Abstract
Molecular imaging is a rapidly emerging research tool and clinical discipline aimed at noninvasive, quantitative visualization of in vivo molecular processes occurring at cellular and subcellular levels. At present, advancement of the molecular imaging field is driven by the development of improved imaging hardware for use in preclinical and clinical settings, the identification and validation of new, biologically relevant imaging targets, and the development of improved imaging probes derived from novel chemistries. Of these 3 essential facets, which comprise a majority of current molecular imaging research, hardware development and novel target discovery significantly outpace the development and clinical advancement of new molecular imaging probes, particularly with respect to cancer imaging.
Keywords: molecular imaging, imaging probe discovery, HTS
To date, several imaging probes that aim to measure fundamental biologic processes known to be dysregulated in tumors, including glucose use, proliferation, apoptosis, hypoxia, and angiogenesis, have been described (1). Many of these imaging probes have been used fairly extensively in the preclinical setting, and some have been used to a lesser extent clinically. In reality, however, molecular imaging in clinical oncology typically consists of the assessment of glucose use with 18F-FDG PET. The regulation of glycolysis is a complicated process involving several biologic factors capable of influencing the overall glycolytic pathway; therefore, 18F-FDG PET studies must be interpreted with some caution, because imaging results can vary significantly depending on the tumor type, stage, and cellular response to a therapeutic intervention. Given the current emphasis on the development of sophisticated molecularly targeted and individualized therapeutic regimens for the treatment of cancer, expanding the oncologist’s imaging repertoire to include probes capable of reporting more specific molecular events and relevant downstream cellular physiology may be of considerable clinical relevance. For these reasons, there is significant interest in the development and validation of additional novel imaging probes that have the potential to contribute insights into tumor biology and the therapeutic response that may extend beyond what can be learned with currently existing tracers, such as 18F-FDG.
The development, validation, and subsequent advancement to the clinic of new molecular imaging probes is a time-consuming, expensive, and multidisciplinary process. Accordingly, there exists a considerable bottleneck between the discovery of new targets and the subsequent realization of imaging probes to interrogate them. As a result, alternative technologies, such as high-throughput, small-molecule screening (HTS), are being explored as vehicles to accelerate the discovery phase of imaging probe development. Compound-enabling infrastructure, such as combinatorial chemistry, diversity-oriented synthesis, and phage display techniques, has matured rapidly in recent years and has led to the assembly of vast, chemically diverse, small-molecule libraries, many containing hundreds of thousands of representative compounds. Paired with state-of-the-art HTS technology, these systems have been widely used in the pharmaceutical industry and, more recently, academia to identify potential drug candidates, although similar methodology also could be applied to molecular imaging probe discovery.
The overall aims of this minireview are to provide a brief introduction to the salient features of HTS and to illustrate the possibilities that may exist for the use of HTS as a platform for accelerating imaging probe discovery. Because the specific implementation of HTS tends to be dictated by the fluidity of rapidly evolving technology, this discussion generally emphasizes the more universal aspects of HTS, such as the types of screening assays possible and typical model systems that may be used, rather than specific details of hardware and instrumentation.
COMPOUNDS AND COMPOUND LIBRARIES
Given their generally favorable biodistribution properties and ability to modulate numerous physiologic processes invitro and in vivo, small molecules are attractive candidates for the development of new drugs as well as potential new imaging probes. Significant progress has been made in recent years toward the rapid assembly of vast libraries of relatively small biopolymers, such as peptides and small, druglike organic molecules. Solid-phase parallel-synthesis methodology, as pioneered by Merrifield (2), initially led to the assembly of peptide libraries and demonstrated the feasibility of high-throughput chemical synthesis. On the basis of this methodology, in more recent years, combinatorial approaches, such as split-pool synthesis, which enable tens of thousands of peptides to be synthesized simultaneously rather than individually in parallel, have increased the rate at which extremely large, diverse peptide libraries can be prepared (3). In a similar fashion, combinatorial methodology has been elegantly applied to chemically diverse nonpeptidic small molecules, as illustrated by the work of Schreiber (4) and Wong et al. (5). In particular, increasing emphasis has been placed on synthesizing structurally complex compounds that more closely reflect the chemical diversity of natural products as a means to improve the quality of compound collections and the likelihood of generating hits. As a result of these developments in chemistry, the rate at which suitably diverse and biologically relevant compound libraries can be assembled and the prospects of identifying meaningful compounds from these collections are now quite high.
BIOLOGIC PLATFORMS FOR HTS
Many model systems that emulate a wide variety of in vivo molecular events can be adapted for HTS. Factors that contribute to the selection of an HTS model system include the intended use of identified candidate molecules (e.g., drug, model compound, or imaging probe), the availability of appropriately validated biologic models (e.g., purified target substrates, cell lines, or model organisms), and cost. Contemporary HTS usually involves biologic models that can be defined broadly as protein-based, cell-based, and organism-based systems (6). Although the specific details of individual assays, such as scale and method of detection, can vary widely, the 3 classes of systems generally are distinguished by their level of reductionism. Screening can be approached with either forward or reverse bias. In forward screening, assays are established to identify candidate compounds with specificity for a phenotype of interest, rather than focusing on a specific, predefined target. Alternatively, reverse screening is designed to seek candidate compounds with specificity for a known target. By convention, reverse screening is more focused than forward screening; this fact can have advantages and drawbacks, depending on the hypothesis being tested. Given that the selection of an HTS model system typically involves some level of compromise among relevance, feasibility, and cost, the salient features and inherent strengths and weaknesses of each class are considered (Fig. 1).
FIGURE 1.
Schematic illustration showing various attributes of HTS models. Adapted from (7).
Protein-Based Assays
Among the most basic of model formats for HTS are protein-based assays. These assays typically are designed to detect protein–ligand interactions with the overall goal of identifying candidate compounds with specificity for a target protein. These target-based assays can be deployed in several ways, but typically candidate compounds and the target protein are dispensed freely in solution or, more commonly, either the compounds or the protein is immobilized on a solid support, such as an inert polymer bead, glass surface, or multiwell plate. For the latter, significant progress has been made toward developing sophisticated microarray technologies that involve the use of either surface-bound compounds, which can be either small molecules or peptides, or surface-bound target proteins, which are immobilized on spatially discrete microspot arrays (6). In these systems, screening takes place via incubation of fabricated microarrays with mobile binding partners, and hits are detected with a variety of techniques, ranging from fluorescence to surface plasmon resonance.
The pure simplicity and comparatively low cost of protein-based screening may be attractive for assays in which a validated target in a disease state has been identified and there is significant understanding of the target biochemistry in solution or on an immobilized support. An inherent drawback to these simple model assays is that they ignore the biologic complexity of intact cells or organ systems, so that there is considerable potential for hits derived from these types of assays to be irrelevant. For example, compounds demonstrating an excellent affinity in biochemical assays may have significant toxicity or be insufficiently internalized to access intracellular targets. Therefore, apart from the intangible biologic milieu of cell-based and organism-based systems, protein-based models are less capable of predicting the ultimate on-target activity of candidate compounds in vivo than are more sophisticated models.
Cell-Based Assays
HTS assays with intact cells are necessarily more complicated and costly than protein-based assays. However, a significant advantage of cell-based screening over protein-based screening is predicated on the fact that many genomic and proteomic events that are known to occur in disease states in vivo can be recapitulated, to some extent, in cultured cells. Accordingly, the interplay that occurs between targets of HTS and other, related and unrelated intracellular and extracellular milieus may afford a significantly higher degree of biologic relevance to cell-based screening, in comparison with the purely biochemical nature of protein-based screening. Cell-based screening can be approached with either forward or reverse bias and can be implemented with several formats, although most incorporate the use of multiwell plates as miniaturized propagation vessels. Cell-based screening routinely incorporates the use of reporter genes, immunofluorescence, or optical imaging assays to detect morphologic or phenotypic responses to candidate compounds.
Limitations associated with cell-based screening include the requirement for suitable cell lines expressing the target of interest. Additionally, cultured cell lines propagated on plastic surfaces as model systems neglect the effects of surrounding stroma and microenvironment on disease and therefore may not properly capture important aspects of the in vivo setting.
Organism-Based Assays
HTS assays with model organisms of various types are rapidly gaining favor for small-molecule discovery because they represent a bridge between potentially irrelevant in vitro screening and expensive, low-throughput screening in mammalian animal models. Despite the fact that organism-based screening is the most complicated and expensive of the HTS assays, candidate compounds identified by such screening are potentially among the most biologically relevant because, unlike the situation with protein-based or cell-based assays, disease states can be screened in an appropriate physiologic context. Model organisms appropriate for use in HTS include invertebrates, such as Drosophila melanogaster (fly) and Caenorhabditis elegans (worm) (7), and vertebrate species, such as Danio rerio (zebrafish) (8). Key advantages of these model systems include rapid reproduction and scalability, feasibility of complex genetic manipulations, and relatively low maintenance and propagation costs. Furthermore, numerous biologic processes and genes are conserved in mammals and these model organisms. Limitations attendant to using these model organisms in HTS include pharmacologic delivery of molecules to the appropriate tissue or target compartment and absolute assessment of the localized concentration of a compound in the organism or tissue of interest.
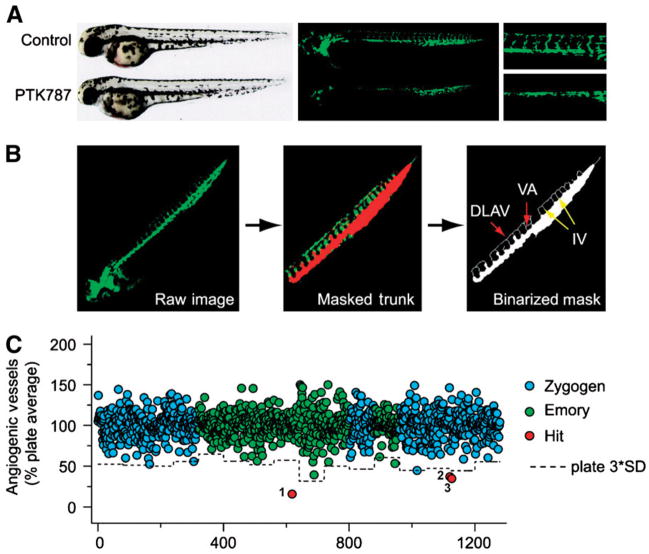
Currently, one of the most widely used model organisms in HTS is the zebrafish. Like D. melanogaster and C. elegans, zebrafish have historically been used to elucidate basic developmental biology. As a vertebrate species, zebrafish are relatively simple to maintain and extremely amenable to genetic and molecular manipulations. Adults mature fairly rapidly (~12 wk) and demonstrate robust fecundity; a single adult breeding pair can generate ~100 eggs per clutch twice a week (8). Screening assays typically involve the use of zebrafish embryos, which are easily manipulated with HTS instrumentation and are optically transparent, enabling a wealth of HTS experiments and detection strategies. Unlike invertebrates, zebrafish develop complex organ systems and were recently elegantly used with HTS to screen complex disease states, such as angiogenesis (9,10) and human cancer (11), as well as fundamental molecular events, such as apoptosis (12). For example, a strategy that capitalized on both the transparent nature and the genetic manipulability of zebrafish was reported. In this strategy, a novel automated screening assay for antiangiogenic compounds was used with transgenic zebrafish expressing a fluorescent reporter under the control of the vascular endothelial growth factor receptor 2 promoter (Fig. 2) (13).
FIGURE 2.
Screening of small-molecule library for antiangiogenic compounds with transgenic zebrafish. (A) Bright-field and fluorescent images of 2-d–postfertilization zebrafish treated with vehicle control or PTK787 at 10 μmol/L (positive control). Angiogenic blood vessel growth was inhibited in head and trunk of PTK787-treated embryos. (B) For quantification of vessel growth in embryos, trunk was manually isolated from fluorescent image. Automated algorithm counted intersegmental vessels (IV) and various branching arteries. DLAV = dorsal longitudinal anastomotic vessel; VA = vertebral artery. (C) LOPAC1280 library was collaboratively screened by Zygogen (blue points) and Emory University (green points). Positive hits (red points) were defined as compounds that caused inhibition of angiogenic vessel count by more than 3 SDs from plate average. (Reprinted with permission of (13).)
IDENTIFICATION OF HITS
HTS hits can be detected in several ways. In general, suitable methodologies are highly sensitive, quantitative, rapid, and reasonably inexpensive. Because each method has inherent strengths and weaknesses, the appropriateness of a detection strategy must be weighed against the specifics of the screening being performed. For example, an established approach for forward screening in cell- and organism-based assays is to observe phenotype modulation in the target specimen by either direct observation (14) or genetic and proteomic profiling (15). Although observation of phenotype modulation is a powerful approach, it can be limited by expense, low throughput on typical HTS scales, and a nonquantitative nature.
Another approach, more typical of reverse screening in protein- and cell-based assays, includes the modulation of known binding pairs; in this approach, the binding of a labeled, high-affinity probe, such as an antibody, is influenced by compounds that bind to the target of interest. This focused strategy, which is typically assayed with fluorescence or bioluminescence techniques, is highly capable of yielding compounds of interest, but allosteric interactions can confound the interpretation of results.
An alternative approach is to perform the screening with labeled small-molecule libraries that enable hits to be detected spectroscopically (16). A key advantage of this strategy is the potential for quantitative detection. However, the presence of the label can strongly influence the physical properties of the compound and the resultant binding potential. Label-free methodologies, such as mass spectrometry, also have potential for the detection of hits in HTS. For example, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry has been shown to be a powerful analytic tool that incorporates high-speed analysis (seconds per sample) and appropriate sensitivity (approximately hundreds of femtomoles) for the analysis of low-molecular-weight compounds (17). It is likely that sensitive, label-free detection methodologies suitable for automated, high-speed analysis will continue to gain attention for HTS applications.
FROM HTS TO IMAGING PROBES AND BEYOND
It is optimistic to anticipate that most compounds identified during an HTS campaign will result in clinically useful imaging probes. Rather, after the identification of a portfolio of compounds deemed interesting, potential candidates should be subjected to considerable scrutiny, validation, and refinement. Analogs with structural similarity may be included in more focused follow-up screening for comparison with the original candidate compounds and to further establish and refine structure–activity relationships. For candidates identified in forward-screening assays, future characterization might include proteomic and genomic profiling used in conjunction with affinity-labeling strategies to determine the specific target(s) of candidate imaging probes (18). On subsequent refinement, derivatization for molecular imaging should emphasize approaches that require minimal structural modifications to maintain the specific bioactivity exhibited by the unlabeled compounds. In some instances, relatively simple synthetic modifications of candidate compounds can yield PET or SPECT agents via halogen substitution with PET-active (18F) or SPECT-active (123I) radionuclides. Although any changes to the original structure of a candidate compound may affect the affinity of binding to the target, simple isotope substitution has potential for success.
CONCLUSION
Among the most prominent of frontiers in imaging science is the development of new molecular probes. Given that high-throughput chemical synthesis capability has enabled the assembly of tremendous compound collections and new molecular targets are now discovered almost daily, a remaining challenge is the rapid identification of highly specific compounds suitable for development and translation as imaging probes. HTS may have a role to play in addressing this major challenge.
Acknowledgments
We thank Dr. Todd E. Peterson for helpful discussions and Dawn Thornton for editorial commentary. This work was supported by a Career Development Award from the National Cancer Institute (K25 CA127349).
References
- 1.Wester HJ. Nuclear imaging probes: from bench to bedside. Clin Cancer Res. 2007;13:3470–3481. doi: 10.1158/1078-0432.CCR-07-0264. [DOI] [PubMed] [Google Scholar]
- 2.Merrifield B. Concept and early development of solid-phase peptide synthesis. Methods Enzymol. 1997;289:3–13. doi: 10.1016/s0076-6879(97)89040-4. [DOI] [PubMed] [Google Scholar]
- 3.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 4.Schreiber SL. Target-oriented and diversity-oriented organic synthesis in drug discovery. Science. 2000;287:1964–1969. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]
- 5.Wong JC, Sternson SM, Louca JB, Hong R, Schreiber SL. Modular synthesis and preliminary biological evaluation of stereochemically diverse 1, 3-dioxanes. Chem Biol. 2004;11:1279–1291. doi: 10.1016/j.chembiol.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson RL, Welch M, Ladlow M, Spring DR. Small-molecule screening: advances in microarraying and cell-imaging technologies. ACS Chem Biol. 2007;2:24–30. doi: 10.1021/cb600321j. [DOI] [PubMed] [Google Scholar]
- 7.Segalat L. Invertebrate animal models of diseases as screening tools in drug discovery. ACS Chem Biol. 2007;2:231–236. doi: 10.1021/cb700009m. [DOI] [PubMed] [Google Scholar]
- 8.Murphey RD, Zon LI. Small molecule screening in the zebrafish. Methods. 2006;39:255–261. doi: 10.1016/j.ymeth.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 9.Nicoli S, Presta M. The zebrafish/tumor xenograft angiogenesis assay. Nat Protoc. 2007;2:2918–2923. doi: 10.1038/nprot.2007.412. [DOI] [PubMed] [Google Scholar]
- 10.Seng WL, Eng K, Lee J, McGrath P. Use of a monoclonal antibody specific for activated endothelial cells to quantitate angiogenesis in vivo in zebrafish after drug treatment. Angiogenesis. 2004;7:243–253. doi: 10.1007/s10456-004-4181-7. [DOI] [PubMed] [Google Scholar]
- 11.Haldi M, Ton C, Seng WL, McGrath P. Human melanoma cells transplanted into zebrafish proliferate, migrate, produce melanin, form masses and stimulate angiogenesis in zebrafish. Angiogenesis. 2006;9:139–151. doi: 10.1007/s10456-006-9040-2. [DOI] [PubMed] [Google Scholar]
- 12.Pyati UJ, Look AT, Hammerschmidt M. Zebrafish as a powerful vertebrate model system for in vivo studies of cell death. Semin Cancer Biol. 2007;17:154–165. doi: 10.1016/j.semcancer.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Tran TC, Sneed B, Haider J, et al. Automated, quantitative screening assay for antiangiogenic compounds using transgenic zebrafish. Cancer Res. 2007;67:11386–11392. doi: 10.1158/0008-5472.CAN-07-3126. [DOI] [PubMed] [Google Scholar]
- 14.den Hertog J. Chemical genetics: drug screens in zebrafish. Biosci Rep. 2005;25:289–297. doi: 10.1007/s10540-005-2891-8. [DOI] [PubMed] [Google Scholar]
- 15.Dephoure N, Howson RW, Blethrow JD, Shokat KM, O’Shea EK. Combining chemical genetics and proteomics to identify protein kinase substrates. Proc Natl Acad Sci USA. 2005;102:17940–17945. doi: 10.1073/pnas.0509080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim YK, Chang YT. Tagged library approach facilitates forward chemical genetics. Mol Biosyst. 2007;3:392–397. doi: 10.1039/b702321a. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Z, Winant RC, Gambhir SS. A new strategy to screen molecular imaging probe uptake in cell culture without radiolabeling using matrix-assisted laser desorption/ ionization time-of-flight mass spectrometry. J Nucl Med. 2005;46:878–886. [PubMed] [Google Scholar]
- 18.Smukste I, Bhalala O, Persico M, Stockwell BR. Using small molecules to overcome drug resistance induced by a viral oncogene. Cancer Cell. 2006;9:133–146. doi: 10.1016/j.ccr.2006.01.012. [DOI] [PubMed] [Google Scholar]