Abstract
Dietary folate supplementation can dramatically reduce the severity and incidence of several common birth defects and adult diseases that are associated with anomalies in homocysteine and folate metabolism. The common polymorphisms that adversely affect these metabolic pathways do not fully account for the particular birth defects and adult diseases that occur in at-risk individuals. To test involvement of folate, homocysteine, and other pathways in disease pathogenesis and treatment response, we analyzed global and pathway-specific changes in gene expression and levels of selected metabolites after depletion and repletion of dietary folate in two genetically distinct inbred strains of mice. Compared with the C57BL/6J strain, A/J showed greater homeostatic response to folate perturbation by retaining a higher serum folate level and minimizing global gene expression changes. Remarkably, folate perturbation led to systematic strain-specific differences only in the expression profile of the cholesterol biosynthesis pathway and to changes in levels of serum and liver total cholesterol. By genetically increasing serum and liver total cholesterol levels in APOE-deficient mice, we modestly but significantly improved folate retention during folate depletion, suggesting that homeostasis among the homocysteine, folate and cholesterol metabolic pathways contributes to the beneficial effects of dietary folate supplementation.
genetic variants in the homocysteine and folate metabolic pathways, which are involved in methionine synthesis, methylation of DNA, RNA, proteins and lipids, synthesis of purines and pyrimidines, and degradation of histidine (26), are associated with increased risk for neural tube defects (56), congenital heart defects (57), cardiovascular disease (27), stroke (20), colon cancer (55), and Alzheimer's disease (4). High levels of homocysteine and insufficient levels of folate are often found in affected individuals and in mothers of babies with neural tube or congenital heart defects (21, 45). Remarkably, dietary folate supplementation often normalizes homocysteine levels and reduces the severity and incidence of these conditions (11, 34, 39, 58). However, the mechanism for folate rescue for these birth defects and adult diseases is unknown.
Single gene mouse mutants can be used in experimental studies where specific dietary factors and metabolites can be precisely perturbed on defined genetic backgrounds. These studies show that targeted deficiency for several genes involved in homocysteine and folate metabolism genes are sufficient to cause neural tube defects (36) and endothelial dysfunction (9, 13, 28). Moreover, adverse phenotypes in mice with genetic variants involving functions such as Wnt signaling (low density lipoprotein receptor-related protein 6) (6, 7), hypoxia (CBP/P300 transactivator) (3), and others (ALX homeobox1, paired box gene 3) (16, 65) can be rescued with dietary folate supplementation, suggesting that these dietary factors can compensate for anomalies in other signaling and metabolic pathways. Moreover, mutants in genes such as Pax3 and other mutants in Wnt and hedgehog signaling show altered homocysteine and folate metabolism (14), suggesting that anomalies in the homocysteine and folate pathways alone do not fully account for these human diseases. Instead complex and dynamic interactions between distinct pathways determine the pathogenic outcome and response to dietary supplementations.
To test for pathways that interact with the homocysteine and folate metabolism, we performed a global gene expression survey of two inbred strains of mice under dietary folate depletion and repletion. We found that these strains differed significantly in response to dietary folate perturbations with A/J showing greater resilience in their homeostatic response. Depletion and repletion with dietary folate specifically affected cholesterol metabolism, and modulating cholesterol levels enhanced homeostatic responses to folate perturbation, suggesting that cholesterol metabolism contribute to the beneficial effects of dietary folate supplementation.
MATERIALS AND METHODS
Mice and diets.
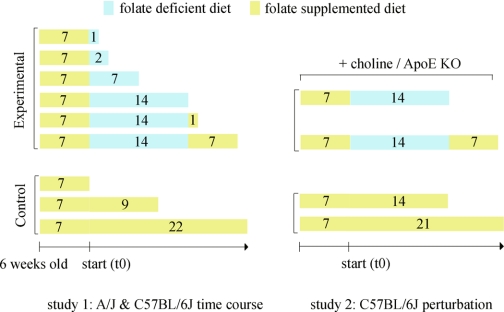
Six-week-old female A/J, C57BL/6J, and B6.129P2-Apoetm1Unc/J (35) were purchased from the Jackson Laboratory. All mice were raised on a control diet containing 4 ppm folic acid (Basal Diet 5755, TestDiet) for 1 wk before the start of studies. Selected mice were then placed on folic acid deficient diet (58C3, TestDiet) containing 1% succinylsulfathiazole, a nonabsorbable antibiotic commonly used to suppress folate production by bacteria in the intestine. All diets contained no more than a trace amount (<50 ppm) of dietary cholesterol and were irradiated by the manufacturer. Treatment plans for studies 1 and 2 (Fig. 1) included eight replicate mice per treatment per strain. A new batch of diet was manufactured for study 2. The folate level in this diet was significantly lower than the previous control diet due to greater loss of folic acid with irradiation of the diet. C57BL/6J mice on choline treatment (study 2) were given water containing 25 mM of choline and 50 mM of saccharin. Saccharin was used to reduce the bitter taste of choline. Folate-depleted C57BL/6J mice (study 2) were placed on water containing 50 mM saccharin to monitor the effect of choline on water consumption.
Fig. 1.
Folate perturbation protocols. Study 1: folate deletion and repletion. Six-week-old A/J and C57BL/6J females were placed on each of the 9 treatment plans with 8 replicate mice per treatment per strain. Study 2: folate depletion and repletion with additional genetic and dietary perturbations in C57BL/6J. C57BL/6J wild type, C57BL/6J wild type on choline supplementation, and ApoE knockout on the C57BL/6J background were all placed on folate depletion and repletion experimental treatments. Wild-type C57BL/6J were also placed on the control diet for both the depletion and repletion period and designated as controls. Eight replicate mice were used for each of the 8 treatment plans.
From each mouse, blood samples were obtained from the retro-orbital sinus and centrifuged. Serum samples were stored at −80°C. Mice were then killed, and tissues were collected, frozen in dry ice, and immediately stored at −80°C. All mice shared the same animal room with controlled temperature, humidity, and 12 h light-dark cycle. Mice were provided food and water ad libitum. The use of mice in these studies was approved by the Case Western Reserve University Institutional Animal Care and Use Committee.
Expression profiling.
For study 1, an equal amount (by weight) of liver tissue from eight replicate mice per treatment plan was pooled as a single sample. For study 2, an equal amount (by weight) of liver tissue from eight replicate mice was separated into two pools of four replicate tissues each. Total RNA from each pool was extracted with TRIzol (Invitrogen, Carlsbad, CA), treated with DNase (Ambion, Austin, TX), and cleaned with RNeasy (Qiagen, Valencia, CA). For study 1 15 μg of pooled total RNA and 15 μg of Universal Mouse Reference RNA (Stratagene, La Jolla, CA) were aminoallyl-labeled with Cy3 and Cy5 in duplicate, with reversing of dyes; and for study 2 15 μg of pooled RNA from treated mice and 15 μg of pooled RNA from control mice for each time point were aminoallyl-labeled with Cy3 and Cy5 in duplicate with reversing of dyes. Cy3- and Cy5-labeled samples were cohybridized to mouse cDNA array representing >25,000 unique genes and expressed sequence tags (ESTs) from NIA mouse 15K set (50) and BMAP mouse cDNA clone set as described (19). Array images were scanned using the GenePix 4000A scanner (Axon Instruments, Union City, CA) and array spot intensities were acquired using TIGR Spotfinder as outlined (62) and normalized using global Lowess regression using MIDAS (44) set at a smoothing parameter of 0.3 and filtered for inconsistent flip-dye pairs at a two-standard deviation cutoff.
Hierarchical clustering.
All microarray data were log2 transformed and filtered to minimize the number of gene expression patterns resulting from noise. Only genes in which the standard deviation across time points was greater than the average standard deviation of replicates were kept. Data were mean centered and scaled (normalized) both across genes and experiments. We performed average linkage hierarchical clustering using Euclidean distance and estimated statistical robustness of each branch with bootstrap resampling of genes (1,000×) using TIGR Multiexperiment Viewer (MeV) (44).
Significant gene expression changes.
To test for significant gene expression changes between two strains across time points (study 1, Fig. 1), we fitted the data using a fixed model ANOVA method previously described (25) using the MAANOVA R package v. 0.98-3. After fitting the effects of array and dye on gene expression variance, we found our null model consisted of variance due to independent effects of strain and folate treatment without interactions between the two effects. Our alternative hypothesis contained the interaction term. The calculated F statistics were compared with the tabulated F distribution on a per-gene basis and adjusted for multiple testing using Bonferroni correction. For the study involving ApoE knockout and choline treatments (study 2, Fig. 1), we performed ANOVA analysis for each time point with treatment as the only variable of interest. To identify gene expression changes specific to each treatment in study 2, we performed postanalysis t-test using tabulated t-statistics with Bonferroni correction for multiple testing and false discovery rate estimation (48).
Pathway analysis.
Each of the microarray clones referenced with a GenBank accession number was converted to Mouse Genome Informatics (MGI) accession number using the TIGR Resourcerer database v.12.0 (52). MGI accession numbers were then used to link each cDNA clone to Gene Ontology (GO) functional annotation (July 2005 monthly repository) (2). We tested for overrepresentation of each GO term in differentially expressed genes using one-tailed Fisher's exact test. We made certain that a gene represented by multiple clones appeared only once when counting the significantly expressed genes as well as when calculating the null distribution from all genes on the array. We tested all terms within the hierarchy of GO functions and used Bonferroni correction for the number of terms examined. We excluded GO terms when the number of observed genes for that term was smaller than three to avoid significance arising from few false positive genes.
Annotation of methylated genes.
For genomic repeats, each of the clone sequences was analyzed against RepBase (24) using RepeatMasker (http://www.repeatmasker.org). Only clones with repeat sequence spanning the entire clone were included. Therefore, clones with partial repeat sequence (hybrid sequence) were excluded from the definition of repeat clones. We obtained a list of imprinted genes from Mouse Imprinting Database at Mammalian Genetics Unit at MRC (http://www.mgu.har.mrc.ac.uk/research/imprinting/). For X-inactivated genes, we assumed for simplicity that all of the genes on the X-chromosome undergo inactivation.
Choline kinase co-regulated gene set.
To identify genes that are co-regulated with choline kinase, we used genes that showed >90% Pearson's correlation with the choline kinase expression profile in C57BL/6J time-course experiment (study 1). We then used these genes to test whether this set of genes were differentially expressed between choline-treated mice vs. mice on control diet using gene set enrichment analysis (GSEA) software v1.0 (33, 49). Given the limited number of four replicates, we performed permutation of gene annotation assignments for each gene to calculate significance scores rather than permutation on sample identities. This could potentially bias the analysis for overestimation of statistical significance. To avoid enrichment of specific terms arising from genes represented by more than one clone, we selected one clone per gene based on the maximum difference between the standard deviation across treatment conditions and the average of standard deviation of replicate measurements.
Serum homocysteine and folate.
Total serum homocysteine level was measured using a liquid chromatographic-tandem mass spectrometric method (30). Serum folate level was measured in duplicate using a microbiological method after deproteination (22).
DNA methylation measurement.
Genomic DNA was extracted from each replicate liver tissue using DNeasy Tissue kit (Qiagen). DNA was treated with methyl-sensitive HpaII and methyl-insensitive MspI restriction enzymes (New England Biolabs, Ipswitch, MA) and cytosine extension assay was performed to quantify the amount of digested DNA (37). The ratio of HpaII to MspI digest indicates the fraction of unmethylated DNA. The DNA digests and cytosine extension assay were performed in duplicate.
Cholesterol measurement.
Serum total cholesterol was quantified by GC/MS (Hewlett Packard 6890 GC with a 5973 mass spectrometer) using selected ion monitoring mode (46) for the A/J and C57BL/6J folate perturbation experiments (study 1, Fig. 1). For experiments involving additional treatments on C57BL/6J (study 2, Fig. 1), serum total cholesterol was measured with an enzymatic assay (1) in duplicate using Infinity Cholesterol Reagent (ThermoElectron, San Jose, CA). Liver cholesterol was extracted with chloroform and methanol as described in Folch et al. (17). Extracted cholesterol from the chloroform layer was air-dried at 50°C and resuspended in 5% Triton X-100 and quantified in duplicate with enzymatic assay as above. The total cholesterol level from liver was standardized against the amount of total protein from the aqueous extract using BCA Protein Assay Kit (Pierce, Rockford, IL).
Statistical analysis of metabolite measurements.
We first used the Kolmogorov-Smirnov test to determine whether data from each analysis group were normally distributed, and we used Bartlett's test to assess homogeneity of variance across sample groups. We used ANOVA for normally distributed data with equal variance and the Kruskal-Wallis test for nonnormal data and unequal variance data to establish statistical difference between groups. To identify the group pairs that showed a significant difference, we used Bonferroni's multiple comparison test for normally distributed data with equal variance and Dunn's multiple comparison test for the nonnormal data or data with unequal variance. All of the statistical analyses were performed using Prism 3.0 (GraphPad Software, San Diego, CA). To plot the percent change in metabolite levels from controls, we extrapolated the three control time points and calculated the percent difference in metabolite level at each sample time point from the extrapolated control.
RESULTS
Overview.
To identify genes, molecules, and pathways that responded to folate perturbation, we characterized expression and metabolite profiles during dietary folate depletion and repletion in two genetically distinct inbred strains of mice (Fig. 1). Time points on the order of days were selected to capture immediate changes and weeks to capture prolonged effects based on published observations that serum folate loss occurs within days after folate depletion (38).
Metabolite responses.
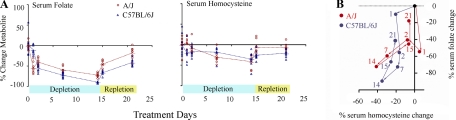
Folate levels differed dramatically between the two strains, both in the immediate response to folate depletion and the ultimate outcome after folate repletion (Fig. 2, A and B). Serum folate declined significantly in A/J after 1 day of folate depletion (54%, Dunn's posttest, rank sum difference = 39.75, P < 0.01) but remained relatively unchanged in C57BL/6J during the same period (10%, Dunn's posttest, rank sum difference = 4.25, P > 0.05). After 2 wk of dietary folate depletion, serum folate loss in A/J was minimized at 74% (Dunn's posttest, rank sum difference = 52.63, P < 0.001), whereas C57BL/6J showed a larger 90% reduction (Dunn's posttest, rank sum difference = 57.38, P < 0.001). After folate repletion, the serum folate level in A/J was restored to within 18% of control (Dunn's posttest, rank sum difference = 11.0, P > 0.05), whereas C57BL/6J still showed a 41% reduction from the control (Dunn's posttest, rank sum difference = 21.0, P > 0.05) (Fig. 2B). We conclude that A/J, despite immediate folate loss, maintained higher serum folate level during dietary depletion and regained higher folate level during repletion compared with C57BL/6J.
Fig. 2.
Serum homocysteine and folate profiles. A: metabolite levels were measured for each of the 8 replicate females per strain per time point. Metabolite levels at each treatment time point were compared with the extrapolated control values (0, 9, 22 days). These differences were normalized to the extrapolated control values and represented as percent change from the control. The metabolite levels of each replicate animal are shown as red circles for A/J and blue triangles for C57BL/6J. The average metabolite values are shown as cross bars and are connected with dotted line across time points. B: plot of serum homocysteine change (x-axis) vs. serum folate change (y-axis). Numbers indicate days under folate perturbation treatment.
In contrast, homocysteine level did not change significantly after 1 day of folate depletion in A/J (5%, Dunn's posttest, rank sum difference = 3.38, P > 0.05) or C57BL/6J (20%, Dunn's posttest, rank sum difference = 20.75, P > 0.05). However, after 2 wk of dietary folate depletion, homocysteine decreased by 35% in A/J (Dunn's posttest, rank sum difference = 35.31, P < 0.01) and 40% in C57BL/6J (Dunn's posttest, rank sum difference = 24.75, P > 0.05). Then, after only 1 day of folate repletion, the homocysteine level was restored to within 6% from its original value in A/J (Dunn's posttest, rank sum difference = 6.81, P > 0.05) vs. 21% in C57BL/6J (Dunn's posttest, rank sum difference = 40.88, P < 0.001). Although both A/J and C57BL/6J reduced serum homocysteine level during folate depletion, A/J restored homocysteine level faster than C57BL/6J during folate repletion and by the end of the study was significantly closer to the control level (Fig. 2B). The stronger resilience in A/J vs. C57BL/6J suggests genetic differences in homeostatic response to perturbations of dietary folate levels.
Global gene expression response.
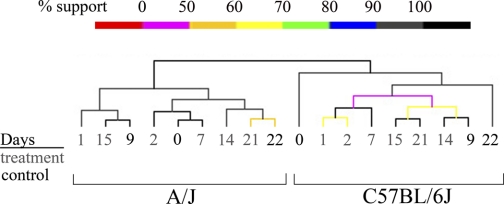
To identify genes and pathways that account for strain-specific response to dietary folate perturbation, we profiled gene expression of total RNA from pooled liver using cDNA arrays consisting of >25,000 unique EST sequences. Liver was selected because the biochemistry of homocysteine and folate metabolism has been extensively studied in this organ (15). We identified dynamic gene expression responses to dietary folate perturbation in the two strains (Fig. 3). Hierarchical clustering of gene expression patterns showed distinct clusters, each of which corresponded exclusively to the time points for each inbred strain, demonstrating that genetic factors had a larger impact than diet on gene expression profiles. Both strains deviated from the initial control during early responses to folate depletion. However, after folate was restored, the last treatment was most similar to the final control in A/J, whereas the last treatment was one of the most distinct from the final control in C57BL/6J. We conclude that the two strains showed distinct gene expression responses to dietary folate perturbation and that A/J showed significantly more resilience to these perturbations than C57BL/6J.
Fig. 3.
Hierarchical clustering of liver gene expression profiles from folate perturbation experiments in A/J and C57BL/6J. Expression profiles were based on 3,950 genes out of 25,000 that showed greater gene expression variance across treatments compared with average variance among replicates. Hierarchical tree shows grouping of expression profiles based on strain. Shown below are pairwise comparisons of each treatment time point within each strain. Darker red indicates similarity between 2 time points.
DNA methylation status.
Folate participates in the transport of methyl-group donors to the homocysteine pathway for methylation of DNA, RNA, proteins, and lipids (26). Changes in DNA methylation can directly impact expression of methylated genes (60), which may account for the lower resilience of C57BL/6J expression to dietary folate perturbations. To test whether dietary folate depletion led to DNA hypomethylation, we measured global DNA methylation levels in both A/J and C57BL/6J across all folate depletion and repletion time points and in the corresponding controls. However, we did not find significant changes in global DNA methylation levels in either strain during folate perturbation (A/J: ANOVA df = 8,62; F = 1.00, P = 0.45, C57BL/6J: ANOVA df = 8,63; F = 0.46, P = 0.88).
Although we did not measure DNA methylation of individual gene loci, expression of intracisternal A particles (IAPs) can serve as markers of DNA hypomethylation during folate depletion (59). IAPs are parasitic sequences in the mouse genome that exist in the order of 1,000 copies and are silenced when methylated. We observed a negative correlation between expression profiles of two IAP probes (Pearson's correlation, nominal P < 0.05) and serum folate profile in C57BL/6J (Fisher's exact test, observed 2, expected 0.2, P = 0.017) but not in A/J (Fisher's exact test, observed 0, expected 0.08, P = 0.99) (Supplemental Table S1).1 We did not observe such correlations in other genomic repeat sequences, imprinted genes, or genes under X-inactivation (Supplemental Tables S1, S2). These data suggest that activation of repetitive sequence expression could contribute to the reduced resilience of C57BL/6J to folate perturbation. Absence of expression response in imprinted and X-inactivated genes suggests that other large classes of methylation-independent genes account for the strain difference in resilience.
Pathway responses.
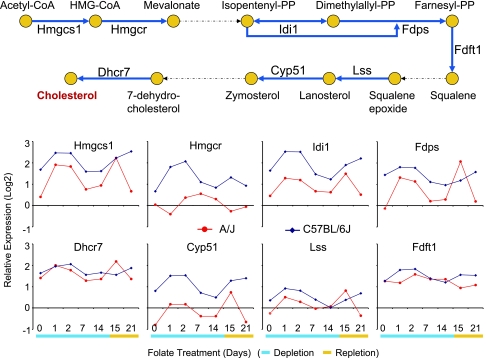
To identify pathways that showed strong strain-specific responses to dietary folate perturbation, we tested for expression changes that showed strain-diet interaction effect using a fixed model ANOVA (25) at P = 0.05 threshold after Bonferroni correction (Supplemental Table S3). Then, using GO annotation (2), we queried specific pathway annotations that were overrepresented among these differentially expressed genes. Remarkably, the only GO annotation that showed significant and systematic overrepresentation was cholesterol biosynthesis and its parent terms (Table 1). The two strains differed in the pattern and magnitude of expression across several cholesterol biosynthesis genes including the rate-limiting enzyme 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) (Fig. 4). Although GO annotations do not contain homocysteine and folate pathways, when we analyzed genes in these pathways (5), we did not find evidence for significant overrepresentation (Fisher's exact test: observed 3, expected 0.63, Bonferroni P > 0.05). Thus dietary folate perturbations resulted specifically and systematically in significant changes in expression patterns for genes involved in cholesterol biosynthesis and related pathways.
Table 1.
Significant overrepresentation of Gene Ontology terms among differentially expressed genes in Study 1
| Pathway | GO Accession No. | All Genes | Observed | Expected | Bonferroni Corrected P Value |
|---|---|---|---|---|---|
| Steroid biosynthesis | 0006694 | 22 | 8 | 0.76 | 0.000161 |
| Lipid biosynthesis | 0008610 | 65 | 12 | 2.25 | 0.000679 |
| Sterol biosynthesis | 0016126 | 14 | 6 | 0.48 | 0.00168 |
| Lipid metabolism | 0006629 | 164 | 18 | 5.67 | 0.00429 |
| Sterol metabolism | 0016125 | 25 | 7 | 0.86 | 0.00662 |
| Cholesterol biosynthesis | 0006695 | 11 | 5 | 0.38 | 0.00825 |
| Steroid metabolism | 0008202 | 38 | 8 | 1.31 | 0.0155 |
| Isoprenoid biosynthesis | 0008299 | 3 | 3 | 0.10 | 0.0187 |
| Cholesterol metabolism | 0008203 | 21 | 6 | 0.73 | 0.0248 |
| Cellular lipid metabolism | 0044255 | 140 | 15 | 4.84 | 0.0339 |
| Alcohol metabolism | 0006066 | 81 | 11 | 2.80 | 0.0391 |
Column ‘All Genes’ refers to genes on the array annotated with the particular Gene Ontology (GO) term.
Fig. 4.
Expression profiles of genes involved in cholesterol biosynthesis (GO: 0006695). Relative expression changes from the Universal Mouse Reference RNA (Stratagene) are represented in log2 scale. Red lines indicate A/J and blue lines indicate C57BL/6J profiles. Genes with significant expression changes that showed interaction effects involving strain and folate perturbation after fixed model ANOVA at P = 0.05 (Bonferroni correction) were Hmgcs1, Hmgcr, Idi1, Fdps, and Dhcr7.
Changes in cholesterol levels.
To test whether expression changes in cholesterol biosynthesis and related pathways corresponded to changes in cholesterol metabolites, we measured liver and serum total cholesterol levels (Fig. 5, A and B). Liver cholesterol level decreased 28% in A/J (Dunn's posttest, rank sum difference = 23.50, P > 0.05) after 1 day of folate depletion but increased 27% in C57BL/6J (Dunn's posttest, rank sum difference = 30.75, P < 0.05) (Fig. 5A). Liver cholesterol in A/J increased to 57% by 14 days of folate depletion (Dunn's posttest, rank sum difference = 40.38, P < 0.001), whereas C57BL/6J level stayed at 32% (Dunn's posttest, rank sum difference = 37.75, P < 0.001). Although we detected strain differences in liver cholesterol profile, we were unable to identify a strong or direct correlation between cholesterol metabolite profile to homocysteine and folate metabolite profiles.
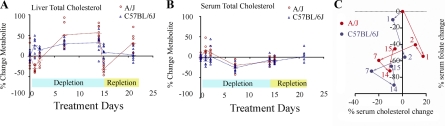
Fig. 5.
Serum and liver cholesterol profiles. A, B: metabolite levels were measured for each of the 8 replicate females per strain per time point. Metabolite levels at each treatment time point were compared with the extrapolated control values (0, 9, 22 days). These differences were normalized to the extrapolated control values and represented as percent change from the control. For the A/J serum total cholesterol level, we did not have data for day 21 of treatment and day 22 of control. Therefore, we plotted treatment days 14 and 15 data with extrapolated control from day 0 and day 9 control. The metabolite levels of each replicate animal are shown as red circles for A/J and blue triangles for C57BL/6J. The average metabolite values are shown as cross bar and are connected with dotted line across time points. C: plot of serum total cholesterol change (x-axis) vs. serum folate change (y-axis). Numbers indicate days under folate perturbation treatment.
Serum cholesterol level increased 17% (Dunn's posttest, rank sum difference = 22.38, P < 0.05) in A/J after 1 day of folate depletion, but there was no increase in C57BL/6J (8% decrease, Dunn's posttest, rank sum difference = 3.63, P > 0.05) (Fig. 5B). In both strains, serum cholesterol levels declined thereafter before returning to control levels by the 14th day of depletion. We then compared the serum folate and serum cholesterol profiles and discovered that the C57BL/6J metabolite profile closely followed the A/J profile with a difference primarily in timing (Fig. 5C). In A/J, an initial increase in serum total cholesterol accompanied loss of serum folate at day 1, whereas this response was delayed an additional day in C57BL/6J. Thereafter the changes in serum cholesterol and folate levels were remarkably similar, suggesting the strains differed primarily in the timing of their initial response to folate depletion.
Do cholesterol levels affect responses to folate perturbation?
We hypothesized that an elevated serum cholesterol level in A/J was associated with higher serum folate maintenance. We tested this hypothesis by elevating serum cholesterol level in C57BL/6J, a strain that showed a delayed and diminished serum cholesterol response. We used mice with targeted deficiency of apolipoprotein E (APOE) on the C57BL/6J inbred background (B6.129P2-Apoetm1Unc/J) to elevate the serum cholesterol level and performed dietary folate perturbations. We sampled 14 days of folate depletion and 14 days of depletion followed by 7 days of repletion (day 21), two time points in A/J that showed improved folate retention compared with C57BL/6J. Although the elevation in serum cholesterol was transient in A/J, chronic elevation in serum cholesterol level in ApoE-deficient mice allowed us to obtain more robust metabolite change. APOE-deficient mice under folate depletion achieved a significant increase in serum total cholesterol (532%, Dunn's posttest, rank sum difference = 28.38, P < 0.05) compared with wild-type mice under folate depletion (Table 2). The magnitude was similar to the fourfold increase in serum cholesterol with ApoE-deficiency alone under normal diet (32). Interestingly, APOE-deficient mice maintained 53% higher serum folate level compared with wild-type mice during folate depletion (Dunn's posttest, rank sum difference = 6.13, P > 0.05). Moreover, after 1 wk of folate repletion, ApoE-deficient mice achieved 37% higher serum folate level than wild-type mice under the same folate perturbation (Dunn's posttest, rank sum difference = 16.00, P > 0.05). We also tested whether higher serum folate level in ApoE-deficient mice would minimize gene expression changes induced by folate depletion. However, gene expression changes in APOE-deficient mice were much larger than the wild-type mice during folate depletion (Table 2). These genes included four out of seven glutathione S-transferase μ-genes, which are involved in oxidative stress response as well as the ApoE gene itself. We conclude that increased cholesterol levels in APOE-deficient mice led to retention of a higher level of serum folate during folate depletion but did not lead to resilience in gene expression profiles.
Table 2.
Effect of ApoE knockout and dietary choline supplements on metabolite and gene expression profiles in C57BL/6J females
|
Treatment |
Metabolites
|
Expression Change
|
|||||
|---|---|---|---|---|---|---|---|
| Time, days | Diet | Additional Perturbation | Serum Folate | Serum Total Cholesterol | Liver Total Cholesterol | FWER P = 0.05 | FDR q =0.01 |
| 14 | folate + | none | 19.1±4.3 | 122±7 | 234±20 | ||
| folate − | none | 5.4±1.6a | 137±13 | 257±28 | 2 | 1 | |
| folate − | ApoE KO | 8.3±3.7 | 729±107a,b | 289±30 | 8 | 27 | |
| folate − | choline | 3.2±1.1a | 152±20 | 299±55 | 7 | 38 | |
| 14/7 | folate +/+ | none | 18.4±2.8 | 127±19 | 235±20 | ||
| folate −/+ | none | 16.4±4.5 | 142±16 | 282±51 | 0 | 0 | |
| folate −/+ | ApoE KO | 22.5±3.9 | 872±61a,b | 335±38a | 3 | 4 | |
| folate −/+ | choline | 16.6±1.8 | 143±23 | 295±42 | 2 | 3 | |
Units for metabolites are pmol/ml serum folate, mg/dl serum total cholesterol, mg total liver cholesterol/g liver protein. Average and SD values are shown. Significant gene expression changes were computed at family-wise error rate (FWER) P = 0.05 and false discovery rate (FDR) q =0.01 using fixed model ANOVA with gene-specific tabulated F statistic. Each treatment was compared to mice on control diet for postanalysis using t-test corrected for multiple testing with the same corresponding thresholds as ANOVA. Significant differences in metabolite levels from C57BL/6J on the control diet
(P < 0.05) and from C57BL/6J on folate treatment diet
(P < 0.05) were calculated based on Kruskal-Wallis test with Dunn's multiple comparison posttest. All mice were on the C57BL/6J background. Folate-treated mice without additional perturbation were maintained on 50 mM saccharin water, whereas folate-treated, choline-supplemented mice were on 50 mM saccharin and 25 mM choline water.
Metabolic functions that link folate perturbation to cholesterol homeostasis.
We observed that dietary folate perturbation induced changes in liver and serum total cholesterol levels. However, homocysteine and folate pathways are not directly linked to the cholesterol biosynthesis pathway and therefore the link between these two pathways is unclear. A possible link is through endoplasmic reticulum (ER) stress. Folate depletion and elevated serum homocysteine level are known to elevate ER stress in the liver. ER stress activates sterol regulatory element binding protein (SREBP), which activates transcription of cholesterol biosynthesis genes (61). However, unlike cholesterol biosynthesis genes, none of the common ER stress genes (GRP78, CHOP, ATF4) showed significant expression changes involving strain and diet interaction (not shown), suggesting that folate depletion did not trigger ER stress response within the timeframe that we examined.
Another possible link between folate perturbation and cholesterol change is through choline. Choline serves as an alternate methyl donor for homocysteine metabolism (12) as well as a precursor for choline phospholipids synthesis required for lipoprotein secretion and transmembrane signaling (64). Therefore, an increased demand for choline due to dietary folate depletion could limit its availability for lipoprotein secretion, thereby altering the cholesterol profile. To test the role of choline in cholesterol pathway responses, we supplemented drinking water with choline and performed the same dietary folate perturbation on C57BL/6J as in the APOE study (Fig. 1). We used saccharin to reduce the bitter taste of choline and found that mice drank 16% less choline-water compared with mice on control saccharin-water (Mann-Whitney U-test 3.50, P = 0.10). Choline supplementation slightly increased liver cholesterol level (11%, Dunn's posttest, rank sum difference = 15.75, P > 0.05) and serum cholesterol level (16%, Dunn's posttest, rank sum difference = 10.00, P > 0.05) compared with nonsupplemented mice during folate depletion (Table 2). However, choline treatment resulted in 41% greater decrease in serum folate level during folate depletion compared with mice without choline supplement (Dunn's posttest, rank sum difference = 7.75, P > 0.05). Choline also induced more gene expression changes compared with mice without choline treatment (Table 2). Moreover, expression of choline kinase, a gene that shuttles choline toward homocysteine methylation and away from choline phospholipids synthesis, was downregulated during folate depletion in C57BL/6J but not in A/J. This gene was further downregulated with choline supplementation (day 14: 1.55-fold decrease, t = 5.64, nominal P = 0.0013, day 21: 1.63-fold decrease, t = 4.92, nominal P = 0.0027) along with its coexpressed genes (day 14: enrichment score = 0.59, Bonferroni P = 0.026, day 21: enrichment score = 0.63, Bonferroni P = 0.027), suggesting that choline supplementation exacerbates the C57BL/6J expression response to folate perturbation rather than revert the expression profile back to control values. We conclude that despite slightly increasing liver and serum cholesterol levels, choline supplementation did not significantly improve serum folate retention and further enhanced expression response away from control profiles.
DISCUSSION
We performed dietary folate perturbation in two distinct inbred strains of mice to identify pathways that interact with homocysteine and folate metabolism. We found that two strains differed in cholesterol metabolism response and that increasing cholesterol levels using APOE-deficient mice improved serum folate retention. Although the effect of retention was modest, modulating a single component will not usually recapitulate the complete response found in the original perturbation study, unless the tested component is a rate-limiting step or a tipping point. The experimental test therefore verified a dependence between cholesterol and folate metabolism.
Previous studies offer insight into why cholesterol biosynthesis was important to folate retention. Cholesterol facilitates clustering of folate receptors on the cell membrane and regulates the rate of cellular folate import (29, 47). Kidney epithelial cells continually cultured in cholesterol deficient media to lower cellular cholesterol level show decreased folate import (8). Additionally, fibroblasts from Smith-Lemli Opitz syndrome (SLOS) patients that are unable to produce sufficient cholesterol show decreased folate uptake and a reduced cell proliferation rate (54). Together, these studies suggest cells could improve folate uptake efficiency by increasing cholesterol biosynthesis during low folate availability.
Folate is transported into cells by two distinct mechanisms (43). The first involves “high capacity/low affinity” system that is driven by an anion gradient. The enzyme is encoded by the reduced folate carrier (RFC) gene. The second system is a “low capacity/high affinity” system involving receptor-mediated phagocytosis of folate by folate receptors (FOLR). When folate level is low, FOLR are used. The effects of cholesterol mentioned above are specific only to FOLR. Interestingly, knockout mice for FOLR show neural tube defects (36), whereas knockout mutants for RFC are embryonic lethal and show deficiency in erythropoiesis upon maternal folate supplement (66). Moreover, none of the other knockout models involving genes in homocysteine and folate metabolism (9, 13, 28) show neural tube defects, suggesting that mechanisms specific to FOLR, perhaps involving cholesterol metabolism, provide clues to mechanisms of neural tube defect.
Studies of human genetics and mouse models have already established that defects in cholesterol metabolism lead to neural tube defects. Mouse knockouts of cholesterol biosynthesis and transport genes, squalene synthase (53) and apolipoprotein B (20), as well as mutations in 7-dehydrocholesterol reductase (DHCR7) in SLOS patients (51), all exhibit low cellular cholesterol levels and neural tube defects. Although a decrease in cholesterol lowers cellular folate uptake, cholesterol also directly affects other pathways associated with neural tube defects. A decrease in cellular cholesterol pharmacologically with cyclodextrin or genetically with Dhcr7 knockout mice impairs hedgehog signaling pathway (10). Mutations in hedgehog signaling in humans including sonic hedgehog (HPE3) (40), patched1 (HPE7) (31), and gli3 (HPE9) (41) lead to holoprosencephaly (HPE). Finally, a recent study showed that mutations in hedgehog signaling gene gli3 and Wnt signaling genes alter homocysteine metabolite and expression profiles (14), suggesting that while cholesterol metabolism can affect folate transport and hedgehog signaling in parallel, homocysteine and folate metabolism also act downstream of these signaling pathways.
Thus far, we have focused on dietary folate depletion and folate retention but not on changes in homocysteine level. Previous mouse studies showed that dietary folate depletion leads to an increase in serum homocysteine. In our study, however, dietary folate depletion mildly decreased serum homocysteine level. An explanation is that we did not use methionine loading, which has often been used in other studies (61). Excess methionine, after being used for methylation, is converted to homocysteine. In the absence of folate, homocysteine cannot be remethylated efficiently and continues to accumulate. Another explanation is that homocysteine was converted to glutathione, an antioxidant, through transsulfuration pathway rather than being remethylated. This reaction would decrease serum homocysteine level in exchange for glutathione production. Previous studies showed that dietary folate depletion induces oxidative stress and reduces serum homocysteine level (42), consistent with activation of transsulfuration pathway. Additionally, the duration of our study was also much shorter than other studies that performed dietary folate depletion to elevate serum homocysteine level (28). Therefore, our study is specific only to cellular stress induced with dietary folate depletion and not to cellular stress induced by elevated homocysteine level.
We explored two potential pathways that might mediate signaling between folate depletion and changes in cholesterol profiles. First, we hypothesized that choline, which has dual function in remethylation of homocysteine and lipoprotein secretion, is limited in supply during folate depletion. Choline supplementation elevated serum and liver cholesterol levels during folate depletion. Human studies also support our observation. Low folate intake decreases plasma choline level (23) and low choline intake decreases serum cholesterol level (63). However, choline did not improve folate retention. It is possible that we supplied insufficient choline to detect a beneficial effect on folate retention, as reflected by modest changes in cholesterol.
We also hypothesized that folate depletion induces ER stress, which in turn activates SREBP and cholesterol biosynthesis. However, we did not detect changes in expression of ER stress response genes. In addition, although elevated homocysteine level is often associated with ER stress in the liver, folate depletion was not accompanied elevated serum homocysteine level. The duration of folate depletion required to induce ER stress is weeks to months, whereas we observed changes in cholesterol profile within days. Although we only found support for choline-mediated changes in cholesterol profiles, ER stress response highlight that dietary folate depletion might affect cholesterol metabolism through other pathways.
We found that cholesterol metabolism contributes significantly to strain differences in response to folate depletion. We identified a homeostatic role of cholesterol metabolism in which enhancing serum and liver cholesterol levels improved folate retention. Strain differences in perturbation response may also translate to human populations. Therefore, cholesterol metabolism could affect beneficial outcome of folate supplement and affect variability in patient response to folate supplementation. Our study highlights the importance of pathway interactions in analyzing complex diseases and the interacting pathways identified in mouse studies could help in designing future human studies.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute Grant HL-58982 (J. H. Nadeau), HL-66580 (J. Quackenbush), HL-73712 (J. Quackenbush), and a gift from the Charles B. Wang Foundation (J. H. Nadeau).
Supplementary Material
Acknowledgments
Data deposition: GSE9245.
Address for reprint requests and other correspondence: J. H. Nadeau, Dept. of Genetics, Case Western Reserve Univ. School of Medicine, 10900 Euclid Ave., Cleveland, OH 44106-4955 (e-mail: jhn4@case.edu).
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Allain CC, Poon LS, Chan CS, Richmond W, Fu PC. Enzymatic determination of total serum cholesterol. Clin Chem 20: 470–475, 1974. [PubMed] [Google Scholar]
- 2.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbera JP, Rodriguez TA, Greene ND, Weninger WJ, Simeone A, Copp AJ, Beddington RS, Dunwoodie S. Folic acid prevents exencephaly in Cited2 deficient mice. Hum Mol Genet 11: 283–293, 2002. [DOI] [PubMed] [Google Scholar]
- 4.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet 39: 17–23, 2007. [DOI] [PubMed] [Google Scholar]
- 5.Carmel R, Jacobsen DW. Homocysteine in Health and Disease. Cambridge, UK: Cambridge University Press, 2001.
- 6.Carter M, Ulrich S, Oofuji Y, Williams DA, Ross ME. Crooked tail (Cd) models human folate-responsive neural tube defects. Hum Mol Genet 8: 2199–2204, 1999. [DOI] [PubMed] [Google Scholar]
- 7.Carter M, Chen X, Slowinska B, Minnerath S, Glickstein S, Shi L, Campagne F, Weinstein H, Ross ME. Crooked tail (Cd) model of human folate-responsive neural tube defects is mutated in Wnt coreceptor lipoprotein receptor-related protein 6. Proc Natl Acad Sci USA 102: 12843–12848, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang WJ, Rothberg KG, Kamen BA, Anderson RG. Lowering the cholesterol content of MA104 cells inhibits receptor-mediated transport of folate. J Cell Biol 118: 63–69, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z, Karaplis AC, Ackerman SL, Pogribny IP, Melnyk S, Lussier-Cacan S, Chen MF, Pai A, John SW, Smith RS, Bottiglieri T, Bagley P, Selhub J, Rudnicki MA, James SJ, Rozen R. Mice deficient in methylenetetrahydrofolate reductase exhibit hyperhomocysteinemia and decreased methylation capacity, with neuropathology and aortic lipid deposition. Hum Mol Genet 10: 433–443, 2001. [DOI] [PubMed] [Google Scholar]
- 10.Cooper MK, Wassif CA, Krakowiak PA, Taipale J, Gong R, Kelley RI, Porter FD, Beachy PA. A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat Genet 33: 508–513, 2003. [DOI] [PubMed] [Google Scholar]
- 11.Czeizel AE, Dudas I. Prevention of the first occurrence of neural-tube defects by periconceptional vitamin supplementation. N Engl J Med 327: 1832–1835, 1992. [DOI] [PubMed] [Google Scholar]
- 12.Da Costa KA, Gaffney CE, Fischer LM, Zeisel SH. Choline deficiency in mice and humans is associated with increased plasma homocysteine concentration after a methionine load. Am J Clin Nutr 81: 440–444, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dayal S, Devlin AM, McCaw RB, Liu ML, Arning E, Bottiglieri T, Shane B, Faraci FM, Lentz SR. Cerebral vascular dysfunction in methionine synthase-deficient mice. Circulation 112: 737–744, 2005. [DOI] [PubMed] [Google Scholar]
- 14.Ernest S, Christensen B, Gilfix BM, Mamer OA, Hosack A, Rodier M, Colmenares C, McGrath J, Bale A, Balling R, Sankoff D, Rosenblatt DS, Nadeau JH. Genetic and molecular control of folate-homocysteine metabolism in mutant mice. Mamm Genome 13: 259–267, 2002. [DOI] [PubMed] [Google Scholar]
- 15.Finkelstein JD Methionine metabolism in mammals. J Nutr Biochem 1: 228–237, 1990. [DOI] [PubMed] [Google Scholar]
- 16.Fleming A, Copp AJ. Embryonic folate metabolism and mouse neural tube defects. Science 280: 2107–2109, 1998. [DOI] [PubMed] [Google Scholar]
- 17.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226: 497–509, 1957. [PubMed] [Google Scholar]
- 18.Furie KL, Kelly PJ. Homocyst(e)ine and stroke. Semin Neurol 26: 24–32, 2006. [DOI] [PubMed] [Google Scholar]
- 19.Hegde P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Hughes JE, Snesrud E, Lee N, Quackenbush J. A concise guide to cDNA microarray analysis. Biotechniques 29: 548–550, 2000. [DOI] [PubMed] [Google Scholar]
- 20.Homanics GE, Maeda N, Traber MG, Kayden HJ, Dehart DB, Sulik KK. Exencephaly and hydrocephaly in mice with targeted modification of the apolipoprotein B (Apob) gene. Teratology 51: 1–10, 1995. [DOI] [PubMed] [Google Scholar]
- 21.Homocysteine Studies Collaboration. Homocysteine and risk of ischemic heart disease and stroke: a meta-analysis. JAMA 288: 2015–2022, 2002. [DOI] [PubMed] [Google Scholar]
- 22.Horne DW Microbiological assay of folates in 96-well microtiter plates. Methods Enzymology 281: 38–43, 1997. [DOI] [PubMed] [Google Scholar]
- 23.Jacob RA, Jenden DJ, Allman-Farinelli MA, Swendseid ME. Folate nutriture alters choline status of women and men fed low choline diets. J Nutr 129: 712–717, 1999. [DOI] [PubMed] [Google Scholar]
- 24.Jurka J Repbase update: a database and an electronic journal of repetitive elements. Trends Genet 16: 418–420, 2000. [DOI] [PubMed] [Google Scholar]
- 25.Kerr MK, Martin M, Churchill GA. Analysis of variance for gene expression microarray data. J Comput Biol 7: 819–837, 2000. [DOI] [PubMed] [Google Scholar]
- 26.Kisliuk RL Folate biochemistry in relation to antifolate selectivity. In: Antifolate Drugs in Cancer Therapy, edited by Jackman AL. Totowa: Humana, 1999, p. 13–36.
- 27.Klerk M, Verhoef P, Clarke R, Blom HJ, Kok FJ, Schouten EG. MTHFR 677C–>T polymorphism and risk of coronary heart disease: a meta-analysis. JAMA 288: 2023–2031, 2002. [DOI] [PubMed] [Google Scholar]
- 28.Lentz SR, Erger RA, Dayal S, Maeda N, Malinow MR, Heistad DD, Faraci FM. Folate dependence of hyperhomocysteinemia and vascular dysfunction in cystathionine β-synthase-deficient mice. Am J Physiol Heart Circ Physiol 279: H970–H975, 2000. [DOI] [PubMed] [Google Scholar]
- 29.Mayor S, Sabharanjak S, Maxfield FR. Cholesterol-dependent retention of GPI-anchored proteins in endosomes. EMBO J 17: 4626–4638, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCann SJ, Gillingwater S, Keevil BG, Cooper DP, Morris MR. Measurement of total homocysteine in plasma and blood spots using liquid chromatography-tandem mass spectrometry: comparison with the plasma Abbott IMx method. Ann Clin Biochem 40: 161–165, 2003. [DOI] [PubMed] [Google Scholar]
- 31.Ming JE, Kaupas ME, Roessler E, Brunner HG, Golabi M, Tekin M, Stratton RF, Sujansky E, Bale SJ, Muenke M. Mutations in PATCHED-1, the receptor for SONIC HEDGEHOG, are associated with holoprosencephaly. Hum Genet 110: 297–301, 2002. [DOI] [PubMed] [Google Scholar]
- 32.Moghadasian MH, McManus BM, Nguyen LB, Shefer S, Nadji M, Godin DV, Green TJ, Hill J, Yang Y, Scudamore CH, Frohlich JJ. Pathophysiology of apolipoprotein E deficiency in mice: relevance to apo E-related disorders in humans. FASEB J 15: 2623–2630, 2001. [DOI] [PubMed] [Google Scholar]
- 33.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273, 2003. [DOI] [PubMed] [Google Scholar]
- 34.MRC Vitamin Study Research Group. Prevention of neural tube defects: results of the Medical Research Council Vitamin Study MRC Vitamin Study Research Group. Lancet 338: 131–137, 1991. [PubMed] [Google Scholar]
- 35.Piedrahita JA, Zhang SH, Hagaman JR, Oliver PM, Maeda N. Generation of mice carrying a mutant apolipoprotein E gene inactivated by gene targeting in embryonic stem cells. Proc Natl Acad Sci USA 89: 4471–4475, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piedrahita JA, Oetama B, Bennett GD, van Waes J, Kamen BA, Richardson J, Lacey SW, Anderson RG, Finnell RH. Mice lacking the folic acid-binding protein Folbp1 are defective in early embryonic development. Nat Genet 23: 228–232, 1999. [DOI] [PubMed] [Google Scholar]
- 37.Pogribny I, Yi P, James SJ. A sensitive new method for rapid detection of abnormal methylation patterns in global DNA and within CpG islands. Biochem Biophys Res Commun 262: 624–628, 1999. [DOI] [PubMed] [Google Scholar]
- 38.Raghunathan K, Schmitz JC, Priest DG. Impact of schedule on leucovorin potentiation of fluorouracil antitumor activity in dietary folic acid deplete mice. Biochem Pharmacol 53: 1197–1202, 1997. [DOI] [PubMed] [Google Scholar]
- 39.Rimm EB, Willett WC, Hu FB, Sampson L, Colditz GA, Manson JE, Hennekens C, Stampfer MJ. Folate and vitamin B6 from diet and supplements in relation to risk of coronary heart disease among women. JAMA 279: 359–364, 1998. [DOI] [PubMed] [Google Scholar]
- 40.Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, Tsui LC, Muenke M. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet 14: 357–360, 1996. [DOI] [PubMed] [Google Scholar]
- 41.Roessler E, Du YZ, Mullor JL, Casas E, Allen WP, Gillessen-Kaesbach G, Roeder ER, Ming JE, Ruiz i Altaba A, Muenke M. Loss-of-function mutations in the human GLI2 gene are associated with pituitary anomalies and holoprosencephaly-like features. Proc Natl Acad Sci USA 100: 13424–13429, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogers EJ, Chen S, Chan A. Folate deficiency and plasma homocysteine during increased oxidative stress. N Engl J Med 357: 421–422, 2007. [DOI] [PubMed] [Google Scholar]
- 43.Rosenblatt DS, Fenton WA. Inherited disorders of folate and cobalamin transport and metabolism. In: The Metabolic and Molecular Bases of Inherited Disease, edited by Scriver CR, Beaudet AL, Sly WS, Valle D. New York: McGraw-Hill, 2001, p. 3897–3933.
- 44.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378, 2003. [DOI] [PubMed] [Google Scholar]
- 45.Seshadri S, Beiser A, Selhub J, Jacques PF, Rosenberg IH, D'Agostino RB, Wilson PW, Wolf PA. Plasma homocysteine as a risk factor for dementia and Alzheimer's disease. N Engl J Med 346: 476–483, 2002. [DOI] [PubMed] [Google Scholar]
- 46.Singer JB, Hill AE, Burrage LC, Olszens KR, Song J, Justice M, O'Brien WE, Conti DV, Witte JS, Lander ES, Nadeau JH. Genetic dissection of complex traits with chromosome substitution strains of mice. Science 304: 445–448, 2004. [DOI] [PubMed] [Google Scholar]
- 47.Smart EJ, Mineo C, Anderson RG. Clustered folate receptors deliver 5-methyltetrahydrofolate to cytoplasm of MA104 cells. J Cell Biol 134: 1169–1177, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, Doi H, Wood WH 3rd, Becker KG, Ko MS. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci USA 97: 9127–9132, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tint GS, Irons M, Elias ER, Batta AK, Frieden R, Chen TS, Salen G. Defective cholesterol biosynthesis associated with the Smith-Lemli-Opitz syndrome. N Engl J Med 330: 107–113, 1994. [DOI] [PubMed] [Google Scholar]
- 52.Tsai J, Sultana R, Lee Y, Pertea G, Karamycheva S, Antonescu V, Cho J, Parvizi B, Cheung F, Quackenbush J. RESOURCERER: a database for annotating and linking microarray resources within and across species. Genome Biol 2: SOFTWARE0002, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tozawa R, Ishibashi S, Osuga J, Yagyu H, Oka T, Chen Z, Ohashi K, Perrey S, Shionoiri F, Yahagi N, Harada K, Gotoda T, Yazaki Y, Yamada N. Embryonic lethality and defective neural tube closure in mice lacking squalene synthase. J Biol Chem 274: 30843–30848, 1999. [DOI] [PubMed] [Google Scholar]
- 54.Tulenko TN, Boeze-Battaglia K, Mason RP, Tint GS, Steiner RD, Connor WE, Labelle EF. A membrane defect in the pathogenesis of the Smith-Lemli-Opitz syndrome. J Lipid Res 47: 134–143, 2006. [DOI] [PubMed] [Google Scholar]
- 55.Ulrich CM, Curtin K, Potter JD, Bigler J, Caan B, Slattery ML. Polymorphisms in the reduced folate carrier, thymidylate synthase, or methionine synthase and risk of colon cancer. Cancer Epidemiol Biomarkers Prev 14: 2509–2516, 2005. [DOI] [PubMed] [Google Scholar]
- 56.Van der Linden IJ, den Heijer M, Afman LA, Gellekink H, Vermeulen SH, Kluijtmans LA, Blom HJ. The methionine synthase reductase 66A>G polymorphism is a maternal risk factor for spina bifida. J Mol Med 84: 1047–1054, 2006. [DOI] [PubMed] [Google Scholar]
- 57.Verkleij-Hagoort A, Bliek J, Sayed-Tabatabaei F, Ursem N, Steegers E, Steegers-Theunissen R. Hyperhomocysteinemia and MTHFR polymorphisms in association with orofacial clefts and congenital heart defects: a meta-analysis. Am J Med Genet A 143: 952–960, 2007. [DOI] [PubMed] [Google Scholar]
- 58.Wang X, Qin X, Demirtas H, Li J, Mao G, Huo Y, Sun N, Liu L, Xu X. Efficacy of folic acid supplementation in stroke prevention: a meta-analysis. Lancet 369: 1876–1882, 2007. [DOI] [PubMed] [Google Scholar]
- 59.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23: 5293–5300, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waterland RA, Lin JR, Smith CA, Jirtle RL. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum Mol Genet 15: 705–716, 2006. [DOI] [PubMed] [Google Scholar]
- 61.Werstuck GH, Lentz SR, Dayal S, Hossain GS, Sood SK, Shi YY, Zhou J, Maeda N, Krisans SK, Malinow MR, Austin RC. Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. J Clin Invest 107: 1263–1273, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang IV, Chen E, Hasseman JP, Liang W, Frank BC, Wang S, Sharov V, Saeed AI, White J, Li J, Lee NH, Yeatman TJ, Quackenbush J. Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol 3: research0062, 2002. [DOI] [PMC free article] [PubMed]
- 63.Zeisel SH, Da Costa KA, Franklin PD, Alexander EA, Lamont JT, Sheard NF, Beiser A. Choline, an essential nutrient for humans. FASEB J 5: 2093–2098, 1991. [PubMed] [Google Scholar]
- 64.Zeisel SH, Blusztajn JK. Choline and human nutrition. Annu Rev Nutr 14: 269–296, 1994. [DOI] [PubMed] [Google Scholar]
- 65.Zhao Q, Behringer RR, de Crombrugghe B. Prenatal folic acid treatment suppresses acrania and meroanencephaly in mice mutant for the Cart1 homeobox gene. Nat Genet 13: 275–283, 1996. [DOI] [PubMed] [Google Scholar]
- 66.Zhao R, Russell RG, Wang Y, Liu L, Gao F, Kneitz B, Edelmann W, Goldman ID. Rescue of embryonic lethality in reduced folate carrier-deficient mice by maternal folic acid supplementation reveals early neonatal failure of hematopoietic organs. J Biol Chem 276: 10224–10228, 2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.