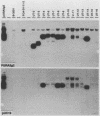
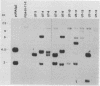
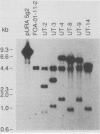
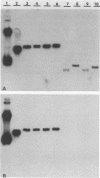
Abstract
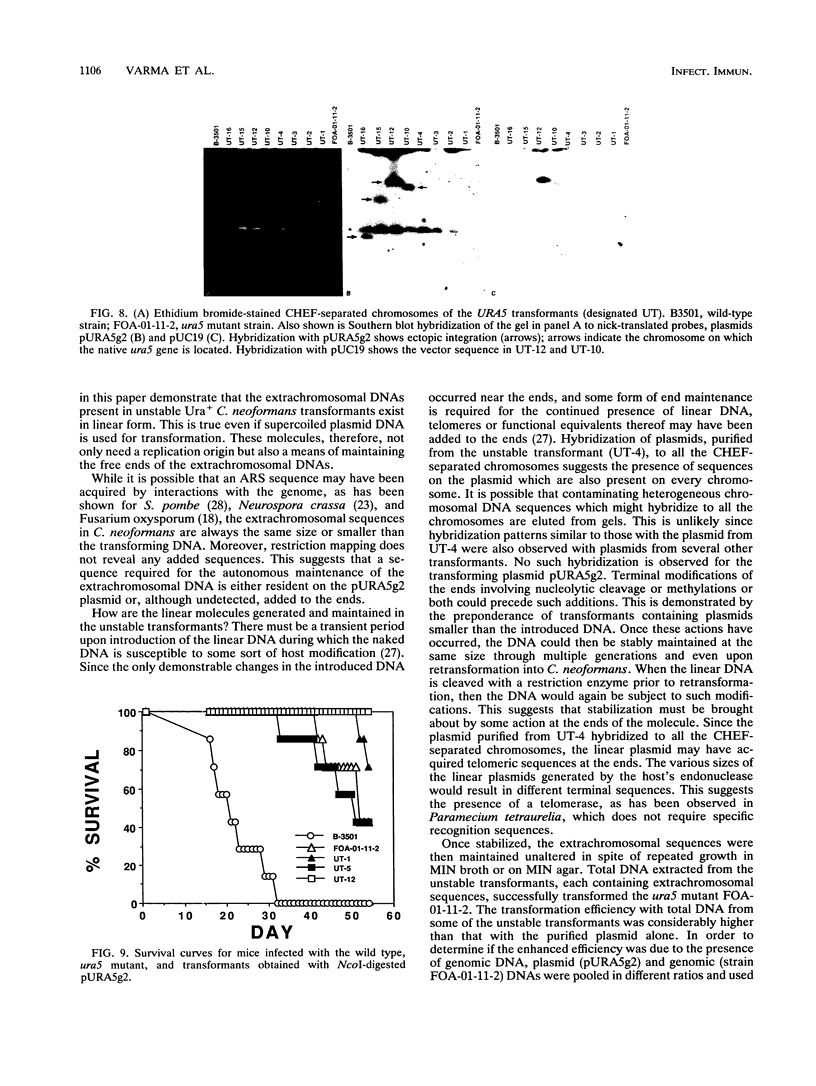
Cryptococcus neoformans var. neoformans ura5 mutants were transformed with linearized or circular plasmids containing the C. neoformans orotidine monophosphate pyrophosphorylase gene. Following electroporation, randomly isolated transformants were analyzed for the mitotic and meiotic stability of uracil prototrophy. All stable transformants tested showed nonspecific ectopic integration. Uracil prototrophy in these transformants was stable through meiosis. Some of the stable transformants showed integration of both URA5 and vector sequences, while others lacked any vector sequences. Unstable transformants exhibited the presence of an autonomously replicating plasmid which had undergone significant sequence rearrangement. The autonomously replicating plasmid in the transformants was observed to be the same size or smaller than the transforming plasmid, was maintained in a linear form, and had acquired a genomic sequence(s) with homology to a sequence(s) on all the chromosomes. The conservation of a 300-bp sequence at the 5' end of the URA5 gene was observed in all the rearranged plasmids. These results suggest mechanisms of plasmid maintenance in C. neoformans that are different from those reported for other yeasts. The ura5 mutant was significantly less virulent than the wild type. The transformants did not recover virulence regardless of prototrophic stability.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Binninger D. M., Skrzynia C., Pukkila P. J., Casselton L. A. DNA-mediated transformation of the basidiomycete Coprinus cinereus. EMBO J. 1987 Apr;6(4):835–840. doi: 10.1002/j.1460-2075.1987.tb04828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock P., Forrester W., Botchan M. DNA sequence studies of simian virus 40 chromosomal excision and integration in rat cells. J Mol Biol. 1984 Mar 25;174(1):55–84. doi: 10.1016/0022-2836(84)90365-6. [DOI] [PubMed] [Google Scholar]
- Case M. E. Genetical and molecular analyses of qa-2 transformants in Neurospora crassa. Genetics. 1986 Jul;113(3):569–587. doi: 10.1093/genetics/113.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawale S. S., Marzluf G. A. Transformation of Neurospora crassa with circular and linear DNA and analysis of the fate of the transforming DNA. Curr Genet. 1985;10(3):205–212. doi: 10.1007/BF00798750. [DOI] [PubMed] [Google Scholar]
- Edman J. C., Kwon-Chung K. J. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol Cell Biol. 1990 Sep;10(9):4538–4544. doi: 10.1128/mcb.10.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faugeron G., Rhounim L., Rossignol J. L. How does the cell count the number of ectopic copies of a gene in the premeiotic inactivation process acting in Ascobolus immersus? Genetics. 1990 Mar;124(3):585–591. doi: 10.1093/genetics/124.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N. V. About maize transposable elements and development. Cell. 1989 Jan 27;56(2):181–191. doi: 10.1016/0092-8674(89)90891-x. [DOI] [PubMed] [Google Scholar]
- Fincham J. R. Transformation in fungi. Microbiol Rev. 1989 Mar;53(1):148–170. doi: 10.1128/mr.53.1.148-170.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folger K. R., Thomas K., Capecchi M. R. Nonreciprocal exchanges of information between DNA duplexes coinjected into mammalian cell nuclei. Mol Cell Biol. 1985 Jan;5(1):59–69. doi: 10.1128/mcb.5.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D., Preer J. R., Jr, Aufderheide K. J., Polisky B. Autonomous replication and addition of telomerelike sequences to DNA microinjected into Paramecium tetraurelia macronuclei. Mol Cell Biol. 1988 Nov;8(11):4765–4772. doi: 10.1128/mcb.8.11.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyon C., Faugeron G. Targeted transformation of Ascobolus immersus and de novo methylation of the resulting duplicated DNA sequences. Mol Cell Biol. 1989 Jul;9(7):2818–2827. doi: 10.1128/mcb.9.7.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y., Marzluf G. A. Transformation of Neurospora crassa with the trp-1 gene and the effect of host strain upon the fate of the transforming DNA. Curr Genet. 1988;13(1):65–70. doi: 10.1007/BF00365758. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung K. J., Rhodes J. C. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect Immun. 1986 Jan;51(1):218–223. doi: 10.1128/iai.51.1.218-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chevanton L., Leblon G., Lebilcot S. Duplications created by transformation in Sordaria macrospora are not inactivated during meiosis. Mol Gen Genet. 1989 Sep;218(3):390–396. doi: 10.1007/BF00332400. [DOI] [PubMed] [Google Scholar]
- Powell W. A., Kistler H. C. In vivo rearrangement of foreign DNA by Fusarium oxysporum produces linear self-replicating plasmids. J Bacteriol. 1990 Jun;172(6):3163–3171. doi: 10.1128/jb.172.6.3163-3171.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Cambareri E. B., Jensen B. C., Haack K. R. Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell. 1987 Dec 4;51(5):741–752. doi: 10.1016/0092-8674(87)90097-3. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Garrett P. W. DNA sequence duplications trigger gene inactivation in Neurospora crassa. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6870–6874. doi: 10.1073/pnas.85.18.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stinchcomb D. T., Struhl K., Davis R. W. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979 Nov 1;282(5734):39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- Stohl L. L., Akins R. A., Lambowitz A. M. Characterization of deletion derivatives of an autonomously replicating Neurospora plasmid. Nucleic Acids Res. 1984 Aug 10;12(15):6169–6178. doi: 10.1093/nar/12.15.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. The new yeast genetics. 1983 Sep 29-Oct 5Nature. 305(5933):391–397. doi: 10.1038/305391a0. [DOI] [PubMed] [Google Scholar]
- Varma A., Kwon-Chung K. J. Rapid method to extract DNA from Cryptococcus neoformans. J Clin Microbiol. 1991 Apr;29(4):810–812. doi: 10.1128/jcm.29.4.810-812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walmsley R. M. Yeast telomeres: the end of the chromosome story? Yeast. 1987 Sep;3(3):139–148. doi: 10.1002/yea.320030302. [DOI] [PubMed] [Google Scholar]
- Wright A. P., Maundrell K., Shall S. Transformation of Schizosaccharomyces pombe by non-homologous, unstable integration of plasmids in the genome. Curr Genet. 1986;10(7):503–508. doi: 10.1007/BF00447383. [DOI] [PubMed] [Google Scholar]
- Yelton M. M., Hamer J. E., Timberlake W. E. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1470–1474. doi: 10.1073/pnas.81.5.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]