Abstract
The protein encoded by the PPARGC1A gene is expressed at high levels in metabolically active tissues and is involved in the control of oxidative stress via reactive oxygen species detoxification. Several recent reports suggest that the PPARGC1A Gly482Ser (rs8192678) missense polymorphism may relate inversely with blood pressure. We used conventional meta-analysis methods to assess the association between Gly482Ser and systolic (SBP) or diastolic blood pressures (DBP) or hypertension in 13,949 individuals from 17 studies, of which 6,042 were previously unpublished observations. The studies comprised cohorts of white European, Asian, and American Indian adults, and adolescents from South America. Stratified analyses were conducted to control for population stratification. Pooled genotype frequencies were 0.47 (Gly482Gly), 0.42 (Gly482Ser), and 0.11 (Ser482Ser). We found no evidence of association between Gly482Ser and SBP [Gly482Gly: mean = 131.0 mmHg, 95% confidence interval (CI) = 130.5–131.5 mmHg; Gly482Ser mean = 133.1 mmHg, 95% CI = 132.6–133.6 mmHg; Ser482Ser: mean = 133.5 mmHg, 95% CI = 132.5–134.5 mmHg; P = 0.409] or DBP (Gly482Gly: mean = 80.3 mmHg, 95% CI = 80.0–80.6 mmHg; Gly482Ser mean = 81.5 mmHg, 95% CI = 81.2–81.8 mmHg; Ser482Ser: mean = 82.1 mmHg, 95% CI = 81.5–82.7 mmHg; P = 0.651). Contrary to previous reports, we did not observe significant effect modification by sex (SBP, P = 0.966; DBP, P = 0.715). We were also unable to confirm the previously reported association between the Ser482 allele and hypertension [odds ratio: 0.97, 95% CI = 0.87–1.08, P = 0.585]. These results were materially unchanged when analyses were focused on whites only. However, statistical evidence of gene-age interaction was apparent for DBP [Gly482Gly: 73.5 (72.8, 74.2), Gly482Ser: 77.0 (76.2, 77.8), Ser482Ser: 79.1 (77.4, 80.9), P = 4.20 × 10−12] and SBP [Gly482Gly: 121.4 (120.4, 122.5), Gly482Ser: 125.9 (124.6, 127.1), Ser482Ser: 129.2 (126.5, 131.9), P = 7.20 × 10−12] in individuals <50 yr (n = 2,511); these genetic effects were absent in those older than 50 yr (n = 5,088) (SBP, P = 0.41; DBP, P = 0.51). Our findings suggest that the PPARGC1A Ser482 allele may be associated with higher blood pressure, but this is only apparent in younger adults.
Keywords: meta-analysis, gene-environment interaction
identifying the genetic variants that increase the risk of high blood pressure may help optimize preventive strategies, primarily because this information could be used to target high-risk individuals for early treatment and helps elucidate the biological underpinnings of the disease. An important part of this process is to undertake appropriately designed confirmation studies to determine the robustness of previously reported associations. One method to achieve this is to undertake meta-analyses of published and unpublished data.
Since its initial reported association with Type 2 diabetes (7), a common missense variant at exon 8 of the peroxisome proliferator activated receptor-γ coactivator 1α (PPARGC1A) gene has been repeatedly associated with type 2 diabetes (4) and with related traits, including obesity (19), dyslipidemia (9), aerobic fitness (15), and insulin resistance (16). In those studies, the minor Ser482 allele is typically associated with increased susceptibility to disease. However, in several large studies that assessed the association between Gly482Ser and hypertension, the Ser482 allele conferred a protective effect (2, 17).
The protein encoded by the PPARGC1A gene coactivates at least 30 transcription factors involved in various aspects of cellular energy metabolism (22). The gene is expressed in tissues with high metabolic activity, such as heart, liver, kidney, and brown adipose, and could play a role in the regulation of blood pressure by interacting with mineralocorticoid and estrogen receptors (17) or through reactive oxygen species detoxification (23).
The objective of the present study was to assess, using conventional meta-analysis and meta-regression techniques, the relationships between the Gly482Ser variant at PPARGC1A and measures of blood pressure and hypertension risk in published and unpublished material from 17 studies comprising 13,949 individuals. We also assessed the putative effect-modifying roles of age, sex, and body mass index (BMI) on the genotype-blood pressure relationships.
MATERIALS AND METHODS
Selection of Studies for Meta-Analysis
All of the studies published before April 2007 were identified by extended computer-based searches of PubMed databases. The following search terms were used: (“peroxisome proliferator activator receptor gamma coactivator 1” or “PPARGC1A” or “PGC-1” or “PGC-1alpha” or “PGC1alpha”) and “polymorphism” and (“hypertension” or “blood pressure”). The retrieved studies were then read in their entirety to assess their appropriateness for inclusion in the meta-analysis. All studies that tested the Gly482Ser polymorphism for association with blood pressure/hypertension were included in this meta-analysis. All of the references cited in the studies were also reviewed to identify additional published work not indexed by PubMed. Only studies in humans that have used validated genotyping methods were considered. Data from these studies were collated for meta-analysis, and, where necessary, the authors of the original paper were contacted to obtain the original data. We were unable to obtain the necessary data in all cases, because the corresponding author either did not respond to the request for data, or declined to provide access to all necessary original data (5, 6, 17, 18). Previously unpublished materials were included from four studies: three of which comprised white middle-aged adults from the UK, and the fourth comprised adult Pima Indians. The details of these studies are provided below.
Previously Unpublished Material
1) In the Medical Research Council (MRC) Ely study, 694 individuals with the necessary clinical data were genotyped from a population-based sampling frame from and around the city of Ely in Cambridgeshire, UK. The genetic (3) and clinical aspects (8) of the study have been described in detail previously. In brief, blood pressure was measured using an Accutorr automated sphygmomanometer (Datascope, Cambridge, UK) three times at 1-min intervals. The average of the three measurements was used in analyses. Height and weight were assessed using standard methods. A sample of fasting blood was drawn, and DNA was extracted from white blood cells.
2) In Cambridgeshire Type 2 diabetes Case-Control Study (CCCS), 507 individuals from the control group and 514 individuals from the case group of the CCCS underwent a standard clinical examination at Addenbrookes Hospital, Cambridge, UK. The genetic (3) and clinical aspects (12) of the study have been described in detail previously. In brief, systolic (SBP) and diastolic blood pressures (DBP) were measured using an Accutorr automated sphygmomanometer (Datascope, Cambridge, UK) on the right arm with the participant in the supine position. A sample of fasting blood was drawn, and DNA was extracted from white blood cells.
3) The European Prospective Investigation of Cancer (EPIC) Obesity case-cohort study is nested within the EPIC-Norfolk Study, a population-based cohort study of 25,663 Europid men and women, aged 40–79 yr, recruited in Norfolk, UK. The cases (n = 1,111 participants) were randomly selected from the obese individuals within this cohort and are defined as those with a BMI > 30 kg/m2. The control-cohort consists of 2,128 individuals randomly selected from the EPIC-Norfolk study, excluding those from the case group. All participants attended a clinic visit where height, weight, and waist circumference were measured using standard anthropometric techniques.
4) In the National Institutes of Health (NIH) Longitudinal Study of Pima Indians, 1,088 adult American Indians, primarily of Pima or Tohonto O'odam ancestry, in whom the necessary genetic and clinical data were available, were included in this study. The genetic (16) and clinical aspects (10) of the study have been described in detail previously. In brief, height and weight were measured with a rigid stadiometer and calibrated scales. SBP and DBP were measured in the supine position and on the right arm with an appropriately sized cuff for arm circumference. Blood pressures were recorded with a mercury gauge sphygmomanometer to the nearest 2 mmHg at the first and fourth Korotkoff sounds, respectively.
All participants provided written, informed consent, and ethical permission was granted by the local research ethics committees for all of the above-mentioned studies.
Statistical Analysis
Meta-analysis.
Two types of analyses were carried out for the meta-analysis of association between the sodium nitroprusside (SNP) and blood pressure traits. In the initial analyses, we obtained raw data, including age and sex, from 11 studies and tested for the interaction between the SNP and sex in each study and in the combined meta-analysis data set using generalized linear regression models. The means of blood pressures were adjusted for age and sex. We then meta-analyzed the data, including the five studies for which raw data were not available. To do this, we first simulated data points in these five studies without raw data using the mean and SD from each study and each genotype. A meta-analysis (ANOVA) was then undertaken combining the 5 simulated data sets and the 11 raw data sets. A random-effects generalized linear model was used if the heterogeneity statistic among studies was statistically significant; otherwise, a fixed-effects model was used. After all 10,000 simulations, the mean of the 10,000 meta-analysis P values was used to infer the final overall level of statistical significance. Hypertension was defined as ≥140 mmHg for SBPs or ≥90 mmHg for DBPs. The use of anti-hypertensive drug treatment for the definition of hypertension varied across studies. For the continuous trait models, individuals receiving blood pressure medication were excluded (n = 1,041). A meta-analysis for hypertension (a case-control meta-analysis) was performed where data permitted (in 12 studies). Owing to the absence of specific data on measurement methods in some studies and methodological restrictions in modeling these differences, we did not specifically account for measurement difference in these analyses. However, the stratified analyses performed here, combined with the random-effects meta-analysis method, control somewhat for heterogeneous measurement methods across studies. All analyses were undertaken in Stata/SE 9.2 for Windows (StataCorp LP). Power calculations were performed using the Quanto V1.1.1 software (14). The significance threshold was set at P < 0.05.
RESULTS
The sample size used in these analyses yields >95% power (α = 0.01) to detect a per allele association of at least 0.6 and 0.4 mmHg for SBP and DBP, respectively. The effect estimates are consistent with those previously reported in the Gly482Ser association studies on hypertension or blood pressure.
Table 1 shows the clinical characteristics of the study participants included from published and unpublished studies for the meta-analysis. In brief, the MRC Ely study comprised, on average, middle-aged overweight white men and women (∼58% female) with normal glucose control. The CCCS comprised overweight late middle-aged men and women (∼36% female) from Cambridgeshire in the UK, with or without Type 2 diabetes. The EPIC obesity case-cohort study comprised white late middle-aged men and women from the Eastern region of the UK. The NIH longitudinal study included young adult American Indians (∼55% female) from the Gila River Indian Reserve in southern Arizona, many of whom were overweight or obese (10).
Table 1.
Characteristics of the study participants used in the meta-analysis
| Cohort | n (Male/Female) | Age, yr | BMI, kg/m2 | SBP, mmHg | DBP, mmHg | Genotype Frequency (n) | Genotype Proportion, % | MAF, % |
|---|---|---|---|---|---|---|---|---|
| Danish whites | ||||||||
| Study 1 (Ref. 8)*† | 459/554 | 57.23 (7.08) | 25.9 (3.98) | 133.12 (17.98) | 84.07 (10.86) | 454/442/117 | 44.8/43.6/11.6 | 33.4 |
| Study 3 (Ref. 8)*† | 75/82 | 65.87 (5.12) | 25.22 (3.82) | 134.75 (19.09) | 77.65 (11.23) | 63/69/25 | 40.1/44.0/15.9 | 37.9 |
| Study 4 (Ref. 8)*† | 430/268 | 59.3 (9.65) | 29.3 (5.04) | 158.28 (23.38) | 95.14 (13.93) | 281/316/101 | 40.2/45.3/14.5 | 37.1 |
| Study 5 (Ref. 8)*† | 256/216 | 46.53 (12.38) | 24.26 (3.08) | 134.44 (19.12) | 76.57 (9.81) | 212/195/65 | 44.9/41.3/13.8 | 34.4 |
| UK whites | ||||||||
| MRC Ely† | 285/409 | 54.55 (10.3) | 26.41 (4.06) | 126.65 (16.59) | 76.41 (10.83) | 269/341/84 | 38.8/49.1/12.1 | 36.7 |
| CCCS cases† | 328/186 | 63.56 (7.82) | 29.71 (5.22) | 144.49 (17.05) | 86.53 (10.67) | 230/226/58 | 44.7/44/11.3 | 33.3 |
| CCCS controls† | 324/183 | 63.85 (7.83) | 27.27 (4.14) | 138.41 (17.68) | 84.93 (11.23) | 211/215/81 | 41.6/42.4/16 | 37.2 |
| Pima Indians† | 495/593 | 20.73 (13.9) | 25.66 (8.21) | 116.26 (20.4) | 68.16 (14.53) | 735/318/35 | 67.6/29.2/3.2 | 17.8 |
| French Canadians (Ref. 2)† | 77/189 | 42.4 (10.15) | 52.83 (9.98) | 142.66 (18.71) | 86.13 (11.87) | 105/122/39 | 39.5/45.9/14.6 | 37.6 |
| EPIC | ||||||||
| Obese† | 484/627 | 59.43 (8.77) | 32.90 (2.87) | 140.93 (17.3) | 86.45 (11.25) | 480/492/139 | 43.2/44.3/12.5 | 34.7 |
| Cohort† | 995/1,133 | 58.96 (8.94) | 26.57 (4.01) | 135.76 (17.56) | 82.73 (10.8) | 932/960/236 | 43.8/45.1/11.1 | 33.6 |
| Asian Indians | ||||||||
| Cases (Ref. 3)‡ | 238/329 | 50.0 (12.0) | 25.1 (4.2) | 128.0 (18.0) | 77.0 (11.0) | 303/212/52 | 53.4/37.4/9.2 | 27.9 |
| Control (Ref. 3)‡ | 399/576 | 44.0 (13.0) | 23.2 (4.4) | 120.0 (17.0) | 75.0 (10.0) | 534/360/81 | 54.8/36.9/8.3 | 26.8 |
| Danish whites (Ref. 4)‡ | 1,108/1,147 | 24.48 (4.27) | 931/1036/288 | 41.3/45.9/12.8 | 35.7 |
Values are unadjusted means (SD); n = 12,455 subjects. An additional 1,504 individuals, in whom quantitative trait data for blood pressure and body mass index (BMI) were collected, were also included in the hypertension meta-analysis. SBP, systolic blood pressure; DBP, diastolic blood pressure; MAF, minor allele frequency; CCCS, Cambridgeshire Type 2 diabetes Case-Control Study; MRC, Medical Research Council; EPIC, European Prospective Investigation of Cancer.
Studies 1, 3, 4, and 5 correspond to the study numbers mentioned in the paper.
Studies with raw data.
Studies from published paper. Mean, SD, and ANOVA P values were obtained from published papers or from authors.
In the pooled data set, the genotype distribution of the Gly482Ser polymorphism was 0.47, 0.42, and 0.11 for Gly482Gly, Gly482Ser, and Ser482Ser genotypes, respectively.
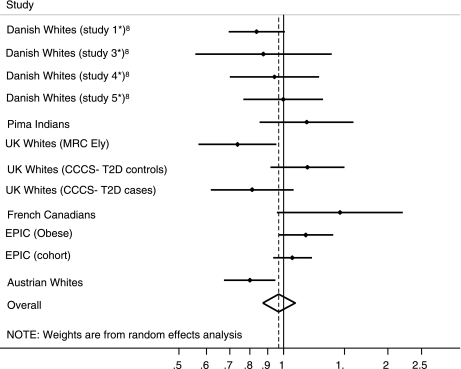
In the meta-analysis, the adjusted associations between Gly482Ser and blood pressure were found to be inverse (DBP: β = −0.13 mmHg per copy of the Ser482 allele, P = 0.333; SBP: β = −0.27 mmHg per copy of the Ser482 allele, P = 0.329), but were not statistically significant. The Gly482Ser variant was not associated with hypertension (Table 2), the overall odds ratio was 0.97, and the 95% confidence interval was 0.87–1.08 (P = 0.585; Fig. 1). The interactions between the SNP and sex from the 11 studies (Tables 3 and 4) were not statistically significant (SBP: P = 0.966; DBP: P = 0.715).
Table 2.
The distribution of Gly482Ser genotypes in people with and without hypertension, and odds ratios and heterogeneity results for the hypertension analyses
| Cohort | Genotype Frequencies |
OR | 95% CI | P Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Without Hypertension |
With Hypertension | |||||||||
| Gly/Gly | Gly/Ser | Ser/Ser | Gly/Gly | Gly/Ser | Ser/Ser | Lower | Upper | |||
| Danish whites | ||||||||||
| Study 1 (Ref. 8)*† | 242 | 253 | 73 | 212 | 189 | 44 | 0.84 | 0.70 | 1.01 | 0.061 |
| Study 3 (Ref. 8)*† | 33 | 47 | 13 | 30 | 22 | 12 | 0.88 | 0.56 | 1.38 | 0.565 |
| Study 4 (Ref. 8)*† | 45 | 42 | 20 | 236 | 274 | 81 | 0.94 | 0.70 | 1.27 | 0.694 |
| Study 5 (Ref. 8)*† | 127 | 121 | 38 | 85 | 74 | 27 | 1.00 | 0.77 | 1.30 | 0.992 |
| Pima Indians† | 641 | 272 | 29 | 94 | 46 | 6 | 1.17 | 0.86 | 1.59 | 0.331 |
| UK whites | ||||||||||
| MRC Ely† | 184 | 251 | 68 | 85 | 90 | 16 | 0.66 | 0.47 | 0.92 | 0.013 |
| CCCS cases† | 66 | 79 | 21 | 164 | 147 | 37 | 0.81 | 0.62 | 1.07 | 0.138 |
| CCCS controls† | 102 | 104 | 31 | 109 | 111 | 50 | 1.17 | 0.92 | 1.50 | 0.203 |
| French Canadians (Ref. 2)† | 30 | 31 | 5 | 75 | 91 | 34 | 1.46 | 0.96 | 2.21 | 0.079 |
| EPIC | ||||||||||
| Obese† | 180 | 163 | 44 | 300 | 329 | 95 | 1.16 | 0.97 | 1.39 | 0.111 |
| Cohort† | 490 | 482 | 119 | 442 | 478 | 117 | 1.06 | 0.93 | 1.21 | 0.355 |
| Austrian whites (Ref. 7) | 286 | 315 | 103 | 231 | 231 | 47 | 0.80 | 0.67 | 0.95 | 0.010 |
| Overall | 0.97 | 0.87 | 1.08 | 0.585 | ||||||
n = 4,711 people with hypertension; n = 5,150 people without hypertension. OR, odds ratio; CI, confidence interval. P value for heterogeneity = 0.006.
Studies 1, 3, 4, and 5 correspond to the study numbers mentioned in the paper.
Studies with raw data.
Fig. 1.
The horizontal axis shows odds ratio estimates with the corresponding 95% confidence interval (CI) for the Gly482Ser allele and the risk of hypertension (n = 9,861). The 95% CIs of pooled estimates are displayed as a horizontal line through the dots. *Studies 1, 3, 4, and 5 correspond to the study numbers mentioned in the paper by Franks et al. (8). T2D, Type 2 diabetes; MRC, Medical Research Council; CCCS, Cambridgeshire Type 2 diabetes Case Control Study; EPIC, European Prospective Investigation of Cancer.
Table 3.
Meta-analysis results for DBP stratified by PPARGC1A Gly482Ser genotypes
| Cohort | DBP, mmHg |
ANOVA, P Value | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype Frequencies |
Gly/Gly | Gly/Ser | Ser/Ser | Significance of Interaction | |||||||||
| Gly/Gly | Gly/Ser | Ser/Ser | Mean | 95% CI | Mean | 95% CI | Mean | 95% CI | Gene-Sex | Gene-Age | Gene-BMI | ||
| Danish whites | |||||||||||||
| Study 1 (Ref. 8)*† | 454 | 442 | 117 | 84.4 | 83.4–85.4 | 84.1 | 83.1–85.1 | 83.0 | 81.1–84.9 | 0.467 | 0.575 | 0.759 | 0.359 |
| Study 3 (Ref. 8)*† | 63 | 69 | 25 | 78.7 | 75.9–81.5 | 76.8 | 74.1–79.5 | 77.4 | 73.0–81.8 | 0.601 | 0.053 | 0.659 | 0.776 |
| Study 4 (Ref. 8)*† | 281 | 316 | 101 | 95.3 | 93.7–96.9 | 95.7 | 94.2–97.2 | 92.7 | 90.0–95.4 | 0.159 | 0.613 | 0.711 | 0.279 |
| Study 5 (Ref. 8)*† | 212 | 195 | 65 | 76.7 | 75.4–78 | 75.7 | 74.4–77.0 | 78.7 | 76.4–81.0 | 0.080 | 0.463 | 0.971 | 0.467 |
| Pima Indians† | 735 | 318 | 35 | 67.9 | 67.1–68.7 | 68.5 | 67.2–69.8 | 70.5 | 66.6–74.4 | 0.363 | 0.924 | 0.593 | 0.504 |
| UK whites | |||||||||||||
| MRC Ely† | 233 | 306 | 73 | 77.0 | 75.7–78.3 | 75.9 | 74.8–77.0 | 73.2 | 70.9–75.5 | 0.018 | 0.251 | 0.532 | 0.397 |
| CCCS cases† | 175 | 176 | 43 | 86.3 | 84.8–87.8 | 85.1 | 83.6–86.6 | 85.8 | 82.7–88.9 | 0.541 | 0.236 | 0.214 | 0.327 |
| CCCS controls† | 194 | 197 | 75 | 83.9 | 82.4–85.4 | 84.6 | 83.0–86.2 | 87.0 | 84.5–89.5 | 0.131 | 0.113 | 0.266 | 0.593 |
| French Canadians (Ref. 2)† | 64 | 73 | 19 | 84.6 | 82.1–87.1 | 87.5 | 85.2–89.8 | 89.0 | 84.4–93.6 | 0.137 | 0.085 | 0.4 | 0.19 |
| EPIC | |||||||||||||
| Obese† | 364 | 339 | 96 | 86.3 | 85.2–87.4 | 86.1 | 85.0–87.2 | 86.0 | 83.9–88.1 | 0.973 | 0.051 | 0.188 | 0.854 |
| Cohort† | 762 | 789 | 201 | 81.9 | 81.2–82.6 | 82.0 | 81.3–82.7 | 82.5 | 81.1–83.9 | 0.805 | 0.878 | 0.59 | 0.858 |
| Danish whites (Ref. 4)‡ | 931 | 1,036 | 288 | 81.8 | 81.1–82.5 | 82.3 | 81.7–82.9 | 82.0 | 80.7–83.3 | 0.39 | |||
| Asian Indians | |||||||||||||
| Controls (Ref. 3)‡ | 534 | 360 | 81 | 76.0 | 75.2–76.8 | 75.0 | 74.0–76.0 | 75.0 | 72.8–77.2 | 0.6 | |||
| Cases (Ref. 3)‡ | 303 | 212 | 52 | 79.0 | 77.8–80.2 | 77.0 | 75.5–78.5 | 75.0 | 72.3–77.7 | 0.007 | |||
| Slovenian whites | |||||||||||||
| Cases (Ref. 5)‡ | 114 | 118 | 29 | 84.7 | 83–86.4 | 85.7 | 83.8–87.6 | 85.0 | 82.5–87.5 | 0.74 | |||
| Controls (Ref. 5)‡ | 14 | 15 | 1 | 83.8 | 78.1–89.5 | 84.8 | 78.5–91.1 | 84.0 | 0.7 | ||||
| Overall | 5,433 | 4,961 | 1,301 | 80.3 | 80–80.6 | 81.5 | 81.2–81.8 | 82.1 | 81.5–82.7 | 0.651§ | 0.715 | 0.00004 | 0.953 |
| Standard error | 0.005§ | ||||||||||||
| Cohort heterogeneity P | 0.021§ | ||||||||||||
| Ethnicity heterogeneity P | 0.414§ | ||||||||||||
| Caucasian only | |||||||||||||
| Overall | 3,861 | 4,071 | 1,133 | 83.4 | 83.0–83.7 | 83.3 | 83.0–83.7 | 83.3 | 82.7–83.9 | 0.659§ | 0.700 | 0.789 | 0.348 |
| Heterogeneity P | 0.109§ | ||||||||||||
Studies 1, 3, 4, and 5 correspond to the study numbers mentioned in the paper.
Studies with raw data. The main effect and interaction analyses are adjusted for age and sex.
Studies from published paper. Mean, SD, and ANOVA P values were obtained from published papers or from authors.
Based on 10,000 simulations. A random-effect generalized linear model is applied if P < 0.05 for heterogeneity; otherwise, a fixed model is applied.
Table 4.
Meta-analysis results for SBP stratified by PPARGC1A Gly482Ser genotypes
| Cohort | SBP, mmHg |
ANOVA, P Value | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype Frequencies |
Gly/Gly | Gly/Ser | Ser/Ser | Significance of Interaction | |||||||||
| Gly/Gly | Gly/Ser | Ser/Ser | Mean | 95% CI | Mean | 95% CI | Mean | 95% CI | Gene-Sex | Gene-Age | Gene-BMI | ||
| Danish whites | |||||||||||||
| Study 1 (Ref. 8)*† | 454 | 442 | 117 | 133.9 | 132.3–135.5 | 133.3 | 131.7–134.9 | 129.6 | 126.5–132.7 | 0.055 | 0.189 | 0.571 | 0.891 |
| Study 3 (Ref. 8)*† | 63 | 69 | 25 | 136.2 | 131.6–140.8 | 131.6 | 127.2–136 | 139.8 | 132.4–147.2 | 0.131 | 0.116 | 0.477 | 0.581 |
| Study 4 (Ref. 8)*† | 281 | 316 | 101 | 157.5 | 154.9–160.1 | 160.0 | 157.6–162.4 | 154.9 | 150.6–159.2 | 0.101 | 0.106 | 0.988 | 0.574 |
| Study 5 (Ref. 8)*† | 212 | 195 | 65 | 135.0 | 132.6–137.4 | 133.7 | 131.2–136.2 | 134.8 | 130.5–139.1 | 0.748 | 0.653 | 0.305 | 0.113 |
| Pima Indians† | 735 | 318 | 35 | 115.8 | 114.6–117 | 116.8 | 114.9–118.7 | 121.6 | 116–127.2 | 0.103 | 0.082 | 0.901 | 0.036 |
| UK whites | |||||||||||||
| MRC Ely† | 233 | 306 | 73 | 127.6 | 125.7–129.5 | 125.2 | 123.5–126.9 | 121.2 | 117.8–124.6 | 0.005 | 0.203 | 0.123 | 0.632 |
| CCCS cases† | 175 | 176 | 43 | 138.4 | 135.9–140.9 | 137.4 | 134.9–139.9 | 141.4 | 136.3–146.5 | 0.197 | 0.031 | 0.336 | 0.439 |
| CCCS controls† | 194 | 197 | 75 | 145.5 | 143.1–147.9 | 143.7 | 141.3–146.1 | 143.8 | 140–147.6 | 0.484 | 0.147 | 0.292 | 0.791 |
| French Canadians (Ref. 2)† | 64 | 73 | 19 | 142.0 | 137.4–146.6 | 144.6 | 140.3–148.9 | 137.8 | 129.4–146.2 | 0.131 | 0.625 | 0.173 | 0.031 |
| EPIC | |||||||||||||
| Obese† | 364 | 339 | 96 | 138.8 | 137.2–140.4 | 141.0 | 139.3–142.7 | 139.1 | 135.9–142.3 | 0.183 | 0.043 | 0.015 | 0.695 |
| Cohort† | 762 | 789 | 201 | 134.1 | 133–135.2 | 134.0 | 132.9–135.1 | 134.4 | 132.2–136.6 | 0.947 | 0.804 | 0.556 | 0.654 |
| Danish whites (Ref. 4)‡ | 931 | 1,036 | 288 | 128.4 | 127.2–129.6 | 129.6 | 128.4–130.8 | 129.5 | 127.1–131.9 | 0.09 | |||
| Asian Indians | |||||||||||||
| Controls (Ref. 3)‡ | 534 | 360 | 81 | 120.0 | 118.6–121.4 | 121.0 | 119.2–122.8 | 119.0 | 115.7–122.3 | 0.42 | |||
| Cases (Ref. 3)‡ | 303 | 212 | 52 | 130.0 | 127.9–132.1 | 126.0 | 123.8–128.2 | 124.0 | 119.7–128.3 | 0.006 | |||
| Slovenian whites | |||||||||||||
| Cases (Ref. 5)‡ | 114 | 118 | 29 | 144.6 | 140.5–148.7 | 146.5 | 142.5–150.5 | 141.0 | 132.8–149.2 | 0.45 | |||
| Controls (Ref. 5)‡ | 14 | 15 | 1 | 143.8 | 133.1–154.5 | 145.2 | 133.1–157.3 | 142.0 | 0.6 | ||||
| Overall | 5,433 | 4,961 | 1,301 | 131.0 | 130.5–131.5 | 133.1 | 132.6–133.6 | 133.5 | 132.5–134.5 | 0.409§ | 0.966 | 0.026 | 0.202 |
| Standard error | 0.006§ | ||||||||||||
| Cohort heterogeneity P | 0.003§ | ||||||||||||
| Ethnicity heterogeneity P | 0.649§ | ||||||||||||
| Caucasian only | |||||||||||||
| Overall | 3,861 | 4,071 | 1,133 | 135.6 | 135.0–136.1 | 135.8 | 135.2–136.3 | 135.3 | 134.2–136.4 | 0.419§ | 0.960 | 0.804 | 0.138 |
| Heterogeneity P | 0.035§ | ||||||||||||
Studies 1, 3, 4, and 5 correspond to the study numbers mentioned in the paper.
Studies with raw data. The main effect and interaction analyses are adjusted for age and sex.
Studies from published paper. Mean, SD, and ANOVA P values were obtained from published papers or from authors.
Based on 10,000 simulations. A random-effect generalized linear model is applied if P < 0.05 for heterogeneity; otherwise, a fixed model is applied.
We observed significant between-study heterogeneity for both SBP (P = 0.003) and DBP (P = 0.021), which was attributable to three of the cohorts [Danish whites (2) (study 5), UK whites (CCCS cases), and Pima Indians]. Excluding these studies from the meta-analysis resolved the between-study heterogeneity, but the associations between the Gly482Ser genotype and SBP or DBP remained nonsignificant (data not shown).
Ethnic-Specific Analyses
To reduce the possibility that our results were confounded by population stratification, we undertook subanalyses in whites only. In these models, we found no evidence of association for any blood pressure trait (Tables 2–4).
Interaction Analyses
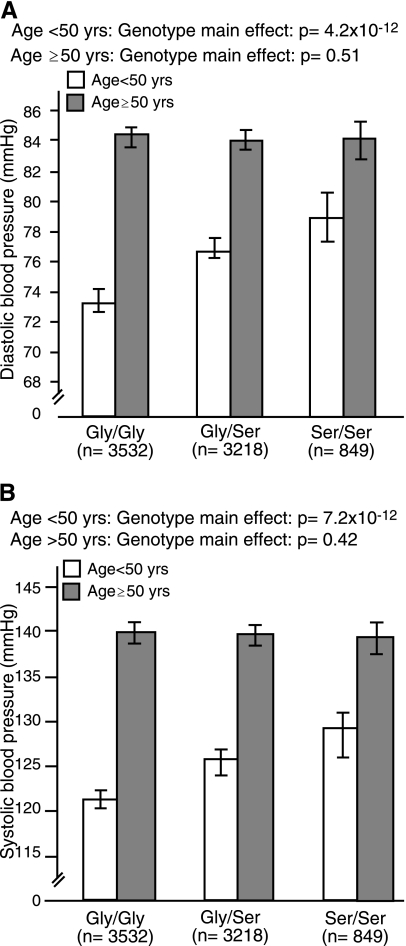
Previous studies have suggested that the effects of PPARGC1A may be modified by age, sex, or obesity. Therefore, we tested genotype interaction terms for age, sex, and BMI. The results of these analyses are shown in Tables 3 and 4. We found no evidence of genotype interactions with sex or BMI. However, within the entire collection of cohorts, we observed a highly statistically significant interaction between genotype and age on DBP (P < 0.0001), and a nominal statistical interaction between genotype and age on SBP (P = 0.026). We proceeded by stratifying the data sets by below (n = 2,511) and above (n = 5,088) age 50 yr and repeating the main effects models. In these analyses, strong genotype associations were evident for SBP [Gly482Gly: 121.4 (120.4, 122.5), Gly482Ser: 125.9 (124.6, 127.1), Ser482Ser: 129.2 (126.5, 131.9), P = 7.20 × 10−12] and DBP [Gly482Gly: 73.5 (72.8, 74.2), Gly482Ser: 77.0 (76.2, 77.8), Ser482Ser: 79.1 (77.4, 80.9), P = 4.20 × 10−12] in younger individuals, and no statistical association was evident in older individuals [SBP: Gly482Gly: 139.9 (139.1, 140.7), Gly482Ser: 139.6 (138.8, 140.4), Ser482Ser: 139.2 (137.7, 140.8), P = 0.41; DBP: Gly482Gly: 84.7 (84.1, 85.2), Gly482Ser: 84.4 (83.8, 84.9), Ser482Ser: 84.5 (83.5, 85.5), P = 0.51] (Fig. 2). When these analyses were repeated in whites only, the interaction terms were no longer statistically significant, which may reflect lower power or confounding by population stratification.
Fig. 2.
Association between the Gly482Ser genotype and diastolic (A) and systolic (B) blood pressure in younger (<50 yr; n = 2,511) and older (>50 yr; n = 5,088) individuals.
DISCUSSION
In European whites, a significant inverse association between the Ser482 allele and hypertension risk has been reported (2, 17). By contrast, the Ser482 allele was associated with an increased risk of hypertension in French men (6) and in Argentinean adolescents (21), and no statistically significant association was evident in Chinese (5), Asian Indians (24), or in Danish whites with the metabolic syndrome (1). To clarify the nature of the relationship between the PPARGC1A Gly482Ser genotype and blood pressure, we undertook a meta-analysis of 13,949 individuals.
Our study included more than 6,000 previously unreported observations and was sufficiently powered to detect effects of lesser magnitude than those previously reported. In our study, we did not observe direct evidence of association between the Gly482Ser genotype and measures of blood pressure. However, a robust statistical gene-age interaction was observed for DBP, and a nominal statistical interaction was observed for SBP, suggesting that the effect of Gly482Ser on blood pressure diminishes with age. It is possible that these observations represent interactions between the PPARGC1A gene and factors associated with younger age, which may include physical activity, a factor that biologically interacts with PPARGC1A (11). It is also possible that genetic effects are more evident at younger ages, owing to the accumulative exposure to environmental risk factors for high blood pressure with age. Alternatively, it may be that cohort effects influence the association of Gly482Ser with blood pressure. Finally, it is possible that survivor bias or treatment effect may inhibit the detection of genetic effects in older individuals.
It is important to highlight that, when the gene × age interaction analyses were conducted in whites only, no statistical evidence of interaction was observed; this may reflect the lower level of statistical power in this subgroup or that the gene-age interaction was attributable to population stratification. The latter is a type of confounding that can occur when genetic associations are tested in pooled samples of diverse ethnic groups. Confounding by population stratification can emerge when disease frequencies differ substantially between ethnic groups, and the genotype being studied is associated with ethnicity. It is possible in this scenario that an apparent association between genotype and the disease is attributable purely to confounding by ethnicity. Thus some prior reports of association between Gly482Ser and hypertension may be limited by this factor. The meta-analytic approach differs from conventional tests of genetic association because the point estimates are first calculated within each subgroup before being combined. By consequence, confounding by population stratification is less likely in a meta-analysis than in a conventional pooled-sample analysis. Thus it is plausible that the absence of an interaction effect in the present study when excluding non-whites from the analyses is due to reduced statistical power rather than confounding by ethnic factors.
Despite moderate-to-high heritability estimates for blood pressure, the recently published genomewide scans have failed to detect, with any degree of robustness, genetic risk factors for this trait (26). This suggests that, although blood pressure is no doubt under genetic control, the specific nature of the trait and/or the way it is assessed in epidemiological studies has seriously hindered the elucidation of the trait's genetic mechanisms. The methodological factors that may impede the detection of genetic associations with blood pressure are many. For example, high blood pressure is a commonly treated condition; if people carrying the Gly482 allele are truly more likely to have elevated blood pressure, these people will also be more likely to be treated for the condition. The differential blood pressure treatment by Gly482Ser genotype would reduce the magnitude of the association between genotype and blood pressure. Furthermore, blood pressure is a highly biologically variable trait and is hence prone to regression dilution, a feature that inherently limits statistical power to detect genetic associations with this trait (13). It is also worth considering that the publications reporting the results from two studies that found significant associations between Gly482Ser and hypertension (18, 25) did not include sufficient continuous trait data for those studies to be included in the blood pressure meta-analysis. Thus we cannot be certain that, with the inclusion of data from those studies, the results would remain the same. A further key consideration, which is supported by the significant gene-age interactions in this study, is that interactions between genetic and environmental factors inhibit the detection of genetic effects on blood pressure.
This is the largest study to date examining the association between the Gly482Ser genotype and blood pressure. Our analyses suggest that Gly482Ser may be associated with blood pressure in younger but not older individuals, such that Ser482 allele homozygotes have DBP and SBP roughly 5 and 9 mmHg higher, respectively, than Gly482 allele homozygotes. These observations contrast the findings of several recent blood pressure studies but are consistent with studies of this locus in relation to other metabolic traits. The recently published results from genomewide association studies have revealed no convincing genetic risk factors for high blood pressure (26). This contrasts the successful detection of genetic risk factors for many other complex disease traits (26). Animal functional studies and human heritability studies suggest that blood pressure is under genetic control. Therefore, it is likely that studies that are specifically designed to overcome the limitations inherent in blood pressure research will be necessary if the genetic basis of this disease trait is to be elucidated.
DISCLOSURES
I. Barroso owns stock in Glaxo SmithKline and Incyte.
GRANTS
The National Institutes of Health Longitudinal Study of Pima Indians was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases. The Medical Research Council (MRC) Ely Study and the Cambridgeshire Case Control Study were support with grants from the UK MRC and the Wellcome Trust (to N. J. Wareham). Several additional aspects of this study were supported by the Wellcome Trust, Västerbottens ALF Committee (Strategic appointment 2006–2009 to P. W. Franks), Novo Nordisk (370579201 to P. W. Franks), the Swedish Heart and Lung Foundation (20070633 to P. W. Franks), and the Swedish Diabetes Association (DIA2006-013 to P. W. Franks).
Acknowledgments
We thank the volunteers who took part in the studies included in this report.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Ambye L, Rasmussen S, Fenger M, Jorgensen T, Borch-Johnsen K, Madsbad S, Urhammer SA. Studies of the Gly482Ser polymorphism of the peroxisome proliferator-activated receptor gamma coactivator 1alpha (PGC-1alpha) gene in Danish subjects with the metabolic syndrome. Diabetes Res Clin Pract 67: 175–179, 2005. [DOI] [PubMed] [Google Scholar]
- 2.Andersen G, Wegner L, Jensen DP, Glumer C, Tarnow L, Drivsholm T, Poulsen P, Hansen SK, Nielsen EM, Ek J, Mouritzen P, Vaag A, Parving HH, Borch-Johnsen K, Jorgensen T, Hansen T, Pedersen O. PGC-1alpha Gly482Ser polymorphism associates with hypertension among Danish whites. Hypertension 45: 565–570, 2005. [DOI] [PubMed] [Google Scholar]
- 3.Barroso I, Luan J, Middelberg RP, Harding AH, Franks PW, Jakes RW, Clayton D, Schafer AJ, O'Rahilly S, Wareham NJ. Candidate gene association study in type 2 diabetes indicates a role for genes involved in beta-cell function as well as insulin action. PLoS Biol 1: E20, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barroso I, Luan J, Sandhu M, Franks PW, Crowley V, Schafer A, O'Rahilly S, Wareham N. Meta-analysis of the Gly482Ser variant in PPARGC1A in type 2 diabetes and related phenotypes. Diabetologia 49: 501–505, 2006. [DOI] [PubMed] [Google Scholar]
- 5.Chen S, Yan W, Huang J, Yang W, Gu D. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha polymorphism is not associated with essential hypertension and type 2 diabetes mellitus in Chinese population. Hypertens Res 27: 813–820, 2004. [DOI] [PubMed] [Google Scholar]
- 6.Cheurfa N, Reis AF, Dubois-Laforgue D, Bellanne-Chantelot C, Timsit J, Velho G. The Gly482Ser polymorphism in the peroxisome proliferator-activated receptor-gamma coactivator-1 gene is associated with hypertension in type 2 diabetic men. Diabetologia 47: 1980–1983, 2004. [DOI] [PubMed] [Google Scholar]
- 7.Ek J, Andersen G, Urhammer SA, Gaede PH, Drivsholm T, Borck-Johnsen K, Hansen T, Pedersen O. Mutation analysis of peroxisome proliferator-activated receptor-gamma coactivator-1 (PGC-1) and relationships of identified amino acid polymorphisms to Type II diabetes mellitus. Diabetologia 44: 2220–2226, 2000. [DOI] [PubMed] [Google Scholar]
- 8.Franks PW, Bhattacharyya S, Luan J, Montague C, Brennand J, Challis B, Brage S, Ekelund U, Middelberg RP, O'Rahilly S, Wareham NJ. Association between physical activity and blood pressure is modified by variants in the G-protein coupled receptor 10. Hypertension 43: 224–228, 2004. [DOI] [PubMed] [Google Scholar]
- 9.Franks PW, Ekelund U, Luan J, Brage S, Schaeffer PJ, O'Rahilly S, Barroso I, Wareham NJ. PPARGC1A coding variation may initiate impaired non-esterified fatty acid clearance during glucose challenge. Diabetologia 50: 569–573, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franks PW, Knowler WC, Nair S, Koska J, Lee YH, Lindsay RS, Walker BR, Looker HC, Permana PA, Tataranni PA, Hanson RL. Interaction between an 11betaHSD1 gene variant and birth era modifies the risk of hypertension in Pima Indians. Hypertension 44: 681–688, 2004. [DOI] [PubMed] [Google Scholar]
- 11.Franks PW, Loos RJ. PGC-1alpha gene and physical activity in type 2 diabetes mellitus. Exerc Sport Sci Rev 34: 171–175, 2006. [DOI] [PubMed] [Google Scholar]
- 12.Franks PW, Luan J, Barroso I, Brage S, Sanchez JLG, Ekelund U, Rios MS, Schafer AJ, O'Rahilly S, Wareham NJ. Variation in the eNOS gene modifies the association between total energy expenditure and glucose intolerance. Diabetes 54: 2795–2801, 2005. [DOI] [PubMed] [Google Scholar]
- 13.Franks PW White-coat hypertension and risk of stroke: do the data really tell us what we need to know? Hypertension 45: 183–184, 2005. [DOI] [PubMed] [Google Scholar]
- 14.Gauderman WJ, Morrison JM. QUANTO 1.1: a computer program for power and sample size calculations for genetic-epidemiology studies (Online). http://hydra.usc.edu/gxe [2006].
- 15.Lucia A, Gomez-Gallego F, Barroso I, Rabadan M, Bandres F, San Juan AF, Chicharro JL, Ekelund U, Brage S, Earnest CP, Wareham NJ, Franks PW. PPARGC1A genotype (Gly482Ser) predicts exceptional endurance capacity in European men. J Appl Physiol 99: 344–348, 2005. [DOI] [PubMed] [Google Scholar]
- 16.Muller YL, Bogardus C, Pedersen O, Baier L. A Gly482Ser missense mutation in the peroxisome proliferator-activated receptor gamma coactivator-1 is associated with altered lipid oxidation and early insulin secretion in Pima Indians. Diabetes 52: 895–898, 2003. [DOI] [PubMed] [Google Scholar]
- 17.Oberkofler H, Holzl B, Esterbauer H, Xie M, Iglseder B, Krempler F, Paulweber B, Patsch W. Peroxisome proliferator-activated receptor-gamma coactivator-1 gene locus: associations with hypertension in middle-aged men. Hypertension 41: 368–372, 2003. [DOI] [PubMed] [Google Scholar]
- 18.Petrovic MG, Kunej T, Peterlin B, Dovc P, Petrovic D. Gly482Ser polymorphism of the peroxisome proliferator-activated receptor-gamma coactivator-1 gene might be a risk factor for diabetic retinopathy in Slovene population (Caucasians) with type 2 diabetes and the Pro12Ala polymorphism of the PPARgamma gene is not. Diabetes Metab Res Rev 21: 470–474, 2005. [DOI] [PubMed] [Google Scholar]
- 19.Ridderstrale M, Johansson LE, Rastam L, Lindblad U. Increased risk of obesity associated with the variant allele of the PPARGC1A Gly482Ser polymorphism in physically inactive elderly men. Diabetologia 49: 496–500, 2006. [DOI] [PubMed] [Google Scholar]
- 20.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316: 1331–1336, 2007. [DOI] [PubMed] [Google Scholar]
- 21.Sookoian S, Garcia SI, Porto PI, Dieuzeide G, Gonzalez CD, Pirola CJ. Peroxisome proliferator-activated receptor gamma and its coactivator-1 alpha may be associated with features of the metabolic syndrome in adolescents. J Mol Endocrinol 35: 373–380, 2005. [DOI] [PubMed] [Google Scholar]
- 22.Soyal S, Krempler F, Oberkofler H, Patsch W. PGC-1alpha: a potent transcriptional cofactor involved in the pathogenesis of type 2 diabetes. Diabetologia 49: 1477–1488, 2006. [DOI] [PubMed] [Google Scholar]
- 23.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 127: 397–408, 2006. [DOI] [PubMed] [Google Scholar]
- 24.Vimaleswaran KS, Radha V, Ghosh S, Majumder PP, Deepa R, Babu HN, Rao MR, Mohan V. Peroxisome proliferator-activated receptor-gamma co-activator-1alpha (PGC-1alpha) gene polymorphisms and their relationship to Type 2 diabetes in Asian Indians. Diabet Med 22: 1516–1521, 2005. [DOI] [PubMed] [Google Scholar]
- 25.Vohl MC, Houde A, Lebel S, Hould FS, Marceau P. Effects of the peroxisome proliferator-activated receptor-gamma co-activator-1 Gly482Ser variant on features of the metabolic syndrome. Mol Genet Metab 86: 300–306, 2005. [DOI] [PubMed] [Google Scholar]
- 26.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]