Abstract
The Caenorhabditis elegans oocyte is a highly amenable system for forward and reverse genetic analysis of receptor-mediated endocytosis. We describe the use of transgenic strains expressing a vitellogenin::green fluorescent protein (YP170::GFP) fusion to monitor yolk endocytosis by the C. elegans oocyte in vivo. This YP170::GFP reporter was used to assay the functions of C. elegans predicted proteins homologous to vertebrate endocytosis factors using RNA-mediated interference. We show that the basic components and pathways of endocytic trafficking are conserved between C. elegans and vertebrates, and that this system can be used to test the endocytic functions of any new gene. We also used the YP170::GFP assay to identify rme (receptor-mediated endocytosis) mutants. We describe a new member of the low-density lipoprotein receptor superfamily, RME-2, identified in our screens for endocytosis defective mutants. We show that RME-2 is the C. elegans yolk receptor.
INTRODUCTION
Receptor-mediated endocytosis, an essential process in all eukaryotes, is required for general cellular functions, including uptake of nutrients (e.g., low-density lipoprotein [LDL] or transferrin) and recycling of membranes and membrane proteins (Mukherjee et al., 1997). Yolk uptake by growing oocytes is a dramatic example of the receptor-mediated endocytosis pathway in many species, including invertebrates such as the nematode Caenorhabditis elegans and vertebrates such as the chicken (Schneider, 1996). C. elegans yolk is secreted from its site of synthesis, the intestine, into the pseudoceolomic space (body cavity) and is ultimately taken up into vesicles within the growing oocytes (Kimble and Sharrock, 1983; Hall et al., 1999). Yolk transport in vertebrates such as the chicken follows a similar route, from liver to bloodstream to ovum (Schneider, 1996).
Yolk is a lipoprotein particle composed of lipids and lipid-binding proteins called vitellogenins. Vitellogenins are among the most abundant proteins found in developing embryos (Sharrock, 1983). Lipids and proteins derived from yolk are thought to provide essential nutrients required to support the rapid development of the embryo. C. elegans vitellogenins YP170, YP115, and YP88 share sequence homology with vertebrate vitellogenins and with ApoB-100, a core component of mammalian LDL particles (Baker, 1988; Spieth et al., 1991). Endocytosis of yolk particles into membrane-bound vesicles of the oocyte is mediated by receptors of the LDL receptor superfamily in C. elegans (this work), insects, and vertebrates. Yolk and yolk receptor endocytic trafficking is thought to proceed through pathways very similar to those used by LDL in somatic cells (Goldstein et al., 1985; Schneider, 1996).
Clathrin-coated pit (CCP) and endosomal trafficking systems, such as those mediating LDL endocytosis, have been intensively studied since Roth and Porter (1964) first discovered coated-pits in the mosquito oocyte plasma membrane. Extensive ligand and receptor tracer studies have since produced a model describing general endocytic trafficking routes in the cell (Mukherjee et al., 1997). Ligand–receptor complexes are thought to cluster in CCPs, which pinch off from the surface as clathrin-coated vesicles, and shed their coats. Uncoated vesicles then fuse with each other and with early endosomes. Acidificaton of the early endosome leads to dissociation of ligands from receptors. After sorting, receptors recycle to the cell surface, whereas ligands are transported to late endosomes and ultimately lysosomes. Yolk storage vesicles are thought to be the oocyte equivalent of late endosomes or lysosomes but with low proteolytic activity (Schneider, 1996).
Although great strides have been made in our understanding of endocytic trafficking, many questions remain about the mechanisms underlying this process. Secretion, which now stands as the best understood trafficking pathway, has been largely elucidated by a combination of mammalian biochemistry and yeast genetics (Rothman and Wieland, 1996; Schekman and Orci, 1996). Endocytosis has been most widely studied by biochemical methods, in large part using physical association or copurification techniques (Mellman, 1996). Some progress has been made using in vitro functional assays, such as early endosome fusion, but many parts of the endocytic pathway, such as receptor recycling, have not been reconstituted (Mukherjee et al., 1997). Genetic analysis in a number of systems, most notably yeast, has also made important contributions to our understanding of endocytosis. For example, genetic analysis in yeast has identified new connections between the actin cytoskeleton and the endocytic pathway (Wendland et al., 1998).
Genetic analysis in metazoan animals such as C. elegans, Drosophila, and mice is one of the least explored, but potentially most fruitful, areas of endocytosis research. Because endocytosis is a mechanism by which cells interact with their environments, important aspects of endocytosis may differ between unicellular and multicellular organisms. For example, lipoprotein uptake, such as LDL or yolk endocytosis, is an adaptation required for nutrient transport and cellular homeostasis within a multicellular organism. Other highly adapted endocytic systems specific to multicellular organisms include growth factor receptor regulation during development, synaptic vesicle recycling in the nervous system, and antigen processing in the immune system (Mellman, 1996).
Genetic analysis in C. elegans and Drosophila has already provided important insights into trafficking mechanisms, even though few studies have specifically targeted endocytosis. For instance, mutants in the Drosophila dynamin homologue shibire were originally identified because of their generally impaired nervous system. Further analysis of these mutants revealed the importance of dynamin in pinching off clathrin-coated vesicles (de Camilli et al., 1995). Similarly, while studying general synaptic function genetically in C. elegans, a role for synaptotagmin was identified in synaptic vesicle recycling (Nonet et al., 1993; Jorgensen et al., 1995).
Here, we describe the use of a YP170::green fluorescent protein (GFP) (Chalfie et al., 1994) reporter to visualize yolk endocytosis in vivo in the C. elegans oocyte. We used this YP170::GFP assay to probe the functions in yolk endocytosis of predicted C. elegans homologues of well-known vertebrate endocytosis genes using RNA-mediated interference (RNAi). This approach showed that the basic components and pathways of endocytic trafficking are well conserved between C. elegans and vertebrates, and that this system can be used to test the endocytic functions of any new gene. We also used the YP170::GFP assay in a classical mutagenesis scheme to identify new rme (receptor-mediated endocytosis) genes encoding endocytosis components. In this paper we describe RME-2, a member of the LDL receptor superfamily, and provide several lines of evidence showing that RME-2 is the C. elegans yolk receptor. The completely described cell lineage, simple anatomy, and advanced knowledge of developmental processes in C. elegans offer the potential to integrate future studies of endocytosis and intracellular trafficking with investigations of related processes such as cell polarity, cell–cell signaling, and excitable cell function.
MATERIALS AND METHODS
General Methods and Strains
Methods for the handling and culturing of C. elegans were essentially those described by Brenner (1974). All strains were grown at 20°C unless otherwise stated. The wild-type parent for all strains was C. elegans var. Bristol strain N2 except for those experiments involving DP13 C. elegans var. Bergerac strain BO or NL917 mut-7(pk204), a Bristol/Bergerac hybrid strain (Nigon, 1949; Williams, 1995). Mutations used were LGII, sqt-1(sc103) (Kramer and Johnson, 1993); LGIII, mut-7(pk204) (van Luenen and Plasterk, 1997) and unc-16(e109) (Brenner, 1974); LGIV, dpy-13(e184) (Brenner, 1974), unc-5(e53) (Brenner, 1974), rme-2(b1005), rme-2(b1008), and rme-2(b1026) (this work); LGX, dyn-1(ky51) (Clark et al., 1997), lin-15(n765ts) (Ferguson and Horvitz, 1985), and bIs1[vit-2::GFP, rol-6(su1006)] (this work); and unmapped, bIs2[vit-2::GFP, rol-6(su1006)] (this work).
Plasmids and Transgenic C. elegans
The plasmid V2B3 encodes a functional VIT-2(YP170B)::GFP fusion protein expressed under vit-2 promoter control. The plasmid was made by ligating a PCR product encoding vit-2 genomic sequences, including 1 kb of promoter and the complete gene lacking a stop codon, into the GFP plasmid pPD95.85 (Fire, Xu, Ahnn, and Seydoux, personal communication), cut with XmaI and KpnI. The vit-2 gene was PCR amplified with primers V2F (TCCCCCCGGGTCCACGGACATTTCTGGGTCATTTG) and V2R (CGGGGTACCAGATAAGCGACGCAGGCGGTTGGGAC), containing engineered XmaI and KpnI sites (underlined), using cosmid DNA (C42D8) as a template. V2B3, at 50 μg/ml, was coinjected with rol-6(d) marker pRF4, at 100 μg/ml, into N2 to make transformed lines using standard methods (Fire, 1986; Mello et al., 1991). Ten of 10 roller lines showed green YP170::GFP fluorescence in embryos, the intestine, and nearly full-grown oocytes of the adult hermaphrodite. Oocyte fluorescence was generally restricted to the one to four late-stage oocytes in each gonad arm, depending on the age of the hermaphrodite. Pseudocoelomic YP170::GFP fluorescence was observed primarily in very young adults just beginning oocyte production or very old hermaphrodites no longer producing fertilized eggs. No expression of YP170::GFP was observed in males. Four integrated lines (bIs1–bIs4) were produced by standard methods and back-crossed twice to N2 (Grant and Greenwald, 1997). YP170::GFP fluorescence in the integrated arrays appeared identical to that of the extrachromosomal arrays. bIs1 was found to map near dyn-1 on LGX. bIs2 proved unlinked to dyn-1 but was not mapped further.
Mutant Isolation and Gene Mapping
Two mutant screens resulted in the isolation of rme-2 alleles. In the first, 10 gravid hermaphrodites of the mutator strain DH1069 mut-7(pk204); bIs1[vit-2::GFP, rol-6(su1006)] were transferred to individual 9-cm Petri dishes and allowed to self-fertilize for 3–4 d, until ∼100–200 gravid F1 progeny were produced. F2 eggs were collected by alkaline hypochlorite digestion and were plated in groups of 1000–2000 onto 15-cm Petri dishes. When the F2 progeny reached adulthood, they were examined with a Leica (Nussloch, Germany) 8Z stereomicroscope equipped with epifluorescence for rare animals displaying little or no YP170::GFP fluorescence in the oocytes and embryos but showing bright YP170::GFP fluorescence in the pseudocoelom. Such animals were considered rme mutants. True breeding rme mutants with morphologically normal gonads were back-crossed first to unc-16(e109) to remove mut-7(pk204) from the strain. Then an unmutagenized bIs1 chromosome was crossed back into the strain. Only one mutant from each plate was kept for further analysis. rme-2(b1005) and rme-2(b1008) were recovered in this screen. The second screen was similar, except that strain DH1006 bIs1[vit2::GFP, rol-6(su1006)] was mutagenized with freshly prepared 2.0 mM N-ethyl-N-nitrosourea as described (De Stasio et al., 1997), and the F1 and F2 generations were raised at 15°C to allow isolation of temperature-sensitive mutants. F2 animals were shifted to 25°C for 12–24 h before screening. rme-2(b1026) was recovered in this screen.
Mutations were assigned to linkage groups and to chromosomal subregions by sequence tagged site (STS) polymorphism mapping (Williams, 1995). b1005, b1008, and b1026 mapped between stP13 and stP44 on LGIV. These three mutations failed to complement one another. Three-point mapping with dpy-13 and unc-5 identified the following crossovers: dpy-13 (64) rme-2 (5) and unc-5.
Cloning and Molecular Characterization
Genomic rme-2 sequences were PCR amplified from rme-2 mutant lysates using primers T11F8-OF (CATTTCTGTCCCGGCGGGAG) and T11F8-OR (ATCGTGCCAAGACCTAGCGCC). The products of four independent PCR reactions were pooled and submitted for automated sequencing using primers T11F8-OF, rme-2F1 (AAGCAAAGGAATTTGATTGCGG), rme-2F2 (TTCTGGAGGAGATGATGAGGTC), rme-2F3 (GTAATGGGATCAAGGAGTGTCC), rme-2F4 (ATCGATCGACTTCATGCATCGC), rme-2F5 (GGCTAATATGGATGGGTCTCAG), rme-2F6 (ACCCATGCCTTGAACTCGAGTG), rme-2F7 (GGTAGTCGGAATTATTGCCTTCC), and rme2-F8 (TCATACGGAAACCCCATGTACG). Sequence changes were confirmed by resequencing the region of interest with a new set of independent PCR products. rme-2(b1005) contained a 26-bp deletion altering or deleting codons 630–638 (ATGACCCATp-CATCATTCGAGTCCTCAA→A), resulting in a predicted frame shift and truncation. rme-2(b1008) contained adjacent missense and single-base insertion mutations within codon 440 (ATT→TGTT), resulting in a predicted frame shift and truncation. rme-2(b1026) contained a transition in codon 493 (GGG→GAG) resulting in a nonconservative G→E amino acid substitution.
cDNA clones for rme-2 were kindly provided by Dr. Yuji Kohara (National Institute of Genetics, Mishima, Japan). The longest, yk8d2, was sequenced on one strand using primers T3, rme-2F2, rme-2F3, rme-2F4, rme-2F5, rme-2F6, rme-2F7, and T7. yk8d2, yk286d10, and yk238g12 each contained a poly(A) tail at the 3′ end, 84 bp after the stop codon.
We determined the 5′ end of the rme-2 message by 5′ rapid amplification of cDNA ends (RACE) according to the manufacturer’s instructions (Life Technologies, Gaithersburg, MD). One microgram of total RNA was reverse transcribed using primer VLDL-RT (CATGAGTTGCACACTCATC) and was poly(G) tailed. First-round PCR was performed using primers VLDL-R1 (CATCGGCAAGCTTATATCCTTC) and the abridged anchor primer. First-round PCR gave only one band on an agarose gel. Second-round PCR was performed using primers rme2-5P (AGTCCGCTACGTTGTCGCATTG) and UAP with a fraction of the first PCR product. Second round PCR gave only one band on an agarose gel. The purified PCR product was cloned into pGEM-T (Promega, Madison, WI). Five of the largest inserts were sequenced. None contained spliced leader sequences. The longest clone extended 35 bp 5′ of the probable start codon. A similar PCR regimen using VLDL-R1 and either SL1 or SL2 primers produced only non–rme-2-related products.
Antibody Production, Western Blots, and Immunostaining
Anti-vitellogenin antibodies described by Sharrock et al. (1990) were kindly provided by T. Blumenthal (University of Colorado) and S. Strome (University of Indiana). Rat polyclonal antisera anti-YP170, anti-YP115, and anti-YP88 and mouse monoclonal antibodies OIC1 and PIIA3 were used in immunofluoresence experiments on dissected gonads as described below. Each antiserum clearly detected abundant yolk granules in the oocytes of wild-type hermaphrodites but failed to detect any yolk granules in the oocytes of rme-2(b1005) hermaphrodites. These antibodies detected abundant yolk in rme-2 mutant intestines.
We expressed 6-HIS-tagged fragments of RME-2 in Escherichia coli, which were purified by nickel chromatography under denaturing conditions, and injected into rabbits for antibody production as described (Grant and Greenwald, 1997). RME-2-EXT antigen (amino acids 180–670) was expressed from a PCR-amplified region of yk8d2 cloned as a BamHI–XhoI fragment into pET24b (Novagen, Madison, WI). RME-2-INT antigen (amino acids 813–925) was expressed from a PCR-amplified region of yk8d2 cloned as a BamHI–XhoI fragment into pET24b (Novagen). Rabbits R-6739, R-6740, R-6741, and R-6742 were immunized and bled by Charles River PharmServices (Southbridge, MA). Antibodies were affinity purified on RME-2-EXT or RME-2-INT antigen columns as described (Gu et al., 1994). Western blotting was performed on mixed stage populations as described (Grant and Greenwald, 1997), except that affinity-purified anti-RME-2-INT or anti-RME-2-EXT antisera were used at a 1:500 dilution. Whole-mount immunostaining was performed as described (Bettinger et al., 1996). Anti-RME-2 antisera was used at a 1:50 dilution overnight at 4°C. Secondary antibodies conjugated to Alexa-488 (Molecular Probes, Eugene, OR), Alexa-546 (Molecular Probes), or Cy3 (Jackson ImmunoResearch, West Grove, PA) were used at a 1:500 dilution. Gonads were dissected as described (Jones et al., 1996), transferred to siliconized microcentrifuge tubes, and fixed for 10 min in 1% paraformaldehyde and 1× PBS. Primary and secondary incubations were as described above for whole-mount samples. Most immunostained specimens were analyzed with a Zeiss (Thornwood, NY) LSM laser scanning confocal microscope.
Ectopic Expression of RME-2
PCR was used to create plasmid 86DF, a full-length rme-2 cDNA, including N-terminal coding sequences and a novel NheI site, cloned into the NheI and KpnI sites of the myo-3 expression vector pPD95.86 (Fire, Xu, Ahnn, and Seydoux, personal communication). The complete cDNA was sequenced to ensure that no errors had been introduced. 86DF was injected at 20 μg/ml along with a lin-15(+) plasmid at 50 μg/ml into the strain DH1112 lin-15(n765ts); bIs2[vit-2::GFP, rol-6(d)]. Three non-Muv lines were grown at 25°C and were fixed and immunostained as described above with rabbit polyclonal anti-RME-2-INT antisera and mouse monoclonal anti-GFP antibody 3E6 (Quantum Biotechnologies, Montreal, Canada). Three of three lines, analyzed by confocal microscopy, showed highly mosaic expression of RME-2 in body wall and vulval muscle cells. In most cases, cells expressing high levels of RME-2 showed coincident staining for YP170::GFP. Because of the small size and presumably active lysosomal degradation pathway of these cells, we were unable to determine whether any of the YP170::GFP associated with these muscle cells was internalized. Muscle cells expressing little or no RME-2, often directly adjacent to strongly positive cells, never stained with anti-GFP antibodies. No anti-GFP immunoreactivity was observed in larvae, although many muscle cells of larvae expressed abundant RME-2 in these lines.
RNA-mediated Interference
The following cDNA clones, kindly provided by Dr. Yuji Kohara or The Institute for Genomic Research (Rockville, MD), were used to produce double-stranded RNA (dsRNA) for microinjection into the pseudocoelom or intestine as described by Fire et al. (1998) and Montgomery et al. (1998): yk24g12 (clathrin heavy chain, T20G5.1), yk132a1 (α-adaptin, T20B5.1), yk115c10 (β-adaptin, Y71H2), CEESS15 (rab5, F26H9.6), yk101c7 (rab7, W03C9.3), yk51h1 (rab11, F53G12.1), yk52c4 (β′-COP, F38E11.5), yk67d8 (ζ-COP, F59E10.3), and yk112c6 (ARF, B0336.2). Plasmids were rescued from the phage clones according to manufacturer’s instructions (Stratagene, La Jolla, CA), the cDNA inserts were PCR amplified with Bluescript vector primers CM024 (TTGTAAAACGACGGCCAG) and CM025 (CATGATTACGCCAAGCTC), transcribed using large-scale T3 and T7 transcription kits (Novagen or Stratagene), and purified by phenol-chloroform extraction or RNA-Quick purification columns (Qiagen, Hilden, Germany). Groups of 15–20 L4 and young adult hermaphrodites of the strain DH1033 sqt-1(sc103); bIs1[vit-2::GFP, rol-6(d)] were microinjected with the appropriate dsRNA and allowed to recover 18–24 h before direct observation of oocytes and embryos within the P0 by fluorescence microscopy. The pseudo–wild-type sqt-1(0) mutation sc103 suppresses the rol-6(d) twisted body phenotype, allowing easier observation and photography of this strain. Progeny from the transitional period (first 18–24 h), and fully transformed period (>24 h) were scored in single-day cohorts. For anti-RME-2 immunostaining, groups of 100 P0s were injected, recovered for 24 h, and immunostained as above.
RESULTS
The YP170::GFP Assay
We created transgenic C. elegans strains that express the full-length yolk protein YP170 fused to GFP (see MATERIALS AND METHODS). This fluorescent YP170::GFP fusion protein is transported like endogenous yolk, from intestine to oocyte, allowing in vivo analysis of secretion and endocytosis by fluorescence microscopy (Figure 1). In the first set of experiments we asked whether yolk endocytosis in the C. elegans oocyte uses the same endocytic pathway components that have been described in studies of general endocytosis in vertebrates. To accomplish this we applied RNAi to our YP170::GFP-expressing strain to reduce or eliminate expression of a series of specific target genes within oocytes (Fire et al., 1998). We then asked whether YP170::GFP uptake by these oocytes was impaired or altered.
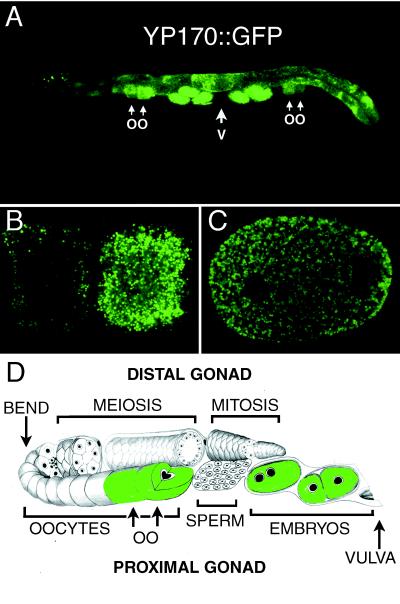
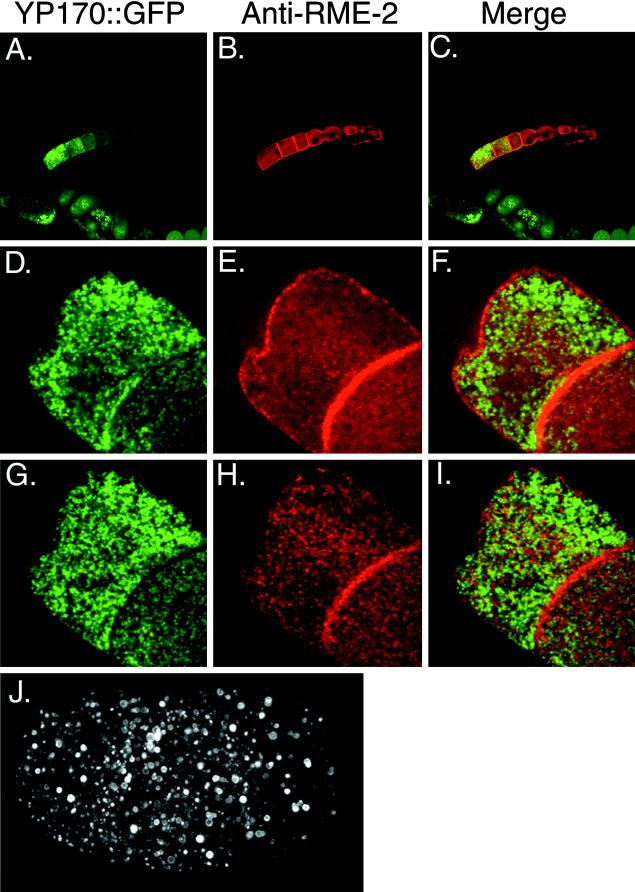
Figure 1.
YP170:: GFP accumulates in vesicles of the oocyte. (A) Whole adult hermaphrodite expressing YP170::GFP. YP170::GFP fluorescence is found in the intestine, late-stage oocytes, and embryos. YP170::GFP localization appeared very similar to endogenous YP170 by fluorescence microscopy (this work) and immuno-electron microscopy (Hall et al., 1999). Immuno-electron microscopic experiments did reveal accumulation of YP170::GFP in enlarged vesicles of the intestine, unlike endogenous YP170 in wild-type worms (Hall et al., 1999). Such accumulation may result from overexpression of YP170 in transgenic strains or properties acquired from the GFP tag. OO, late-stage oocytes; V, vulva. (B) Confocal micrograph of YP170::GFP vesicles within two nearly full-grown oocytes of one gonad arm. (C) Confocal micrograph of YP170::GFP vesicles within a two-cell embryo. (D) Stylized drawing of one gonad arm connected to the spermatheca and the uterus.
RNAi was performed by microinjecting dsRNA homologous to the target gene of interest into L4 or young adult hermaphrodites and later scoring the injected animals and their progeny for induced phenotypes. This technique has been used extensively in C. elegans and faithfully mimics most phenotypes caused by mutations that reduce or eliminate target gene function and is especially effective in germ cells (Fire et al., 1998; Montgomery et al., 1998).
Clathrin-coated Pit Components Are Required for Yolk Endocytosis
Both receptor-bound ligands and extracellular fluid enter animal cells through CCPs (Mellman, 1996). The clathrin cage, composed of clathrin heavy chain and clathrin light chain molecules, forms repeated triskelions that self-assemble into planar lattices associated with clathrin adaptor complexes called AP2, which are associated with clustered transmembrane receptors. These planar lattices are thought to round up to form pits, which pinch off from the membrane to form coated vesicles. Another pit-associated protein, dynamin, is required for pinching off clathrin-coated vesicles from the plasma membrane.
The adaptor complex AP2 is a heterotetramer of four “adaptins” (Pearse and Robinson, 1990). Each AP2 complex contains two large subunits, α- and β-adaptin, a medium chain μ2 (AP50), and a small chain ς2 (AP17). The μ2 subunit of AP2 is thought to mediate association with the tyrosine-based internalization signals of receptor tails (Ohno et al., 1995). The α-adaptin subunit is thought to mediate essential interactions, directly or indirectly, with a host of associated endocytosis factors, including dynamin (Wang et al., 1995). The β-adaptin subunit is thought to connect the AP2 complex to clathrin (Pearse and Robinson, 1990). No specific function has been assigned to the ς2 subunit of AP2.
By scanning the essentially complete C. elegans genome sequence, we were able to identify predicted C. elegans proteins representing probable CCP components: clathrin heavy chain (T20G5.1), α-adaptin (T20B5.1), β-adaptin (Y71H2_389.E), μ2 (R160.1), and ς2 (F02E8.3). C. elegans clathrin heavy chain, α-adaptin, and β-adaptin RNAi all produced similar defects in oocytes and embryos. We found that RNAi of these CCP components strongly inhibited uptake of YP170::GFP into oocytes, resulting in high accumulation of YP170::GFP in the pseudocoelomic space and many fewer YP170::GFP-containing yolk granules in late-stage oocytes and embryos. In all of these cases, late-stage oocytes were smaller than in wild type, and embryos proved inviable. These results are consistent with the proposed roles of clathrin, α-adaptin, and β-adaptin in coated pit and coated vesicle formation.
Unlike the other CCP components we tested, Ce-μ2(RNAi) and Ce- ς2(RNAi) did not result in an apparent reduction in YP170::GFP uptake into the oocyte. YP170::GFP appeared at wild-type levels in oocytes and embryos and did not accumulate in the pseudocoelomic space. All progeny of Ce-μ2(RNAi) and Ce-ς2(RNAi) mothers were severely dumpy (Dpy). The Dpy phenotype is characterized by a very short body and is most commonly associated with defects in cuticle formation (Levy et al., 1993). Reduced body length is probably caused by a partial failure in embryonic elongation, a major event in embryonic morphogenesis. The phenotype was very similar when Ce-μ2 and Ce-ς2 RNAi was performed simultaneously. No synthetic phenotypes of Ce-μ2 and Ce-ς2 were revealed. We also performed RNAi on Ce-μ2 and unc-101, a C. elegans μ1(AP47) gene, simultaneously (Lee et al., 1994). No reduction in YP170::GFP uptake by Ce-μ2(RNAi); unc-101(RNAi) oocytes was observed. We did observe strong Unc and strong Dpy phenotypes in all progeny of these animals, consistent with a simple additive effect of the two dsRNAs. Taken together these results are surprising, perhaps indicating the μ2 and ς2 subunits are not required for yolk endocytosis.
Finally we examined the requirement for dyn-1, the C. elegans dynamin gene, in YP170::GFP endocytosis by oocytes. We used RNAi and a temperature-sensitive mutation, dyn-1(ky51ts), to reduce or eliminate dynamin function (Clark et al., 1997). dyn-1(RNAi) animals and dyn-1(ky51ts) mutants, at the nonpermissive temperature of 25°C, showed high-level accumulation of YP170::GFP in the pseudocoelomic space and strongly reduced accumulation of YP170::GFP within oocytes and embryos. dyn-1(RNAi) animals produced only dead embryos, whereas dyn-1(ky51ts) mutants produced a reduced number of embryos (average = 56; n = 9) at 25°C, 52% of which were inviable. These results are consistent with the proposed role of dynamin in coated vesicle formation.
Endocytic Sorting Pathways Are Required for Normal Yolk Endocytosis
Once molecules have been internalized they enter the endocytic sorting system. Different rab proteins, small GTPases of the ras superfamily, are thought to mediate each unique fusion step in vesicular transport (Mellman, 1996). Genetic studies indicate that rab protein function is often required for vesicle formation as well (Pfeffer, 1994). The rab4 and rab5 proteins are found to associate with early endosomes in mammalian cells. rab4 is important for receptor recycling to the cell surface, whereas rab5 is thought to be required for ligand clustering into coated pits and early endosome fusion (Bucci et al., 1992; van der Sluijs et al., 1992). The rab11 protein associates with early endosomes and the pericentriolar recycling compartment and is thought to be important for transport from early endosomes to pericentriolar recycling endosomes, an important step in recycling for at least some receptors (Ullrich et al., 1996). rab7 protein is associated primarily with late endosomes and is thought to be required for transport of cargo from early to late endosomes (Feng et al., 1995; Press et al., 1998).
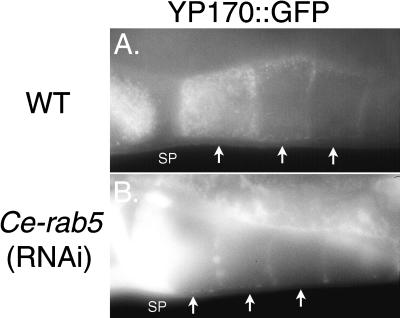
We identified three predicted regulators of endosome function, Ce-rab5 (F26H9.6), Ce-rab7 (W03C9.3), and Ce-rab11(F53G12.1), in the essentially complete genome sequence of C. elegans but failed to identify a homologue of rab4. We reduced or eliminated the expression of these predicted endocytosis pathway components by RNAi and examined the resulting defects in the oocytes in a YP170::GFP-expressing strain (see Table 1). Each of these rabs would be expected to regulate a separate step in endocytic trafficking. Indeed, reducing or eliminating expression of each of these rabs produced profound but distinct phenotypes in the C. elegans oocyte. Blocking expression of Ce-rab5 gave results similar to those of CCP component RNAi, except that the phenotype was stronger; i.e. YP170::GFP uptake by oocytes was completely abolished (Figure 2). All progeny embryos of Ce-rab5(RNAi) hermaphrodites died.
Table 1.
Assay of C. elegans endocytosis genes in yolk uptake into the oocyte
| Gene targeted for RNAi | Predicted protein homology | % Identity with mammalian homologue | YP170∷GFP in pseudocoelom | YP170∷GFP in full-grown oocytes | Phenotype of progeny |
|---|---|---|---|---|---|
| None | NA | NA | − | +++ | Normal |
| T20G5.1 | CHC (Clathrin Heavy Chain) | 70 | +++ | + | Dead embryos |
| T20B5.1 | α-Adaptin | 57 | +++ | + | Dead embryos |
| Y71H2_389.E | β-Adaptin | 53 | +++ | + | Dead embryos |
| R160.1 | μ2(AP50) | 78 | − | +++ | Strong Dpy |
| F02E8.3 | ς2(AP17) | 93 | − | +++ | Strong Dpy |
| C02C6.1 (dyn-1)a | Dynamin | 66 | +++ | + | Dead embryos |
| F26H9.6 | Rab5 | 77 | +++ | − | Dead embryos |
| F53G12.1 | Rab11 | 80 | + | ++ | Dead embryos |
| +++ | Mild Dpy | ||||
| W03C9.3 | Rab7 | 71 | − | Abnormal localization | Large yolk granules throughout L1 body |
Similar results were found for the dyn-1(ky51ts) mutant (see RESULTS).
Figure 2.
Ce-rab5(RNAi) blocks endocytosis of YP170::GFP. (A) Fluorescence micrograph of late-stage wild-type oocytes (arrows). The most full-grown oocyte (far left) is heavily labeled with YP170::GFP vesicles, whereas the two earlier-stage oocytes show less labeling. SP, spermatheca. (B) Fluorescence micrograph of full-grown or nearly full-grown Ce-rab5(RNAi) oocytes (arrows). All YP170::GFP fluorescence is found between oocytes, in the spermatheca, pseudocoelom, or intestine. Extremely high levels of extracellular YP170::GFP are seen in the spermatheca (SP). No uptake of YP170::GFP was observed.
Ce-rab11 RNAi had a milder effect on endocytosis by the oocyte. Although YP170::GFP accumulated in the pseudo-coelom, indicative of reduced yolk uptake, YP170::GFP accumulation within oocytes and embyos was closer to wild type than after Ce-chc(RNAi) or Ce-rab5(RNAi) (Table 1). Nevertheless, all embryos produced by Ce-rab11(RNAi) hermaphrodites died before hatching.
Although Ce-rab7(RNAi) did not appear to significantly impair the uptake of YP170::GFP into oocytes, YP170::GFP was mislocalized in them (see Figure 9M). Rather than small dispersed YP170::GFP vesicles, Ce-rab7(RNAi) oocytes contained fewer vesicles of larger size than wild type that were located more peripherally than in wild type. Such vesicles were often an order of magnitude larger than typical yolk granules. We found that newly hatched Ce-rab7(RNAi) larvae contained these large yolk vesicles throughout their bodies. Larvae and adult progeny of Ce-rab7(RNAi) worms appeared mildly Dpy.
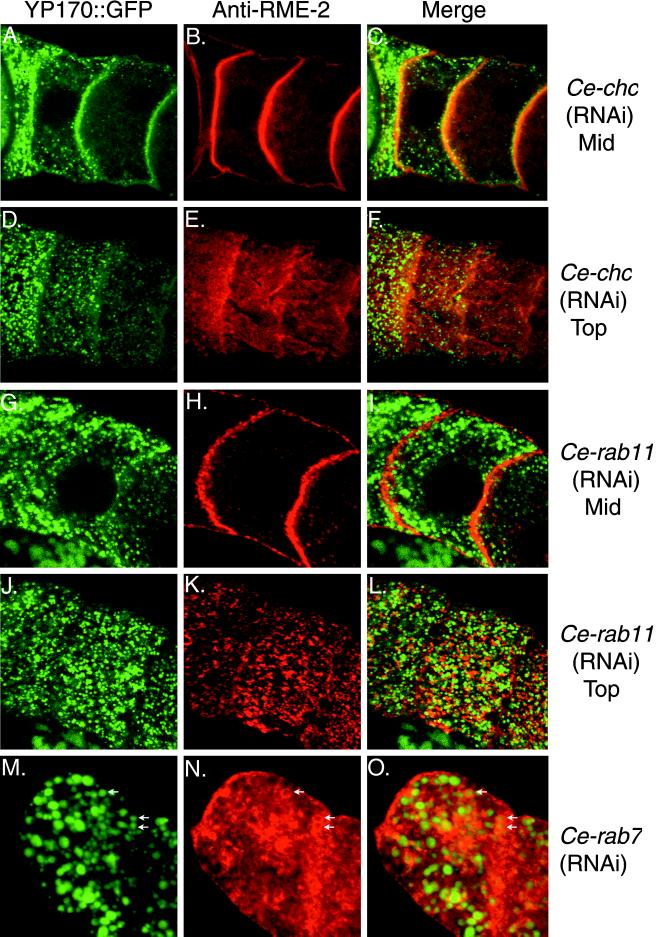
Figure 9.
YP170::GFP and RME-2 localization after RNAi of certain endocytosis factors. Confocal micrographs of oocytes in dissected gonads from RNAi animals show YP170::GFP fluorescence (green) and anti-RME-2 immunostaining (red). Mid, middle focal plane; Top, top focal plane. Ce-clathrin heavy chain (Ce-chc) RNAi (A–C) resulted in accumulation of YP170::GFP and RME-2 at the cell surface and more diffuse surface anti-RME-2 staining (E) than wild type (Figure 6). Ce-rab11(RNAi) oocytes showed reduced internalized YP170::GFP (G) and more punctate, less diffuse RME-2 staining (H and K) than wild-type. Ce-rab7(RNAi) oocytes showed YP170::GFP accumulation in large peripheral vesicles (M) and RME-2 accumulation (N) in these same large vesicles. Some of the vesicles containing both YP170::GFP and RME-2 are marked with arrows.
The Secretory Pathway Is Required for Proper Oocyte Formation and Function
Two types of transport vesicles mediate trafficking within the secretory pathway. COPII-coated vesicles mediate transport from the endoplasmic reticulum (ER) to the Golgi apparatus, whereas COPI-coated vesicles are thought to mediate transport within the Golgi and from the Golgi to the ER for recycling of secretory pathway components (Gaynor et al., 1998; Lowe and Kreis, 1998). Some COPI components have also been associated with early endosome function (Daro et al., 1997). Components of both coats have been shown to be required for efficient transport of secreted and membrane proteins to the cell surface (Gaynor et al., 1998).
We reasoned that phenotypes caused by defects in the secretory pathway might indirectly affect endocytosis in the oocyte by preventing newly synthesized yolk receptors from reaching the cell surface. To compare the phenotypic consequences of a general secretory disruption to specific disruption of endocytic function in the oocyte, we performed RNAi on predicted components of the C. elegans COPI complex, Ce-β′-cop (F38E11.5), Ce-ζ-cop(F59E10.3), and Ce-arf1 (B0336.2).
RNAi of these genes blocked YP170::GFP uptake as expected (Table 2). Ce-β′-cop(RNAi), Ce-ζ-cop(RNAi), and Ce-arf(RNAi) each produced the same array of additional phenotypes, largely distinct from those caused by RNA interference of predicted endocytosis specific genes. Each predicted COPI-coated vesicle RNAi caused many oocytes per gonad arm to take on a rounded appearance with an enlarged nucleus and cytoplasmic streaming, as previously described for mutants defective in the C. elegans Sec61p γ homologue (emo-1; Iwasaki et al., 1996). Unlike emo-1, we observed additional defects: polynucleate oocytes and shell-less or weak-shelled embryos that disintegrated in utero or shortly after being laid (Table 2). Such phenotypes could be caused by defects in secretion of membrane required for efficient cytokinesis during oogenesis and by defects in the secretion of eggshell components after fertilization, respectively. These phenotypes were not observed in the endocytic component RNAi experiments. The differences in phenotypes that we observed when endocytic functions were disrupted as opposed to those observed when secretory functions were disrupted will be helpful in distinguishing between mutants defective in one process versus the other. The exceptions will be genes involved in both processes. In such cases we expect defects in the secretory pathway to mask defects in the endocytosis pathway.
Table 2.
Requirement for predicted COPI secretory pathway components
| Gene targeted for RNAi | Predicted protein homology | % Identity with mammalian homologue | YP170∷GFP in pseudocoelom | YP170∷GFP in full-grown oocytes | Phenotype of progeny |
|---|---|---|---|---|---|
| None | NA | NA | − | +++ | Normal |
| F38E11.5 | β′-COP | 58 | +++ | − | Dead embryos, abnormal oocytes |
| F59E10.3 | ζ-COP | 53 | +++ | − | Dead embryos, abnormal oocytes |
| B0336.2 | ARF1 | 93 | +++ | − | Dead embryos, abnormal oocytes |
Identification and Phenotype of rme-2
Having validated the YP170::GFP endocytosis assay by the RNAi experiments described above, we set out to use this assay in a mutant screen to identify new genes required for endocytosis. In theory such a mutant screen could identify genes specifically required for yolk endocytosis as well as general factors required for all endocytosis. Here we describe the identification of the C. elegans yolk receptor as an example of the effectiveness of this genetic screen in identifying components of the oocyte endocytosis pathway.
We predicted that mutants defective in endocytosis would produce morphologically wild-type oocytes and early embryos devoid of YP170::GFP, and that high levels of YP170::GFP would accumulate in the pseudocoelom of such mutants. Our prediction that endocytosis-defective oocytes and early embryos would appear nearly wild-type by Nomarski optics was based on our RNAi studies of endocytosis genes and studies of the dyn-1 mutant. In addition, studies of sex determination mutants in C. elegans showed that oocytes produced in intersex animals, lacking yolk production, have nearly normal morphology (Doniach and Hodgkin, 1984). Therefore, we screened the F2 generation of a mutagenized YP170::GFP-producing strain for adult hermaphrodites, which contained normal-appearing oocytes that were refractory to YP170::GFP endocytosis. Such mutants show a characteristic accumulation of YP170::GFP fluorescence in the pseudocoelomic space, outlining the internal organs, with reduced or absent YP170::GFP fluorescence in the oocytes and embryos. We isolated three mutant alleles constituting one complementation group, rme-2, in which oocytes of normal appearance are produced but are completely devoid of YP170::GFP fluorescence (Figure 3). The other 11 rme genes identified in this screen will be described elsewhere.
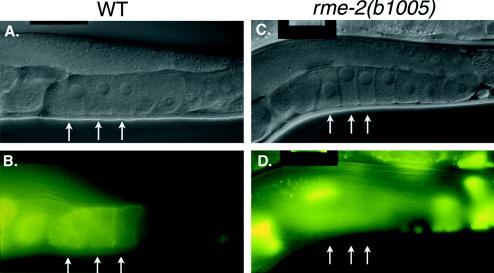
Figure 3.
rme-2 mutants are completely deficient in yolk endocytosis. (A and B) Normarski (A) and fluorescence (B) micrographs of wild-type hermaphrodites expressing YP170::GFP. The three latest-stage oocytes, each with progressively more internalized YP170::GFP, are marked with arrows. (C and D) Nomarski (C) and fluorescence (D) micrographs of rme-2(b1005) hermaphrodites expressing YP170::GFP. Note the absence of YP170::GFP in the three latest stage oocytes (arrows) despite the nearly normal morphology of the rme-2 germ line. All YP170::GFP fluorescence is in the pseudocoelom or in the intestine.
rme-2 mutants are characterized by slightly small oocytes devoid of yolk, otherwise normal germ line morphology, high-level pseudocoelomic yolk accumulation, reduced embryo production, and low embryo viability. We find that rme-2 mutant oocytes lack any detectable endogenous yolk proteins, as assayed by immunofluorescence, including YP170A, YP170B, YP115, and YP88 (see MATERIALS AND METHODS). Embryo production is reduced in rme-2 mutants to an average of 78 (n = 47) compared with ∼300 in wild type. Damaged oocytes and embryos are found in the uterus of rme-2 mutant hermaphrodites, consistent with a defect in ovulation. Defects in ovulation, such as those found in endomitotic (emo) mutants, cause some oocytes to become fragmented by premature closure, or failure to open, the spermathecal valve (Clandinin et al., 1998; McCarter et al., 1999). Time-lapse video recordings of rme-2 mutant ovulations, performed by Tim Schedl (personal communication), indicate a frequent failure of spermathecal dilation as well as common premature closure, which breaks passing oocytes into fragments. A high percentage of rme-2 gonad arms contain one endomitotic-like oocyte proximal to the spermatheca (Schedl, personal communication). Most rme-2 mutant embryos fail to hatch (viability, 23%). Embryos that hatch proceed through normal larval development to produce adult animals with the defects described above.
Molecular Characterization of rme-2
We mapped rme-2 to a position very close to the left of unc-5 on LGIV (see MATERIALS AND METHODS). Upon examination of the predicted genes in this region (C. elegans Sequencing Consortium, 1998), we noticed T11F8.3, a gene that could encode a receptor of the LDL receptor (LDLR) superfamily. Because yolk receptors from insects and vertebrates are members of the LDLR superfamily, we reasoned that rme-2 and T11F8.3 might be the same gene (Stifani et al., 1990; Schonbaum et al., 1995). This hypothesis was strengthend by our subsequent finding that T11F8.3(RNAi) produced phenotypes very similar to those found in rme-2 mutants, including late-stage oocytes and early embryos devoid of YP170::GFP. Sequencing of the T11F8.3 gene from each rme-2 mutant revealed a unique DNA sequence change, which alters the predicted protein product (See below). These results indicate that rme-2 corresponds to the predicted gene T11F8.3.
We determined the sequence of a nearly complete rme-2 cDNA clone, yk8d2. We also determined the 5′ end of the rme-2 mRNA by 5′ RACE. Several RACE products were sequenced, the longest of which extended 153 bp farther 5′ than yk8d2, 35 bases 5′ of a likely AUG start codon. This AUG is the first of the open reading frame and directly precedes a predicted hydrophobic amino acid sequence with features of a secretory signal sequence (von Heijne, 1986). No evidence for a trans-spliced leader on rme-2 messages was obtained. A combination of cDNA and RACE product sequencing indicated a full-length rme-2 mRNA size of 2.9 kb. Northern analysis of poly(A)+ and total RNA from mixed stage N2 worms confirmed the presence of a single rme-2 mRNA species of ∼2.9 kb (our unpublished findings).
RME-2 is an LDLR Family Member
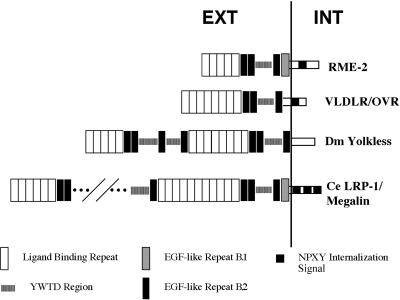
The predicted rme-2 protein is 925 amino acids in length, containing several repeated sequence motifs, with overall similarity to members of the LDLR superfamily of lipoprotein receptors (Figure 4). Hydropathy analysis indicates two highly hydrophobic sequences within RME-2. The first hydrophobic sequence is a likely N-terminal secretory signal sequence, whereas the second located near the C terminus is a likely transmembrane domain. These findings are consistent with a typical type I transmembrane protein topology, with a large N-terminal extracellular domain and a short (110 aa) intracellular domain. C-terminal to the predicted signal sequence are five tandem class A ligand-binding repeats, each containing six cysteine residues with characteristic spacing and typical anionic SDE or DDE sequences (Esser et al., 1988). These are followed by two predicted class B EGF-like cysteine-based repeats (Sudhof et al., 1985). The EGF-like repeats are followed by an intervening region of so-called YWTD repeats, which have been predicted to produce a sixfold symmetrical β-propeller structure (Springer, 1998). Between the YWTD region and the predicted transmembrane domain we find two more class B EGF-like repeats. Finally, within the predicted intracellular domain we find a single predicted internalization signal of the NPXY type (Chen et al., 1990).
Figure 4.
RME-2 is a member of the LDLR superfamily of lipoprotein receptors (GenBank accession number AF185706). Repetitive motifs shown include acidic ligand-binding sequences with six cysteines each, EGF-like repeats with six cysteines each (B.1 and B.2 type; Herz et al., 1988), YWTD repeats, which are thought to form a β-propeller structure (Springer, 1998), and the consensus internalization signal FDNPXY. VLDLR/OVR, very low-density lipoprotein receptor (Gafvels et al., 1993)/chicken yolk receptor (Shen et al., 1993); Dm Yolkless, Drosophila yolk receptor (Schonbaum et al., 1995); Ce LRP-1/Megalin, C. elegans megalin gene product (Yochem and Greenwald, 1993). Only a partial map of lrp-1 motifs is shown because of its large size. For a complete description, see Yochem and Greenwald (1993).
We sequenced PCR products including the complete coding region of the rme-2 gene from each rme-2 mutant. Both rme-2(b1005) and rme-2(b1008) contained DNA sequence alterations producing predicted truncations of the rme-2 protein, deleting part of the YWTD region, EGF-like repeats 3 and 4, and the predicted transmembrane and intracellular domains. Such mutated proteins are unlikely to retain any endocytic function and are predicted null alleles. rme-2(b1026) contained a single nucleotide transition, resulting in a nonconservative amino acid substitution, G→E, in the fourth predicted YWTD repeat.
Analysis of the rme-2 Protein
To analyze the rme-2 protein, we produced affinity-purified polyclonal antisera specific for the predicted extracellular (anti-RME-2-EXT) and intracellular (anti-RME-2-INT) domains of RME-2 (see MATERIALS AND METHODS). Western blots of total protein from mixed stage populations probed with either anti-RME-2 antiserum showed a single band of ∼110 kDa, very close to the predicted size for RME-2 (Figure 5). No protein bands were detected with either anti-RME-2 antisera in lanes containing protein from rme-2(b1005) or rme-2(b1008) mutants, indicating the specificity of the antisera (Figure 5).
Figure 5.
RME-2 Western blot. Total C. elegans proteins were size separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and probed with anti-RME-2(EXT) affinity-purified antibodies. Identical results were observed with anti-RME-2(INT) affinity-purified antibodies (our unpublished findings). Equal protein loading was confirmed by Coomassie blue staining of duplicate gels (our unpublished findings).
We examined the expression pattern of RME-2 in whole-mount mixed populations, mixed stage embryo populations, and dissected adult hermaphrodite gonad preparations by immunofluorescence using our specific anti-RME-2 antisera. We found abundant RME-2 in the proximal germ line of adult hermaphrodites and within all cells of early embryos. We failed to detect RME-2 in any other cells (see MATERIALS AND METHODS).
The adult hermaphrodite gonad consists of two ovaries connected to a central uterus and two spermathecae (Figure 1D). Eggs are laid through the vulva, a ventral opening in the body wall connected to the uterus. Each ovotestis is a reflexed tube with a vulva proximal and vulva distal arm connected by a bend region. The distal arm contains immature germ cell nuclei, with partial plasma membranes, connected by a shared central cytoplasm. Mitotic nuclei are found at the extreme distal end. As germ cells progress in the proximal direction they enter meiosis, reaching diplotene of meiotic prophase I near the bend. In the loop region germ cells become partitioned into individual cells and become differentiated oocytes, increasing in size as they approach the spermatheca and uterus.
RME-2 expression appears to be an early event in oocyte differentiation. Within the germ line, RME-2 first becomes detectable within germ cells in the bend region of the gonad arm. RME-2 first appears in these cells in cytoplasmic puncta reminiscent of vesicles. In later-stage oocytes anti-RME-2 antibodies stain many prominent puncta at or near the plasma membrane in addition to the intracellular puncta (Figure 6B).
Figure 6.
Localization of RME-2 in the gonad and embryo of a YP170::GFP-expressing strain. (A–C) Confocal micrographs of a dissected gonad, showing YP170::GFP (green) and anti-RME-2 (red). Embryo and intestine YP170::GFP fluorescence is evident at the bottom of A and C. Embryos in B do not stain with anti-RME-2 antibodies, because their eggshells are not permeablized by the procedure used to fix and stain the dissected gonad. High-magnification (3000×) confocal micrographs of full-grown oocytes in a middle focal plane (D–F) or top focal plane (G–I) show abundant YP170::GFP and anti-RME-2 immunofluorescence. RME-2 and YP170::GFP signals do not appear to colocalize at steady state. RME-2 immunoreactivity patterns are very different in embryos. All RME-2 appears intracellular in the four-cell embryo (J) and declines progressively during embryogenesis.
As the oocytes grow, they begin to accumulate yolk proteins taken up from the pseudocoelom. The uptake of YP170::GFP into oocytes coincides with the appearance of RME-2 on or near the cell surface (Figure 6A). In nearly full-grown oocytes of a gonad arm, cytoplasmic localization of RME-2 is reduced, and prominent cell surface staining appears. Much of the RME-2 detected by immunostaining in late-stage oocytes appears to be clustered at or near the cell surface, consistent with being localized to CCPs or plasma membrane proximal endosomes (Figure 6, E and H).
Wild-type embryos gave a very different immunostaining pattern from oocytes. All cells of early embryos display prominent anti-RME-2 immunostaining in abundant intracellular vesicular structures but lack any apparent RME-2 in the plasma membrane (Figure 6J). At the earliest time points in embryogenesis we were able to examine (see MATERIALS AND METHODS), all RME-2 was found in these intracellular vesicles. Therefore, this redistribution of RME-2 from the cell surface to intracellular vesicles appears to occur sometime shortly after ovulation, perhaps coincident with the dramatic cellular changes associated with fertilization. As the embryos develop, the number of anti-RME-2–reactive vesicles diminishes, until by hatching little or no RME-2 is detected. The gradual disappearance of RME-2 from these vesicles may indicate degradation in a lysosome-like compartment. We did not find evidence of new RME-2 expression during embryogenesis.
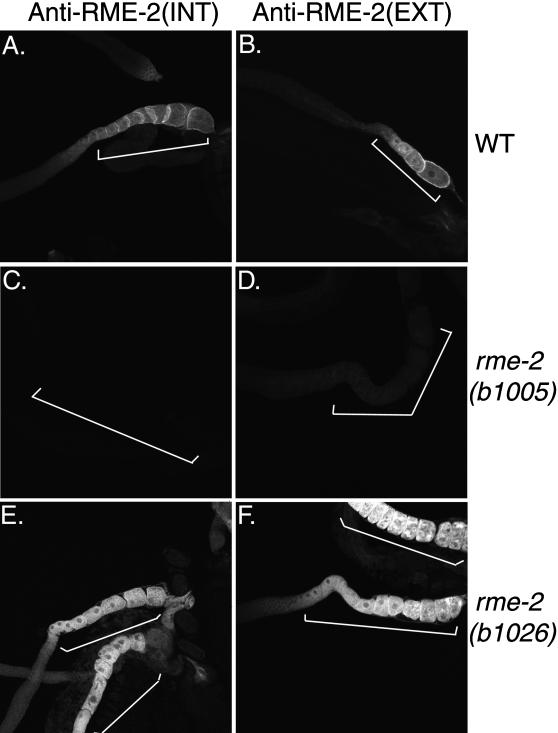
Each of the three rme-2 mutants displays aberrant gonadal immunostaining patterns with anti-RME-2 antisera. Both mutants predicted to produce truncated versions of RME-2, rme-2(b1005) and rme-2(b1008), fail to stain with antisera specific for the RME-2 intracellular domain, and both display very weak or no immunostaining with antisera specific for the RME-2 extracellular domain (Figure 7). Residual staining in these mutants detected with anti-RME-2-EXT antisera is diffuse and is not clearly localized to the oocyte surface membranes. The YWTD missense mutant rme-2(b1026) produces abundant cytoplasmic immunostaining with both anti-RME-2 antisera but lacks defined RME-2 localization at the oocyte plasma membrane (Figure 7). Because rme-2(b1026) contains a missense mutation in its extracellular domain, it may be recognized by the ER quality control machinery as misfolded and prevented from exiting the ER. This effect is commonly found with misfolded proteins passing through the ER. Such ER retention has been seen before in C. elegans in the case of a mutant GLP-1 receptor (Wen and Greenwald, 1999).
Figure 7.
Anti-RME-2 immunostaining of rme-2 mutants. (A and B) Confocal micrographs of wild-type dissected gonads stained with antibodies to the RME-2 intracellular and extracellular domains, respectively. Micrographs were produced using identical scan settings, allowing direct comparison of fluorescent intensities. Oocytes are indicated by brackets. Differences in staining in A and B result from differences in gonad age rather than differences in immunofluoresence pattern between antisera. The gonad in B is from a young adult, whereas that in A is from an older adult. (C and D) rme-2(b1005) mutant oocytes failed to stain with anti-RME-2(INT) antibodies (C) and showed very little reactivity to anti-RME-2(EXT) antibodies (D). rme-2(b1008) mutant oocytes gave very similar results (our unpublished findings). (E and F) rme-2(b1026) mutant oocytes showed strong anti-RME-2 immunofluorescence throughout the cell, possibly indicating ER retention.
RME-2 Is Sufficient to Induce YP170::GFP Binding
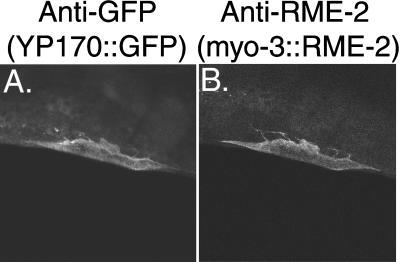
If rme-2 encodes the C. elegans yolk receptor, as indicated by its mutant phenotype and LDLR homology, then ectopic expression of RME-2 in cell types other than the oocyte should lead to the association of yolk with such cells. We tested this hypothesis by expressing RME-2 in body wall muscle cells using the myo-3 promoter (Okkema et al., 1993) and checked these transgenic animals for localization of YP170::GFP to RME-2-expressing muscle cells. In these strains body wall muscle cells expressing high levels of RME-2, as assayed by anti-RME-2 immunostaining, showed significant surface accumulation of YP170::GFP, whereas adjacent muscle cells expressing little or no RME-2 did not accumulate YP170::GFP (Figure 8). Thus, expression of RME-2 in these cells is sufficient to direct the specific binding of yolk particles. We were unable to determine whether YP170::GFP was internalized by RME-2-expressing muscle cells (see MATERIALS AND METHODS).
Figure 8.
Ectopic expression of RME-2 induces ectopic binding of YP170::GFP to RME-2-expressing cells. Shown is anti-GFP (A) and anti-RME-2 (B) immunolabeling of an adult hermaphrodite body wall muscle cell in a strain of the genotype lin-15(n765); bIs1[vit-2::gfp, rol-6(d)]; Ex[myo-3::rme-2, lin-15(+)].
Endocytic Trafficking of RME-2 in the Oocyte
We wished to determine directly the importance of some of the general endocytosis factors described above in RME-2 endocytic cycling and to help establish whether YP170::GFP and RME-2 cosegregate through parts of the endocytic pathway as expected for a ligand–receptor pair. To accomplish these goals we analyzed the subcellular localization of the RME-2 yolk receptor in oocytes by immunofluorescence after RNAi of a select group of general endocytosis genes.
We found that oocytes from Ce-chc(RNAi) animals displayed higher than wild-type accumulation of RME-2 at the oocyte cell surface and showed a concomitant reduction in cytoplasmic RME-2 staining (Figure 9). YP170::GFP and RME-2 colocalized at the oocyte surface. Furthermore, we observed that the prominent RME-2 puncta at or near the surface of wild-type oocytes were reduced or missing in Ce-chc(RNAi) oocytes. Instead, surface RME-2 staining appeared diffuse (Figure 9). Like the Ce-chc(RNAi), Ce-rab5(RNAi) resulted in RME-2 localization at or near the cell surface of the oocyte. RME-2 immunostaining appeared diffuse, failing to produce prominent puncta (our unpublished findings). Ce-rab11(RNAi) resulted in RME-2 immunofluorescence near the cell surface appearing more punctate, with some of the puncta taking on a tubular or mesh-like appearance (Figure 9). Ce-rab11(RNAi) did not result in colocalization of YP170::GFP and RME-2. Ce-rab7(RNAi) oocytes showed reduced RME-2 accumulation at or near the oocyte surface. Surface-proximal RME-2 displayed a normal distribution of puncta (our unpublished findings). Unexpectedly, Ce-rab7(RNAi) oocytes showed prominent accumulation of RME-2 to the same large peripheral vesicles that accumulated YP170::GFP (Figure 9). By analogy to mammalian systems we expect these large vesicular structures to be early endosomes. Accumulation of RME-2 in early endocytic structures in Ce-rab7(RNAi) oocytes may indicate a role for Ce-rab7 in receptor recycling. Colocalization of YP170::GFP and RME-2 to the same endocytic structures in Ce-rab7(RNAi) oocytes is consistent with their identification as a ligand–receptor pair.
DISCUSSION
Yolk Endocytosis: In vivo Endocytic Trafficking in C. elegans
We have developed an in vivo visual assay for the uptake and trafficking of an endocytic ligand, YP170::GFP, by growing oocytes. We tested the importance of conserved components of the general endocytosis pathway for YP170::GFP endocytosis by RNA-mediated interference and found that components of conserved sequence showed conservation of function as well. This new assay system has also allowed us to perform mutant screens to identify new genes required for endocytosis. We expected to identify genes specifically required for yolk uptake, such as the rme-2 gene described here, as well as genes required for endocytosis in many or all cell types. Preliminary analysis of other rme mutants identified in the same screen indicates that many are required more generally for endocytic trafficking (Grant, Hirsh, Pedraza, and Zhang, unpublished observations). Together these experiments describe a general system for reverse and forward genetic analysis of endocytosis in C. elegans.
RME-2: The C. elegans Yolk Receptor
We propose that RME-2 is the C. elegans yolk receptor based on its mutant phenotype, expression pattern, molecular nature, and sufficiency to induce yolk binding in a heterologous cell type. Yolk receptors from insects and vertebrates have been cloned in recent years (Schneider, 1996; Sappington and Raikhel, 1998). All of them are members of the LDLR superfamily of lipoprotein receptors. They vary in size, mainly in their extracellular domains, differing in the number of class A ligand-binding repeats (LRs), class B EGF-like repeats, and YWTD repeat regions (Schneider, 1996). The chicken and Xenopus yolk receptors, like the human VLDL receptor, are eight ligand-binding repeat, or LR8, members of this family and contain typical NPXY internalization motifs in their intracellular domains (Schneider, 1996). The yolk receptors of mosquito and Drosophila are LR13 receptors, with class A repeats divided into an N-terminal group of five and a more membrane-proximal group of eight. Each of these lacks a consensus NPXY signal but instead contains likely dileucine internalization signals (Sappington and Raikhel, 1998). The C. elegans rme-2 gene is the first LR5 member of the LDLR gene family and contains a typical NPXY internalization signal like its vertebrate cousins. It would be interesting to test the affinity of C. elegans, mosquito, and Drosophila yolk receptor N-terminal five class A repeat clusters for yolk binding, because they are unique among the known LDLR superfamily members.
The C. elegans genome contains several other members of the LDLR superfamily, including an LR8 (T13C2.4; Springer, 1998), and two very large receptors related to megalin (lrp-1) and LRP (F47B3.8/T21E3.3). Of these, only lrp-1 has been characterized genetically. LRP-1 is expressed primarily in the hypodermis, and lrp-1 mutants are larval lethals with apparent molting defects (Yochem et al., 1999). No evidence was found for genetic redundancy between rme-2 and lrp-1 in double mutant strains (Yochem, personal communication). Further analysis by C. elegans genetics is likely to reveal more important information about the roles of this conserved metazoan gene family in cellular homeostasis and development.
Yolk Trafficking in the Embryo
After fertilization, yolk granules are segregated approximately equally among progeny blastomeres. Yolk proteins appear to be metabolized slowly during embryogenesis, with many yolk granules remaining in newly hatched larvae. At hatching most of the remaining yolk is found in cells of the intestine. Bossinger and Schierenberg (1992, 1996) showed that selective secretion and reuptake, rather than selective segregation or degradation, are responsible for the visible change in yolk distribution during embryogenesis. Around midembryogenesis, nonintestinal cells resecrete most of the yolk particles they inherited from the oocyte. This resecreted yolk accumulates briefly in the perivitelline space and is then taken up into new storage vesicles by embryonic intestinal cells. One reasonable hypothesis would be that the same receptor responsible for yolk uptake into oocytes from the pseudocoelom would also function in the embryonic intestine for this second uptake event. However, close examination of the RME-2 expression patterns with anti-RME-2 antibodies failed to reveal evidence for RME-2 endocytic function during embryogenesis. In mammals oxidized LDL is taken up by a different receptor than regular LDL. The receptor for oxidized LDL is a member of the lectin family (Sawamura et al., 1997). The C. elegans genome contains many genes encoding proteins similar to lectin-type receptors (Grant, unpublished observation). Perhaps one or more of these receptors is involved in the uptake of yolk into the embryonic intestine.
We have observed that most YP170::GFP remaining in newly hatched larvae is in the intestine, indicating that this fusion protein also undergoes embryonic transport like endogenous yolk. We have also observed that blocking a late transport step in YP170::GFP uptake into the oocyte, by Ce-rab7(RNAi), leads to ectopic localization of enlarged yolk granules throughout the body of newly hatched larvae (our unpublished findings). This may indicate that yolk granules formed from an early endocytic compartment, as opposed to a late endocytic compartment, are not competent for resecretion during embryogenesis. Perhaps this process of resecretion is analogous to the regulated secretion of lysosomal contents by mammalian cells (reviewed by Page et al., 1998) and would be amenable to genetic analysis in C. elegans.
General Endocytic Trafficking Factors
Yolk endocytosis systems in many organisms have been compared with the best studied endocytosis pathway, that of mammalian LDLs (Goldstein et al., 1985). The homology between mammalian ApoB-100 and the vitellogenins as well as that between LDLR and the known vitellogenin receptors has prompted the hypothesis that the mammalian LDL uptake pathway is the most recent modification of the ancient yolk endocytosis apparatus (Schneider, 1996). Like the LDL uptake pathway, vitellogenin receptors bound to yolk particles are thought to be internalized by clathrin-coated vesicles, which uncoat and fuse to form early endosomes. Unlike somatic cells, oocytes are thought to lack lysosomes per se and their high proteolytic capacity (Schneider, 1996). Yolk remains stored in an endosomal compartment, probably equivalent to lysosomes until used during embryogenesis.
The complete genome sequence of C. elegans and the facile reverse genetic technique of RNA-mediated interference allowed us to test some of these hypotheses in our YP170::GFP uptake assay. In general our results correlated well with the predicted order of events in the yolk uptake pathway of C. elegans. C. elegans homologues of CCP components were clearly important for yolk uptake, as was the C. elegans homologue of the early endosomal regulator rab5. Ce-chc(RNAi) and Ce-rab5(RNAi) induced accumulation of ligand (YP170::GFP) and receptor (RME-2) together at or near the oocyte surface. In both cases RME-2 immunofluoresence at or near the cell surface appeared diffuse, lacking the characteristic punctate pattern typical of wild-type oocytes and might represent diffuse plasma membrane localization. Although we currently cannot identify the RME-2 membrane-proximal puncta as cell surface structures such as pits or subsurface structures such as endosomes, either might be expected to lack RME-2 after depletion of clathrin or rab5. Interestingly, a role for rab5–guanine nucleotide dissociation inhibitor complexes in sequestration of ligand–receptor complexes into CCPs, in addition to the well-studied function of rab5 in endosome fusion, was recently shown (Horiuchi et al., 1997; McLauchlan et al., 1998). Our results are consistent with uptake of vitellogenin–RME-2 complexes through CCPs and early endosomes.
In these RNAi experiments, the only predicted CCP components that did not appear to be required for YP170::GFP endocytosis were Ce-μ2 and Ce-ς2. Alone or in combination, RNAi for these genes produced only one phenotype, extremely Dpy progeny. This result is quite surprising, because μ2 is thought to provide the primary link between receptors and the internalization machinery (Ohno et al., 1995). In addition, clathrin adaptor complexes are thought to become unstable if one component of the tetramer is compromised, implying that removal of any one subunit should give the same phenotypes (e.g., Dell’Angelica et al., 1999).
One interesting interpretation of these results would be that Ce-μ2 and Ce-ς2 are not required for yolk endocytosis, or viability, unlike Ce-α-adaptin and Ce-β-adaptin. The similar Dpy phenotypes displayed in Ce-μ2(RNAi) and Ce-ς2(RNAi) animals may indicate defects in hypodermal development. Perhaps medium or small chains from other adaptin complexes in the worm (e.g., AP1 and AP3) can partially substitute for Ce-μ2 and Ce-ς2, as suggested by some overlap in the in vitro affinities of medium chains for sorting signals (Ohno et al., 1998). We did not find evidence for functional redundancy between Ce-μ2 and unc-101, a Ce-μ1. Another possibility is that the RNAi technique did not sufficiently reduce Ce-μ2 and Ce-ς2 function to reveal their null phenotypes. If this is the case, we might have expected Ce-μ2(RNAi); Ce-ς2(RNAi) to show additional defects. Again, these experiments failed to reveal a role for Ce-μ2 and Ce-ς2 in yolk endocytosis. Full exploration of these possibilities awaits the isolation and analysis of Ce-μ2 and Ce-ς2 null mutants.
The Ce-rab11(RNAi) phenotype was consistent with a role for receptor recycling in yolk endocytosis: YP170::GFP uptake was reduced, and RME-2 localization was altered. RME-2 accumulated in surface-proximal structures that appeared more numerous, enlarged, and less regular than puncta prominent in wild-type oocytes. These results are consistent with possible accumulation of RME-2 receptors in early endosomal or recycling structures. We searched the C. elegans genome for rab11 and rab4, regulators of receptor recycling. We found a second gene (W04G5.1) homologous to rab11 but none closely related to rab4. Because of the sequence similarity to one another (80% identical at the mRNA level), it is possible that RNAi against one Ce-rab11 would interfere with expression of both (Fire et al., 1998). Perhaps one or both rab11 homologues serve functions similar to rab4 in mammalian cells.
In mammalian cells rab7 is thought to be required for transport from early to late endosomes (Feng et al., 1995). Mammalian tissue culture cells, expressing a dominant negative form of rab7, accumulate cell surface molecules traversing the endocytic route from cell surface to the lysosome in enlarged early endosomes. We have blocked expression of Ce-rab7 by RNAi and observed accumulation of YP170::GFP in large peripheral vesicles of the oocyte, a phenotype very similar to that described above for mammalian cells (Figure 9).
Unexpectedly, we also observed significant accumulation of the putative receptor, RME-2, in these same large peripheral vesicles of oocytes depleted of Ce-rab7 activity. Expression of dominant negative Rab7 protein was not found to interfere with recycling of receptors from the early endosome to the cell surface in mammalian cells (Feng et al., 1995). Because a large volume of yolk is taken up into oocytes in a relatively short period, and because RME-2 is similar to the rapidly recycled LDLR, it seems likely that RME-2 recycles frequently during oogenesis. Because we do not see RME-2 accumulating in the same storage vesicles as yolk particles in wild-type oocytes, it seems likely that yolk and yolk receptor are sorted from one another after internalization. After sorting, the receptors could be recycled or degraded.
Because oocytes are thought to have very low degradative capacity, we consider two simple models of RME-2 recycling as likely scenarios that could account for these observations. First, RME-2 could be rapidly recycled from early endosomes, in a Ce-rab7-dependent manner. Such a requirement for rab7 activity in recycling from the early endosome could be specific for C. elegans or might have been missed using dominant negative techniques in mammalian cells, which presumably function by titrating out rab7 exchange factors rather than depleting the cell of rab7 itself (Feig, 1999). Alternatively, RME-2 might be recycled from late endosomes, as has been reported for LRP–receptor-associated protein complexes (Czekay et al., 1997), in which case, Ce-rab7(RNAi) could inhibit RME-2 from reaching late endosomes for recycling. It will be interesting to differentiate between these models in future studies.
Much remains to be discovered about the mechanisms of endocytic trafficking. We anticipate that continued analysis of receptor-mediated endocytosis in the C. elegans oocyte, especially in-depth analysis of new rme mutants, identified in screens for defective yolk uptake, will contribute to our understanding of this important cellular process.
ACKNOWLEDGMENTS
We thank J. Fares, I. Greenwald, T. Schedl, H. Wilkinson, and Y. Zhang for helpful discussions during the course of this work; L. Pedraza for excellent technical assistance; T. Blumenthal, A. Fire, Y. Kohara, R. Plasterk, S. Strome, and H.G. van Luenen for reagents; and I. Greenwald, O. Hobert, H. Wilkinson, J. Yochem, T. Schedl, and Y. Zhang for comments on this manuscript. We thank Tim Schedl for unpublished information on the nature of ovulation defects in rme-2 mutants. Many of the strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources of the National Institutes of Health. This work was supported by National Institutes of Health National Service Research Award F32 GM19167-02 to B.G. and March of Dimes grant FY99-583 to D.H..
REFERENCES
- Baker ME. Is vitellogenin an ancestor of apolipoprotein B-100 of human low-density lipoprotein and human lipoprotein lipase. Biochem J. 1988;255:1057–1060. doi: 10.1042/bj2551057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettinger JC, Lee K, Rougvie AE. Stage-specific accumulation of the terminal differentiation factor LIN-29 during Caenorhabditis elegans development. Development. 1996;122:2517–2527. doi: 10.1242/dev.122.8.2517. [DOI] [PubMed] [Google Scholar]
- Bossinger O, Schierenberg E. Cell-cell communication in the embryo of Caenorhabditis elegans. Dev Biol. 1992;151:401–409. doi: 10.1016/0012-1606(92)90180-o. [DOI] [PubMed] [Google Scholar]
- Bossinger O, Schierenberg E. The use of fluorescent marker dyes for studying intercellular communication in nematode embryos. Int J Dev Biol. 1996;40:431–439. [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–804. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Chen WJ, Goldstein JL, Brown MS. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J Biol Chem. 1990;265:3116–3123. [PubMed] [Google Scholar]
- Clandinin TR, DeModena JA, Sternberg PW. Inositol trisphosphate mediates a RAS-independent response to LET-23 receptor tyrosine kinase activation in C. elegans. Cell. 1998;92:523–533. doi: 10.1016/s0092-8674(00)80945-9. [DOI] [PubMed] [Google Scholar]
- Clark SG, Shurland DL, Meyerowitz EM, Bargmann CI, van der Bliek AM. A dynamin GTPase mutation causes a rapid and reversible temperature-inducible locomotion defect in C. elegans. Proc Natl Acad Sci USA. 1997;94:10438–10443. doi: 10.1073/pnas.94.19.10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czekay RP, Orlando RA, Woodward L, Lundstrom M, Farquhar MG. Endocytic trafficking of megalin/RAP complexes: dissociation of the complexes in late endosomes. Mol Biol Cell. 1997;8:517–532. doi: 10.1091/mbc.8.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daro E, Sheff D, Gomez M, Kreis T, Mellman I. Inhibition of endosome function in CHO cells bearing a temperature-sensitive defect in the coatomer (COPI) component epsilon-COP. J Cell Biol. 1997;139:1747–1759. doi: 10.1083/jcb.139.7.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Camilli P, Takei K, McPherson PS. The function of dynamin in endocytosis. Curr Opin Neurobiol. 1995;5:559–565. doi: 10.1016/0959-4388(95)80059-x. [DOI] [PubMed] [Google Scholar]
- De Stasio E, Lephoto C, Azuma L, Holst C, Stanislaus D, Uttam J. Characterization of revertants of unc-93(e1500) in Caenorhabditis elegans induced by N-ethyl-N-nitrosourea. Genetics. 1997;147:597–608. doi: 10.1093/genetics/147.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Angelica EC, Shotelersuk V, Aguilar RC, Gahl WA, Bonifacino JS. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Mol Cell. 1999;3:11–21. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- Doniach T, Hodgkin J. A sex determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev Biol. 1984;106:223–235. doi: 10.1016/0012-1606(84)90077-0. [DOI] [PubMed] [Google Scholar]
- Esser V, Limbird LE, Brown MS, Goldstein JL, Russell DW. Mutational analysis of the ligand binding domain of the low density lipoprotein receptor. J Biol Chem. 1988;263:13282–13290. [PubMed] [Google Scholar]
- Feig LA. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat Cell Biol. 1999;1:E25–E27. doi: 10.1038/10018. [DOI] [PubMed] [Google Scholar]
- Feng Y, Press B, Wandinger-Ness A. Rab 7: an important regulator of late endocytic membrane traffic. J Cell Biol. 1995;131:1435–1452. doi: 10.1083/jcb.131.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson EL, Horvitz HR. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics. 1985;110:17–72. doi: 10.1093/genetics/110.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A. Integrative transformation of Caenorhabditis elegans. EMBO J. 1986;5:2673–2680. doi: 10.1002/j.1460-2075.1986.tb04550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gafvels ME, Caird M, Britt D, Jackson CL, Patterson D, Strauss JFd. Cloning of a cDNA encoding a putative human very low density lipoprotein/apolipoprotein E receptor and assignment of the gene to chromosome 9pter-p23. Somat Cell Mol Genet. 1993;19:557–569. doi: 10.1007/BF01233382. [DOI] [PubMed] [Google Scholar]
- Gaynor EC, Graham TR, Emr SD. COPI in ER/Golgi and intraGolgi transport: do yeast COPI mutants point the way? Biochim Biophys Acta. 1998;1404:33–51. doi: 10.1016/s0167-4889(98)00045-7. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS, Anderson RG, Russell DW, Schneider WJ. Receptor-mediated endocytosis: concepts emerging from the LDL receptor system. Annu Rev Cell Biol. 1985;1:1–39. doi: 10.1146/annurev.cb.01.110185.000245. [DOI] [PubMed] [Google Scholar]
- Grant B, Greenwald I. Structure, function, and expression of SEL-1, a negative regulator of LIN-12 and GLP-1 in C. elegans. Development. 1997;124:637–644. doi: 10.1242/dev.124.3.637. [DOI] [PubMed] [Google Scholar]
- Gu J, Stephenson CG, Iadarola MJ. Recombinant proteins attached to a nickel-NTA column: use in affinity purification of antibodies. Biotechniques. 1994;17:257–262. [PubMed] [Google Scholar]
- Hall DH, Winfrey VP, Blaeuer G, Hoffman LH, Furuta T, Rose KL, Hobert O, Greenstein D. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev Biol. 1999;212:101–123. doi: 10.1006/dbio.1999.9356. [DOI] [PubMed] [Google Scholar]
- Herz J, Hamann U, Rogne S, Myklebost D, Gausepohl H, Stanley KK. Surface localization and high affinity for calcium for a 500-kd liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 1988;7:4119–4127. doi: 10.1002/j.1460-2075.1988.tb03306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi H, et al. A novel rab5 GDP/GTP exchange factor complexed to rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- Iwasaki K, McCarter J, Francis R, Schedl T. emo-1, a Caenorhabditis elegans Sec61p gamma homologue, is required for oocyte development and ovulation. J Cell Biol. 1996;134:699–714. doi: 10.1083/jcb.134.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Francis R, Schedl T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev Biol. 1996;180:165–183. doi: 10.1006/dbio.1996.0293. [DOI] [PubMed] [Google Scholar]
- Jorgensen EM, Hartwieg E, Schuske K, Nonet ML, Jin Y, Horvitz HR. Defective recycling of synaptic vesicles in synaptotagmin mutants of Caenorhabditis elegans. Nature. 1995;378:196–199. doi: 10.1038/378196a0. [DOI] [PubMed] [Google Scholar]
- Kimble J, Sharrock WJ. Tissue-specific synthesis of yolk proteins in C. elegans. Dev Biol. 1983;96:189–196. doi: 10.1016/0012-1606(83)90322-6. [DOI] [PubMed] [Google Scholar]
- Kramer JM, Johnson JJ. Analysis of mutations in the sqt-1 and rol-6 collagen gene of Caenorhabditis elegans. Genetics. 1993;135:1035–1045. doi: 10.1093/genetics/135.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Jongeward GD, Sternberg PW. unc-101, a gene required for many aspects of Caenorhabditis elegans development and behavior, encodes a clathrin-associated protein. Genes & Dev. 1994;8:60–73. doi: 10.1101/gad.8.1.60. [DOI] [PubMed] [Google Scholar]
- Levy AD, Yang J, Kramer JM. Molecular genetic analysis of the Caenorhabditis elegans dpy-2 and dpy-10 collagen genes: a variety of molecular alterations affect organismal morphology. Mol Biol Cell. 1993;4:803–817. doi: 10.1091/mbc.4.8.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe M, Kreis TE. Regulation of membrane traffic in animal cells by COPI. Biochim Biophys Acta. 1998;1404:53–66. doi: 10.1016/s0167-4889(98)00046-9. [DOI] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol. 1999;205:111–128. doi: 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- McLauchlan H, Newell J, Morrice N, Osborne A, West M, Smythe E. A novel role for Rab5-GDI in ligand sequestration into clathrin-coated pits. Curr Biol. 1998;8:34–45. doi: 10.1016/s0960-9822(98)70018-1. [DOI] [PubMed] [Google Scholar]
- Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery MK, Xu S, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Ghosh RN, Maxfield FR. Endocytosis. Physiol Rev. 1997;77:759–803. doi: 10.1152/physrev.1997.77.3.759. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Grundahl K, Meyer BJ, Rand JB. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- Nigon V. Les modalites de la reproduction et le determinisme du sexe chez quelques nematodes libres. Ann Sci Nat. 1949;11:1–132. [Google Scholar]
- Ohno H, Aguilar RC, Yeh D, Taura D, Saito T, Bonifacino JS. The medium subunits of adaptor complexes recognize distinct but overlapping sets of tyrosine-based sorting signals. J Biol Chem. 1998;273:25915–25921. doi: 10.1074/jbc.273.40.25915. [DOI] [PubMed] [Google Scholar]
- Ohno H, Stewart J, Fournier MC, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino JS. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A. Sequence requirements for myosin gene-expression and regulation in Caenorhabditis elegans. Genetics. 1993;135:385–404. doi: 10.1093/genetics/135.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page LJ, Darmon AJ, Uellner R, Griffiths GM. L is for lytic granules: lysosomes that kill. Biochim Biophys Acta. 1998;1401:146–156. doi: 10.1016/s0167-4889(97)00138-9. [DOI] [PubMed] [Google Scholar]
- Pearse BM, Robinson MS. Clathrin, adaptors, and sorting. Annu Rev Cell Biol. 1990;6:151–171. doi: 10.1146/annurev.cb.06.110190.001055. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Rab GTPases: master regulators of membrane trafficking. Curr Opin Cell Biol. 1994;6:522–526. doi: 10.1016/0955-0674(94)90071-x. [DOI] [PubMed] [Google Scholar]
- Press B, Feng Y, Hoflack B, Wandinger-Ness A. Mutant Rab7 causes the accumulation of cathepsin D and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J Cell Biol. 1998;140:1075–1089. doi: 10.1083/jcb.140.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TF, Porter KR. Yolk protein uptake in the oocyte of the mosquito Aedes aegypti. J Cell Biol. 1964;20:313–332. doi: 10.1083/jcb.20.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- Sappington TW, Raikhel AS. Molecular characteristics of insect vitellogenins and vitellogenin receptors. Insect Biochem Mol Biol. 1998;28:277–300. doi: 10.1016/s0965-1748(97)00110-0. [DOI] [PubMed] [Google Scholar]
- Sawamura T, et al. An endothelial receptor for oxidized low-density lipoprotein. Nature. 1997;386:73–77. doi: 10.1038/386073a0. [DOI] [PubMed] [Google Scholar]
- Schekman R, Orci L. Coat proteins and vesicle budding. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- Schneider WJ. Vitellogenin receptors: oocyte-specific members of the low-density lipoprotein receptor supergene family. Int Rev Cytol. 1996;166:103–137. doi: 10.1016/s0074-7696(08)62507-3. [DOI] [PubMed] [Google Scholar]
- Schonbaum CP, Lee S, Mahowald AP. The Drosophila yolkless gene encodes a vitellogenin receptor belonging to the low density lipoprotein receptor superfamily. Proc Natl Acad Sci USA. 1995;92:1485–1489. doi: 10.1073/pnas.92.5.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock WJ. Yolk proteins of C. elegans. Dev Biol. 1983;96:182–188. doi: 10.1016/0012-1606(83)90321-4. [DOI] [PubMed] [Google Scholar]
- Sharrock WJ, Sutherlin ME, Leske K, Cheng TK, Kim TY. Two distinct yolk lipoprotein complexes from Caenorhabditis elegans. J Biol Chem. 1990;265:14422–14431. [PubMed] [Google Scholar]
- Shen X, Steyrer E, Retzek H, Sanders EJ, Schneider WJ. Chicken oocyte growth: receptor-mediated yolk deposition. Cell Tissue Res. 1993;272:459–471. doi: 10.1007/BF00318552. [DOI] [PubMed] [Google Scholar]
- Spieth J, Nettleton M, Zucker-Aprison E, Lea K, Blumenthal T. Vitellogenin motifs conserved in nematodes and vertebrates. J Mol Evol. 1991;32:429–438. doi: 10.1007/BF02101283. [DOI] [PubMed] [Google Scholar]
- Springer TA. An extracellular beta-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J Mol Biol. 1998;283:837–862. doi: 10.1006/jmbi.1998.2115. [DOI] [PubMed] [Google Scholar]
- Stifani S, Barber DL, Nimpf J, Schneider WJ. A single chicken oocyte plasma membrane protein mediates uptake of very low density lipoprotein and vitellogenin. Proc Natl Acad Sci USA. 1990;87:1955–1959. doi: 10.1073/pnas.87.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC, Goldstein JL, Brown MS, Russell DW. The LDL receptor gene: a mosaic of exons shared with different proteins. Science. 1985;228:815–822. doi: 10.1126/science.2988123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluijs P, Hull M, Webster P, Male P, Goud B, Mellman I. The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell. 1992;70:729–740. doi: 10.1016/0092-8674(92)90307-x. [DOI] [PubMed] [Google Scholar]
- van Luenen HGAM, Plasterk RHA. Transposons. In: Blumenthal T, Riddle DL, Meyer BJ, Priess JR, editors. C. elegans. II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 97–116. [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Sudhof TC, Anderson RG. The appendage domain of alpha-adaptin is a high affinity binding site for dynamin. J Biol Chem. 1995;270:10079–10083. doi: 10.1074/jbc.270.17.10079. [DOI] [PubMed] [Google Scholar]
- Wen C, Greenwald I. p24 proteins and quality control of LIN-12 and GLP-1 trafficking in Caenorhabditis elegans. J Cell Biol. 1999;145:1165–1175. doi: 10.1083/jcb.145.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B, Emr SD, Riezman H. Protein traffic in the yeast endocytic and vacuolar protein sorting pathways. Curr Opin Cell Biol. 1998;10:513–522. doi: 10.1016/s0955-0674(98)80067-7. [DOI] [PubMed] [Google Scholar]
- Williams BD. Genetic mapping with polymorphic sequence-tagged sites. Methods Cell Biol. 1995;48:81–96. doi: 10.1016/s0091-679x(08)61384-9. [DOI] [PubMed] [Google Scholar]
- Yochem J, Greenwald I. A gene for a low density lipoprotein receptor-related protein in the nematode Caenorhabditis elegans. Proc Natl Acad Sci USA. 1993;90:4572–4576. doi: 10.1073/pnas.90.10.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J, Tuck S, Greenwald I, Han M. A gp330/megalin-related protein is required in the major epidermis of Caenorhabditis elegans for completion of molting. Development. 1999;126:597–606. doi: 10.1242/dev.126.3.597. [DOI] [PubMed] [Google Scholar]