.19 and 0.20, P < 0.01). Interestingly, aBMD was positively correlated with serum IGF-I and body fat in female mice (R = 0.22 and 0.34, P ≤ 0.001), but not in males (P ≥ 0.11).
DISCUSSION
Maintenance of optimal BMD in adulthood is critical to preventing the onset of osteoporosis; however, the mechanisms involved in maintaining BMD are not well understood. To further elucidate the mechanisms involved in regulating BMD, we utilized a novel approach to begin to identify chromosomes that contribute to variations in BMD in adult mice. Specifically, we evaluated BMD in A/J and B6 mice and determined that BMD is reduced in A/J male and female mice compared with B6. On the basis of this knowledge and the efficiency of the CSS model, we chose to use this method for identification of chromosomes that contribute to this difference in these two inbred strains of mice. Using these unique strains of mice, we have provided the first data demonstrating specific A/J chromosomes involved in regulating BMD and serum IGF-I in adult mice. Not only is this method more efficient than traditional QTL analysis, but it can be used to identify novel QTLs, as well as confirm previous findings in other strains. In addition, due to random segregation and gene-gene interaction, differences can be identified in CSS even if the parental strains are not different. In the past decade, several studies in humans and mice have identified numerous QTLs for various bone-related phenotypes. Many of these studies also identified specific candidate genes for osteoporosis-related phenotypes, which are elaborately summarized by Liu et al. (13, 14). Using our CSS model, we were able to confirm several previously identified QTL for aBMD as discussed below.
In our study we found several A/J chromosomes that contribute to smaller BW in the males. In addition, several chromosomes in the female A/J mice regulate BW in the B6 background, thus suggesting that genes on these chromosomes may be suppressed in the A/J background. Several previous studies have utilized the QTL approach in various inbred strains of mice and identified several chromosomal regions involved in mediating BW and body composition in mice (24). Similar to these studies, we determined that several chromosomes are involved in mediating BW; however, not all are associated with body fat. Of the 10 B.A CSS that have BW different from controls, only 6 have a similar change in body fat. These findings suggest that some of the changes in BW are associated with other factors such as body water, lean, or mineral mass in adult mice. In support of this, further analysis determined that lean mass was positively correlated with body weight (r = 0.8071 and 0.8895 for males and females, respectively; P < 0.0001), however, BMC was not correlated with body weight (P ≥ 0.17). In addition, 6 of the 10 B.A. CSS for BW were also identified for lean body mass (data not shown). A previous meta-analysis of several QTL studies identified chromosomes 1, 2, 7, 11, 15, and 17 as prominent chromosomes involved in regulating BW and body fat (24). Similarly, we identified A/J chromosome 11 as a regulator of BW and body fat in female mice. It would not be expected that all crosses would identify the same QTL for any given phenotype. Since inbred strains vary in their genomic polymorphisms, the QTL discovered in a given cross will be specific to the genetic differences between the progenitor strains.
We identified several chromosomes in male and female A/J mice that regulate aBMD in both a positive and negative manner. These findings are consistent with published QTL studies involving various inbred strain crosses that reveal evidence for multiple genetic loci contributing to BMD variation (13, 14). Similar to a previous study in our laboratory (15), we identified chromosome 9 in both male and female mice as a regulator of aBMD. We also identified chromosome 1 to regulate aBMD in a sex-dependent manner. Other studies have shown evidence for the presence of sex-specific BMD QTL in chromosome 1 (7). However, there are some chromosomes identified in previous studies that we did not determine as regulators of aBMD and vice versa (11, 15). This is not surprising based on BMD QTL studies in various inbred strain crosses of mice that have shown evidence for both common and unique QTL for individual crosses. These differences may be due to the different inbred strains of mice used as well as the different ages of the mice. Alternatively gene-gene interactions may be different depending on the genetic background. To determine if A/J chromosomes contain genetic loci that regulate aBMD independent of BW, body fat, or lean body mass we adjusted aBMD for these factors and identified several chromosomes in both males and females that regulate aBMD in a positive or negative manner (Table 1). Interestingly, we identified chromosome 4 in both males and females as a positive regulator of aBMD independent of BW, body fat, or lean body mass. These findings are consistent with previous reports in other inbred strains of mice (1, 17, 20), thus suggesting that a gene or genes found on this chromosome are critical to maintaining aBMD in mice.
Table 1.
Comparison of chromosomes identified when aBMD is adjusted for BW, LBM, and BF
|
Females |
Males
|
||||||
|---|---|---|---|---|---|---|---|
| Unadjusted | Adjusted for BW | Adjusted for LBM | Adjusted for BF | Unadjusted | Adjusted for BW | Adjusted for LBM | Adjusted for BF |
| A/J | A/J | ||||||
| Chr 1 | |||||||
| Chr 2 | Chr 2 | ||||||
| Chr 3 | Chr 3 | Chr 3 | |||||
| Chr 4 | Chr 4 | Chr 4 | Chr 4 | Chr 4 | Chr 4 | ||
| Chr 6 | |||||||
| Chr 8 | Chr 8 | Chr 8 | |||||
| Chr 9 | Chr 9 | ||||||
| Chr 10 | Chr 10 | Chr 10 | Chr 10 | ||||
| Chr 11 | Chr 11 | Chr 11 | |||||
| Chr 12 | Chr 12 | Chr 12 | |||||
| Chr 14 | |||||||
| Chr 18 | Chr 18 | ||||||
aBMD, areal bone mineral density; BW, body weight; LBM, lean body mass; BF, body fat.
To determine if A/J chromosomes that regulate aBMD and body composition also contribute to variation in serum IGF-I, we evaluated circulating concentrations of IGF-I in CSS mice. Surprisingly, several chromosomes associated with aBMD also contributed to variations in serum IGF-I. Previous studies have identified numerous chromosomes associated with changes in serum IGF-I including chromosomes 1, 2, 3, 4, 5, 6, 7, 8, 10, 12, and 17 (5, 9, 10, 22). We also identified several of these chromosomes, 3, 6, and 8, in male and/or female mice. Surprisingly, we did not identify a contribution by chromosome 10, on which the mouse IGF-I gene is located. Previous studies using B6-C3H/HeJ intercross have narrowed down the chromosome 10 IGF-I QTL region to 18.3 Mb containing 148 genes (5, 22). The serum differences in IGF-I between B6 and C3H/HeJ mice could be due to allelic differences in IGF-I gene itself or other gene(s) within the 18.3 Mb region. If IGF-I is the chromosome 10 QTL gene, we would then predict allelic differences in the regulatory regions of IGF-I gene between B6-C3H/HeJ but not between B6-A/J strain pairs. Our search for single nucleotide polymorphism (SNP) differences among the three strains in the IGF-I gene using available databases (http://phenome.jax.org/pub-cgi/phenome/mpdcgi?rtn=docs/home) revealed four and six SNPs respectively between B6 and C3H/HeJ and between B6 and A/J. However, there was only a single SNP between B6 and AJ and none between B6 and C3H/HeJ in the regulatory region, thus suggesting that other genes located in the QTL region of chromosome 10 may be responsible for regulating serum IGF-I. It is also possible that we did not observe a contribution of chromosome 10 because of the different ages of the mice used in each study.
Several genetic factors can influence the phenotypes observed for each chromosome. For example, several studies have identified sex-specific effects of certain QTLs for BMD and femoral cross-sectional area (6, 7, 12, 19). Therefore it is critical to evaluate males and females separately, as to avoid eliminating or accentuating certain QTLs identified (6). Based on our analysis of males and females separately, we identified several chromosomes which regulate BW, body fat, aBMD, and IGF-I that are sex dependent or independent. In addition to novel genes, these sex effects could be associated with effects of sex steroid regulation. In addition, gene-gene interaction on a particular chromosome can also contribute to differences between these CSS. Further studies are needed to identify specific genes involved in regulating these phenotypes and identify if any gene-gene interaction(s) occurs.
QTL studies not only identify chromosomal regions involved in regulating specific phenotypes, such as aBMD, they also generate potential candidate genes. For example, several well-studied genes associated with osteoporosis, as well as several novel genes identified in previous QTL studies were identified in our CSS (13). To identify specific genes within the loci identified for each chromosome in our model, we plan to backcross selected B.A consomic strains to the B6 host strain to precisely locate the causative genes. However, we have done a preliminary search for potential candidate genes by looking for genes within each chromosome that are known to be involved in regulating aBMD. Specifically, we looked at genes located on chromosome 4 that demonstrated a BW-independent effect on BMD in both male and female mice. We identified seven genes (liver alkaline phosphatase; bone morphogenic protein 8b; disheveled 1; leptin receptor; pregnancy-associated plasma protein a; runt-related transcription factor 1 translocated to 1; and runt-related transcription factor 3) known to be involved in bone maintenance, that contain several SNPs that are different between A/J and B6 mice. These findings demonstrate the effectiveness of our model to identify potential candidate genes.
Because a large number of chromosomes were evaluated in this study, there remains a possibility that one or more of the chromosomes identified for phenotypes studied could represent false positives. We will therefore use a number of criteria to select chromosomes for future studies on gene identification which include: 1) the significance level for the CSS of interest; 2) magnitude of phenotypic change for the CSS of interest; 3) phenotypic change in both sexes; 4) confirmation of chromosome identified by published QTL studies for phenotype of interest; and 5) the extent of SNP and candidate gene information on chromosome of interest. We believe that prioritization of CSS based on multiple well-reasoned criteria will reduce the likelihood of focusing on false positives for future fine mapping and gene identification work.
In summary, the CSS model provides a unique and efficient mechanism to identify potential genes involved in mediating specific phenotypes, such as aBMD. Overall, our findings suggest that multiple genes are involved in regulating adult aBMD, body fat and serum IGF-I. Further studies, in which CSS are crossed with B6 mice, are needed to identify the specific genes involved in regulating aBMD, body fat, and IGF-I.
GRANTS
This work was supported in part by Loma Linda University Seed Grant (to K. E. Govoni and S. Mohan), National Institutes of Health Grants AR-048139 (S. Mohan) and RR-16049 (to L. R. Donahue, P. I.).
Acknowledgments
The authors thank Catrina Alarcon, Joe Rung-Aroon, and Jann Smallwood for technical assistance.
Current address for K. E. Govoni: Dept. of Animal Science, Univ. of Connecticut, Storrs, CT 06269.
Address for reprint requests and other correspondence: S. Mohan, Musculoskeletal Disease Center (151), Jerry L. Pettis Memorial VA Med. Ctr., 11201 Benton St., Loma Linda, CA 92357 (e-mail: Subburaman.Mohan@va.gov).
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Alam I, Sun Q, Liu L, Koller DL, Fishburn T, Carr LG, Econs MJ, Foroud T, Turner CH. Identification of a quantitative trait locus on rat chromosome 4 that is strongly linked to femoral neck structure and strength. Bone 39: 93–99, 2006. [DOI] [PubMed] [Google Scholar]
- 2.Beamer WG, Shultz KL, Ackert-Bicknell CL, Horton LG, Delahunty KM, Coombs HF 3rd, Donahue LR, Canalis E, Rosen CJ. Genetic dissection of mouse distal chromosome 1 reveals three linked BMD QTLs with sex-dependent regulation of bone phenotypes. J Bone Miner Res 22: 1187–1196, 2007. [DOI] [PubMed] [Google Scholar]
- 3.Beamer WG, Shultz KL, Churchill GA, Frankel WN, Baylink DJ, Rosen CJ, Donahue LR. Quantitative trait loci for bone density in C57BL/6J and CAST/EiJ inbred mice. Mamm Genome 10: 1043–1049, 1999. [DOI] [PubMed] [Google Scholar]
- 4.Beamer WG, Shultz KL, Donahue LR, Churchill GA, Sen S, Wergedal JR, Baylink DJ, Rosen CJ. Quantitative trait loci for femoral and lumbar vertebral bone mineral density in C57BL/6J and C3H/HeJ inbred strains of mice. J Bone Miner Res 16: 1195–1206, 2001. [DOI] [PubMed] [Google Scholar]
- 5.Delahunty KM, Shultz KL, Gronowicz GA, Koczon-Jaremko B, Adamo ML, Horton LG, Lorenzo J, Donahue LR, Ackert-Bicknell C, Kream BE, Beamer WG, Rosen CJ. Congenic mice provide in vivo evidence for a genetic locus that modulates serum insulin-like growth factor-I and bone acquisition. Endocrinology 147: 3915–3923, 2006. [DOI] [PubMed] [Google Scholar]
- 6.Donahue LR, Beamer W, Shultz KL, Hurd J, Guido V, Rosen C. QTL for bone and body composition phenotypes are sex specific in a mouse model of GH/IGF-I deficiency. J Bone Miner Res 20: S232, 2005. [Google Scholar]
- 7.Edderkaoui B, Baylink DJ, Beamer WG, Shultz KL, Wergedal JE, Mohan S. Genetic regulation of femoral bone mineral density: complexity of sex effect in chromosome 1 revealed by congenic sublines of mice. Bone 41: 340–345, 2007. [DOI] [PubMed] [Google Scholar]
- 8.Edderkaoui B, Baylink DJ, Beamer WG, Wergedal JE, Porte R, Chaudhuri A, Mohan S. Identification of mouse Duffy antigen receptor for chemokines (Darc) as a BMD QTL gene. Genome Res 17: 577–585, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanlon P, Lorenz WA, Shao Z, Harper JM, Galecki AT, Miller RA, Burke DT. Three-locus and four-locus QTL interactions influence mouse insulin-like growth factor-I. Physiol Genomics 26: 46–54, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harper JM, Wolf N, Galecki AT, Pinkosky SL, Miller RA. Hormone levels and cataract scores as sex-specific, mid-life predictors of longevity in genetically heterogeneous mice. Mech Ageing Dev 124: 801–810, 2003. [DOI] [PubMed] [Google Scholar]
- 11.Ishimori N, Li R, Walsh KA, Korstanje R, Rollins JA, Petkov P, Pletcher MT, Wiltshire T, Donahue LR, Rosen CJ, Beamer WG, Churchill GA, Paigen B. Quantitative trait loci that determine BMD in C57BL/6J and 129S1/SvImJ inbred mice. J Bone Miner Res 21: 105–112, 2006. [DOI] [PubMed] [Google Scholar]
- 12.Klein RF, Turner RJ, Skinner LD, Vartanian KA, Serang M, Carlos AS, Shea M, Belknap JK, Orwoll ES. Mapping quantitative trait loci that influence femoral cross-sectional area in mice. J Bone Miner Res 17: 1752–1760, 2002. [DOI] [PubMed] [Google Scholar]
- 13.Liu YJ, Shen H, Xiao P, Xiong DH, Li LH, Recker RR, Deng HW. Molecular genetic studies of gene identification for osteoporosis: a 2004 update. J Bone Miner Res 21: 1511–1535, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu YZ, Liu YJ, Recker RR, Deng HW. Molecular studies of identification of genes for osteoporosis: the 2002 update. J Endocrinol 177: 147–196, 2003. [DOI] [PubMed] [Google Scholar]
- 15.Masinde GL, Li X, Gu W, Wergedal J, Mohan S, Baylink DJ. Quantitative trait loci for bone density in mice: the genes determining total skeletal density and femur density show little overlap in F2 mice. Calcif Tissue Int 71: 421–428, 2002. [DOI] [PubMed] [Google Scholar]
- 16.Mohan S, Baylink DJ. Development of a simple valid method for the complete removal of insulin-like growth factor (IGF)-binding proteins from IGFs in human serum and other biological fluids: comparison with acid-ethanol treatment and C18 Sep-Pak separation. J Clin Endocrinol Metab 80: 637–647, 1995. [DOI] [PubMed] [Google Scholar]
- 17.Mohan S, Chest V, Chadwick RB, Wergedal JE, Srivastava AK. Chemical mutagenesis induced two high bone density mouse mutants map to a concordant distal chromosome 4 locus. Bone 41: 860–868, 2007. [DOI] [PubMed] [Google Scholar]
- 18.Nadeau JH, Singer JB, Matin A, Lander ES. Analysing complex genetic traits with chromosome substitution strains. Nat Genet 24: 221–225, 2000. [DOI] [PubMed] [Google Scholar]
- 19.Ralston SH, Galwey N, MacKay I, Albagha OM, Cardon L, Compston JE, Cooper C, Duncan E, Keen R, Langdahl B, McLellan A, O'Riordan J, Pols HA, Reid DM, Uitterlinden AG, Wass J, Bennett ST. Loci for regulation of bone mineral density in men and women identified by genome wide linkage scan: the FAMOS study. Hum Mol Genet 14: 943–951, 2005. [DOI] [PubMed] [Google Scholar]
- 20.Robling AG, Li J, Shultz KL, Beamer WG, Turner CH. Evidence for a skeletal mechanosensitivity gene on mouse chromosome 4. FASEB J 17: 324–326, 2003. [DOI] [PubMed] [Google Scholar]
- 21.Rosen CJ, Ackert-Bicknell CL, Adamo ML, Shultz KL, Rubin J, Donahue LR, Horton LG, Delahunty KM, Beamer WG, Sipos J, Clemmons D, Nelson T, Bouxsein ML, Horowitz M. Congenic mice with low serum IGF-I have increased body fat, reduced bone mineral density, and an altered osteoblast differentiation program. Bone 35: 1046–1058, 2004. [DOI] [PubMed] [Google Scholar]
- 22.Rosen CJ, Churchill GA, Donahue LR, Shultz KL, Burgess JK, Powell DR, Ackert C, Beamer WG. Mapping quantitative trait loci for serum insulin-like growth factor-1 levels in mice. Bone 27: 521–528, 2000. [DOI] [PubMed] [Google Scholar]
- 23.Singer JB, Hill AE, Burrage LC, Olszens KR, Song J, Justice M, O'Brien WE, Conti DV, Witte JS, Lander ES, Nadeau JH. Genetic dissection of complex traits with chromosome substitution strains of mice. Science 304: 445–448, 2004. [DOI] [PubMed] [Google Scholar]
- 24.Wuschke S, Dahm S, Schmidt C, Joost HG, Al-Hasani H. A meta-analysis of quantitative trait loci associated with body weight and adiposity in mice. Int J Obes (Lond) 31: 829–841, 2007. [DOI] [PubMed] [Google Scholar]
- 25.Yu H, Mohan S, Edderkaoui B, Masinde GL, Davidson HM, Wergedal JE, Beamer WG, Baylink DJ. Detecting novel bone density and bone size quantitative trait loci using a cross of MRL/MpJ and CAST/EiJ inbred mice. Calcif Tissue Int 80: 103–110, 2007. [DOI] [PubMed] [Google Scholar]