Summary
Capping protein is an integral component of Arp2/3-nucleated actin networks that drive amoeboid motility. Increasing the concentration of capping protein, which caps barbed ends of actin filaments and prevents elongation, increases the rate of actin-based motility in vivo and in vitro. We studied the synergy between capping protein and Arp2/3 using an in vitro actin-based motility system reconstituted from purified proteins. We find that capping protein increases the rate of motility by promoting more frequent filament nucleation by the Arp2/3 complex, and not by increasing the rate of filament elongation as previously suggested. One consequence of this coupling between capping and nucleation is that, while the rate of motility depends strongly on the concentration of capping protein and Arp2/3, the net rate of actin assembly is insensitive to changes in either factor. By reorganizing their architecture, dendritic actin networks harness the same assembly kinetics to drive different rates of motility.
Introduction
Eukaryotic cells construct actin filament networks that control their shape and internal organization and the first step in assembly of these networks is nucleation of new actin filaments. Recent work has identified multiple cellular factors capable of making new actin filaments (reviewed in Baum and Kunda, 2005; Goley and Welch, 2006; Kovar, 2006) and each of these factors appears to be specialized for construction of a particular type of network. One of the most well studied nucleation factors is the Arp2/3 complex, which initiates assembly of new filaments from the sides of preexisting filaments to generate networks of branched filament arrays (Mullins et al., 1998). At present, these branched networks are known to underlie four basic cellular structures: (1) the leading edge of motile, amoeboid cells (Iwasa and Mullins, 2007; Kelleher et al., 1995; Machesky et al., 1997; Welch et al., 1997); (2) the advancing edges of phagocytic and macropinocytic cups (May et al., 2000); (3) actin networks required for late stages of endocytosis (Kaksonen et al., 2003; Merrifield et al., 2004); and (4) ‘comet tails’ that drive intracellular motility of various pathogens (reviewed in Stevens et al., 2006). To generate force and move membranes, the Arp2/3 complex must collaborate with other actin-associated proteins, including capping protein (CP), cofilin and profilin. Together, these factors form a biochemical cycle that assembles and disassembles motile actin networks. In vitro they are sufficient to reconstitute sustained, actin-based motility (Loisel et al., 1999) and, in vivo, they are necessary for construction of leading edge actin networks (Iwasa and Mullins, 2007).
The ‘dendritic’ nucleation activity of the Arp2/3 complex is stimulated by a relatively well-understood sequence of events: (1) An Arp2/3 complex binds to the side of an actin filament. (2) The filament-bound complex interacts with a nucleation-promoting factor (NPF, usually a member of the WASP family of proteins) which induces a conformational change in the complex. (3) An actin monomer bound to the WH2 domain of the NPF is brought into contact with the complex, triggering a further conformational change that results in formation of a stable actin nucleus (Dayel and Mullins, 2004; Goley et al., 2004; Kelly et al., 2006; Marchand et al., 2001). Nucleation is usually studied in vitro, in dilute solutions where all of the components are soluble and free to diffuse and where the rate of nucleation is limited mainly by the concentration of the Arp2/3 complex. To generate force and move cargo, however, the nucleation machinery must be confined to a two-dimensional surface where it encounters high local concentrations of filamentous actin (1–10 mM) and NPFs. The effects of these boundary conditions on Arp2/3 activity are not well understood.
How other components of the actin assembly cycle contribute to the architecture and mechanics of Arp2/3-dependent motile networks is also not well understood. Several studies have shown that CP plays a key role in actin based motility and assembly of dynamic actin networks in vivo (Bear et al., 2002; Hug et al., 1995; Iwasa and Mullins, 2007; Mejillano et al., 2004) and in vitro (Loisel et al., 1999; van der Gucht et al., 2005). These studies share a common, paradoxical observation: that CP, which terminates filament elongation, actually promotes network assembly and accelerates actin-based motility. For example, increasing CP concentrations in Dictyostelium produces faster rates of migration (Hug et al., 1995). Cell migration is a complex process that requires integration of many cellular systems, including: actin-dependent protrusion of the leading edge, adhesion and de-adhesion of the cell to the substrate, and myosin-dependent contraction of the cortex to maintain proper positioning of the nucleus and cell body. In principle, CP could contribute to any of these processes but experimental evidence suggests that its major role in migration is to help construct lamellipodial actin networks at the leading edge. Addition of chemoattractant to Dictyostelium cells, for example, produces a burst of new actin polymerization and a simultaneous recruitment of CP to the cell periphery (Eddy et al., 1997). In Drosophila S2 cells, Xenopus fibroblasts, and mammalian melanoma cells, CP activity is limited primarily to the leading lamellipodial actin network and knockdown of its expression displaces the Arp2/3 complex from the cell periphery and causes complete loss of lamellipodial actin (Iwasa and Mullins, 2007; Mejillano et al., 2004; Miyoshi et al., 2006). Consistent with these in vivo observations, reconstitution of Arp2/3-dependent motility in vitro absolutely requires CP (Loisel et al., 1999).
The most widely accepted explanation of the effect of CP on actin-based motility is the Actin Funneling Hypothesis (Carlier and Pantaloni, 1997). The Funneling Hypothesis rests on the assumption that faster motility requires faster actin filament elongation. Accordingly, Carlier and Pantaloni proposed that CP increases the steady-state actin monomer concentration by capping the fast-growing barbed ends of most of the actin filaments in the cell. The increased actin monomer concentration causes the small number of barbed ends that remain uncapped to elongate faster and drive faster motility. This model provides a straightforward and attractive explanation for the effect of capping protein on actin network assembly but it has never been tested experimentally.
To test the predictions of the Funneling Hypothesis and to better understand how the Arp2/3 complex collaborates with CP, we reconstituted actin-based motility using purified components. Following the ground-breaking work of Loisel et al., we mix polystyrene beads coated with ActA, an Arp2/3-activating protein from Listeria monocytogenes, together with five purified factors: cytoplasmic actin, the Arp2/3 complex, CP, cofilin, and profilin. Cofilin promotes disassembly of actin filaments that have hydrolyzed their bound ATP and profilin catalyzes nucleotide exchange by actin monomers. Together, these proteins promote turnover of the actin network and recycling of the actin monomers. In our motility mixture, ActA-coated beads first assemble spherically symmetrical actin shells and then, within 2–5 minutes, break symmetry and move at speeds of around 5 µm/min. These speeds are comparable to those measured for Listeria in the cytoplasm of infected cells (Dabiri et al., 1990). We studied the synergy between Arp2/3 and CP by varying their concentrations and examining the effect on the morphology, movement, and biochemical composition of the motile actin networks. We draw two major conclusions from these experiments: (1) CP does not funnel actin monomers onto growing barbed ends but rather shunts monomers away from barbed ends and onto the Arp2/3 complex. Thus, CP does not increase the rate of motility by increasing the rate of filament elongation but by promoting more frequent filament nucleation by the Arp2/3 complex. (2) The rate of motility can be uncoupled from the rate of actin network assembly. By reorganizing their architecture in response to changing forces, dendritic actin networks can support different rates of motility with the same overall polymer assembly kinetics.
Results
To initiate actin-based motility, we mix ActA-coated polystyrene beads with actin, the Arp2/3 complex, CP, cofilin, and profilin. Sustained motility occurs only within a narrow window of protein concentrations and progresses through four distinct stages (Figure 1A, panels, and Movie S1). First, spherical actin shells are assembled on the beads. Within five minutes, the beads break free of their actin shells (i.e. “break symmetry”) and produce polarized comet tails. During the first half hour of motility (burst phase) the beads move at their highest speed (4–6 µm/min). Subsequently, the system settles into 2–3 hours of steady-state motility, during which the beads move at a constant, slower velocity (2.2 µm/min using 5 µm beads). This is approximately four times faster than previously reported for similar reconstituted systems (Bernheim-Groswasser et al., 2002). The difference may be due to our use of non-muscle cytoplasmic actin rather than α-skeletal muscle actin (Cameron et al., 2004). In the absence of ATP regeneration, motility eventually winds down: tails shorten and speeds drop. We typically detect movement at speeds greater than 1 µm/min for more than 5 hours (Figure 1A, graph).
Figure 1. Effects of Arp2/3 and CP on steady-state motility.
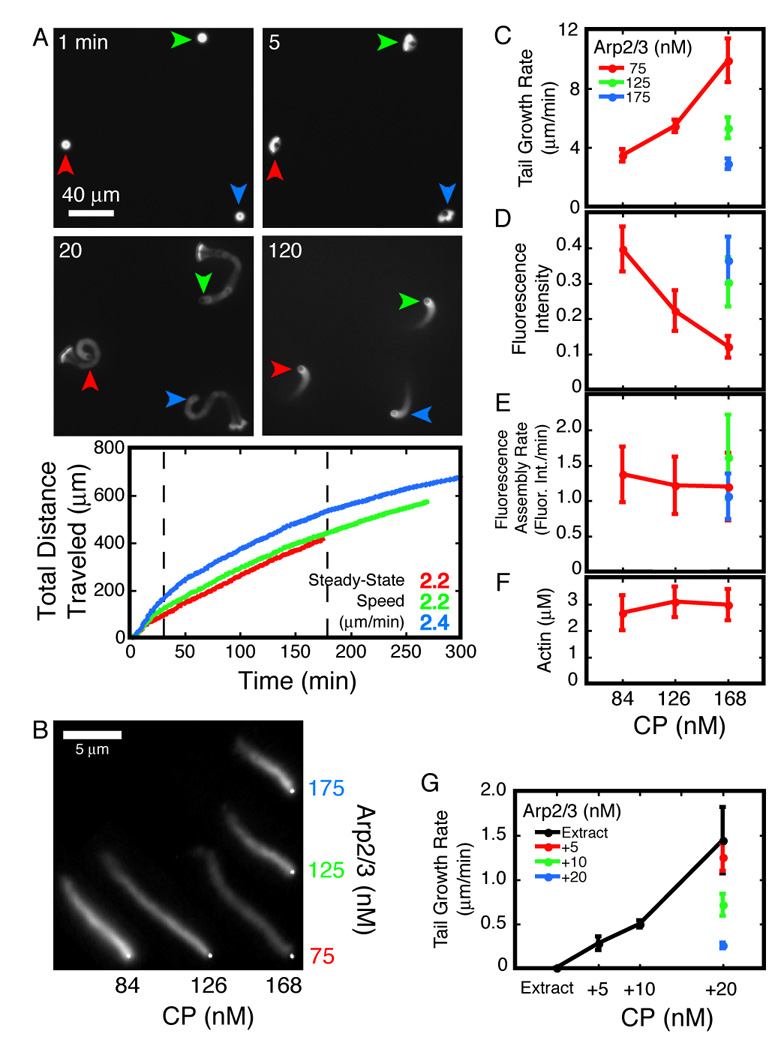
A. Panels: Four stages of motility—shell building, symmetry breaking, transient burst, and steady-state (clockwise, starting from top-left.) Arrowheads track individual beads through the panels. Graph: Total distance traveled by beads in the panels. Curves and speed figures are color-matched to the arrowheads. Steady-state speeds are the slopes of data between the dashed lines (30 to 180 minutes). Conditions: 5 µm ActA-coated beads mixed with 7.5 µM actin (5% TMR-labeled), 5 µM cofilin, 3 µM profilin, 120 nM Arp2/3, 168 nM CP.
B–F. Representative actin comet tails assembled on 220 nm ActA-coated beads in the presence of various concentrations of Arp2/3 and CP as indicated. Images were acquired approximately 30 minutes after mixing. Brightfield bead images and fluorescent comet tails were digitally merged for presentation. Brightness and contrast of the fluorescent images were set to the same values. Conditions: 7.5 µM actin (5% TMR-labeled), 4 µM cofilin, and 3 µM profilin.
C. Rate of motility decreases with [Arp2/3] and increases with [CP]. Each data point is the slope of the line fit to a time series of length measurements. Error bars are slopes of fits to the standard deviation of each time series (average n=140).
D. Actin density in the comet tails increases with [Arp2/3] and decreases with [CP]. Fluorescence intensities along single-pixel spines of actin comet tails were measured. Each data point is the intensity averaged over the first micron length of the tails. Error bars are standard deviations for 20 comet tails.
E. Rate of actin network assembly does not change with [Arp2/3] or [CP]. Rate of fluorescent actin incorporation is the product of tail growth rate (C) and fluorescence intensity (D). Error bars mark the product of the upper and lower error limits of the two terms.
F. Soluble actin concentration at steady-state does not change with [Arp2/3] or [CP]. Error bars mark the standard error for 3 independent measurements.
G. Rate of motility in brain cytosolic extract decreases with [Arp2/3] and increases with [CP]. Rates of tail growth on 220 nm ActA-coated beads were measured as in (C); see text for details. Each data point is the slope of the line fit to a time series of length measurements. Error bars are slopes of fits to the standard deviation of each time series (average n=77). Conditions: Brain cytosolic extract supplemented with 2 µM actin (10% TMR-labeled) and Arp2/3 and CP as indicated.
Effects of Arp2/3 and CP on Motility
All stages of motility are sensitive to changes in the concentrations of Arp2/3 and CP. With 5 µm beads, for example, steady-state motility occurs only within a window of approximately ±20% of the optimum concentrations of both proteins. Outside this narrow concentration range, actin shells fail to break symmetry or actin networks lose polarity soon after the burst phase. Smaller beads establish and maintain polarized networks more readily (Cameron et al., 1999, and our unpublished observations), so we used 220 nm beads to characterize the effects of Arp2/3 and CP on steady-state motility (Figure 1B).
The rate of steady-state motility increases with more CP and decreases with more Arp2/3 complex. Drift and Brownian motion make motility of 220 nm beads difficult to analyze by particle tracking, so we developed a two-color fluorescence method to measure the rate of actin comet tail growth directly. We simultaneously initiated motility in two identical reactions, one doped with 5% Alexa488- (green) labeled actin and the other with TMR- (red) labeled actin. After allowing them to come to steady state, we mixed equal volumes of the two reactions together to generate comet tails in which old and new actin filaments are labeled with different colors. After mixing, we quenched tail growth with latrunculin B and phalloidin. The combined activities of the two molecules arrests both polymerization and depolymerization and preserves the ratio of monomeric to polymeric actin (Supplemental Figure 1A). To calculate tail growth rates for each condition, we imaged comet tails arrested at various time points after mixing and measured the extent of new assembly (Supplemental Figure 1B). Increasing the Arp2/3 concentration from 75 to 175 nM decreases the rate of tail growth more than three-fold (Figure 1C) while an increase in CP concentration produces a comparable increase in the rate of tail growth (Figure 1C). Further increasing the CP concentration produces sparse but very fast growing tails that often lose contact with the bead and do not support efficient motility (data not shown).
The effect of CP on motility is not due to changes in the steady-state concentration of monomeric actin. The Actin Funneling Hypothesis proposes that CP increases the rate of motility by decreasing the number of free barbed ends and increasing the concentration of monomeric actin at steady state (Carlier and Pantaloni, 1997). We tested this model by quantifying the concentration of monomeric actin in high speed supernatants of latrunculin B/phalloidin arrested motility reactions. At steady state, the concentration of soluble actin is constant and insensitive to changes in CP concentration (Figure 1F), arguing strongly against the Funneling Hypothesis. We compared the fluorescence intensity in comet tails at steady state and found that the amount of polymeric actin near the bead surface decreases with CP concentration and increases with Arp2/3 concentration, a mirror image of the effects on tail growth rate (Figure 1D). The product of the average fluorescence intensity and the tail growth rate is proportional to the overall rate of actin assembly at the bead surface. This product is the same for all conditions (Figure 1E), suggesting that neither CP nor the Arp2/3 complex affects the rate at which polymer is assembled on the bead surface. Together, these results suggest that CP and Arp2/3 regulate motility not by altering actin assembly kinetics but instead by reorganizing the architecture of the actin network: Increasing the CP concentration decreases the density of the actin network, while increasing the Arp2/3 concentration increases the network density.
To test whether the results from our six-component system hold true in the more complex environment of the cytoplasm, we studied the effect of CP and the Arp2/3 complex on bead motility in brain cytosolic extract (Laurent et al., 1999; Vignjevic et al., 2003; Yarar et al., 1999). In extract doped with TMR-labeled actin, 40 % of 220 nm, ActA-coated beads produce polarized comet tails and move (Movie S2). Consistent with earlier observations (Laurent et al., 1999), the rate of tail growth is too slow to measure accurately. Both the percentage of motile beads and the rate of tail growth increase monotonically with the addition of exogenous CP. With 20 nM added CP, more than 90 % of the beads produce tails that grow at 1.44±0.37 µm/min (Figure 1G). The Arp2/3 complex has the opposite effect on motility, decreasing the rate of tail growth with increasing concentrations (Figure 1G). The agreement between the reconstituted and the extract-based motility systems argues that the results obtained with pure proteins accurately reflect what happens in the cytoplasm of living cells.
The rate of filament nucleation during steady-state motility increases with CP concentration. The rate of actin polymerization is proportional to the product of the monomer and free barbed end concentrations. Given that neither the assembly rate (Figure 1E) nor the monomer concentration (Figure 1F) changes with CP concentration, we conclude that the density of free barbed ends at the bead surface is also insensitive to CP. This strongly suggests that CP increases the rate of filament nucleation. That is, filament barbed ends lost to capping must be replaced by an increased rate of nucleation. We tested this prediction by measuring the ratio of Alexa488-labeled Arp2/3 to TMR-labeled actin in comet tails during steady-state motility (Figure 2A). Given the constant actin assembly rate (Figure 1E), the Arp2/3-per-actin ratio is a direct readout of the relative nucleation rate. Arp2/3 can associate with actin comet tails in one of two ways: (1) It can be incorporated into the network through dendritic nucleation, or (2) it can decorate actin filaments through side-binding (Mullins et al., 1998). By comparing the fluorescence intensities of comet tails assembled in the presence of Alexa488-labeled Arp2/3 to those incubated with Alexa488-labeled Arp2/3 after assembly with unlabeled Arp2/3, we found that the contribution of side-binding to the total signal was negligible (Supplemental Figure 2A). At three different concentrations of the Arp2/3 complex, the ratio of Arp2/3 to actin increases with increasing CP concentration (Figure 2B). Thus, the rate of nucleation increases with increasing CP concentration. This result is consistent with an earlier study which found that increasing gelsolin concentrations lead to more densely branched actin comet tails (Wiesner et al., 2003). Remarkably, we note that the ratio of Arp2/3:actin is insensitive to the concentration of the Arp2/3 complex, suggesting that the rate of nucleation is independent of the Arp2/3 concentration
Figure 2. CP increases the rate of Arp2/3 nucleation during steady-state motility.
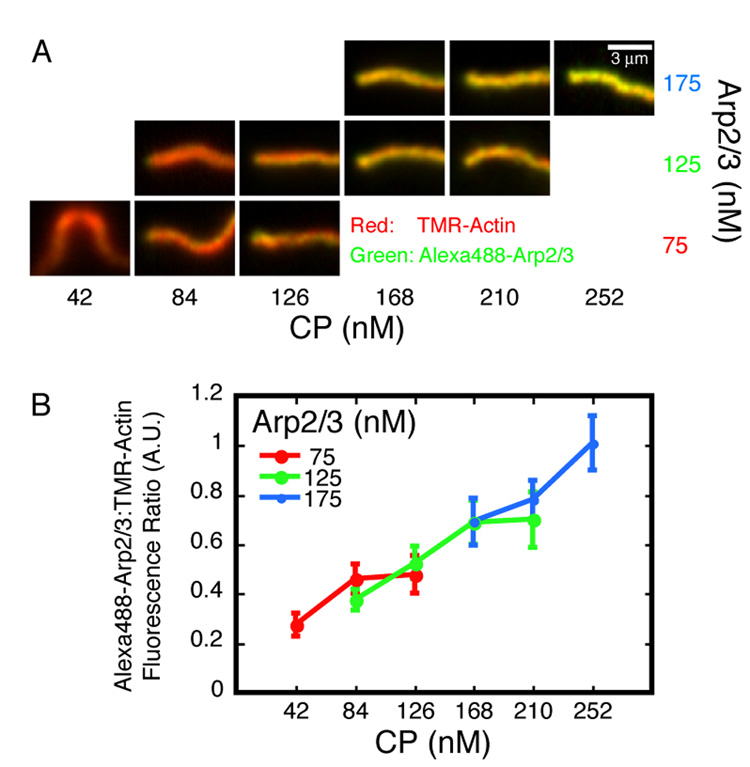
A. Representative actin comet tails assembled on 220 nm ActA-coated beads in the presence of various concentrations of Arp2/3 (10% Alexa488-labeled) and CP as indicated. Images were acquired approximately 30 minutes after mixing. Tails are framed with the beads (not shown) to the left of each panel, except the lower left panel (75 nM Arp2/3, 42 nM CP) where the bead would be top-center. This condition, as well as lower [CP] than that shown for the higher two Arp2/3 concentrations, produces twin tails on every bead. The red (i.e. actin) channel of each image was set to 90% saturation; this value was scaled by the same factor to determine the green (i.e. Arp2/3) channel display limits for all images. Conditions: 7.5 µM actin (5% TMR-labeled), 4 µM cofilin, and 3 µM profilin.
B. Fluorescence intensities along single-pixel spines of actin comet tails were measured. Each data point is the ratio of Alexa488-Arp2/3 to TMR-actin intensity averaged over the first micron length of the tails. Error bars are standard deviations (average n=27).
Effects of Arp2/3 and CP on Shell Growth and Symmetry Breaking
To understand how the Arp2/3 complex and CP regulate the assembly and architecture of motile actin networks, we studied the radially symmetrical growth of actin shells prior to symmetry breaking. Shell growth does not require cofilin and profilin, the two components involved in actin network disassembly and recycling, enabling us to focus exclusively on the roles of CP and the Arp2/3 complex. Because we were not limited to conditions that produce efficient symmetry breaking and sustained motility, we could also examine a larger range of Arp2/3 complex and CP concentrations.
As in motility, actin polymerization during shell growth occurs exclusively at the bead surface. We determined this by following new polymer assembly in a pulse-chase experiment. We initiated shell assembly with Alexa488-labeled (green) actin and then chased with TMR-labeled (red) actin (Figure 3A and Movie S3). The red actin localizes to the bead surface, displacing the older, green actin shell. In addition, the red actin is completely excluded from the green shell, indicating that fast-growing barbed ends are confined to the bead surface and do not elongate in directions away from the bead.
Figure 3. CP is necessary for shell growth and symmetry breaking.
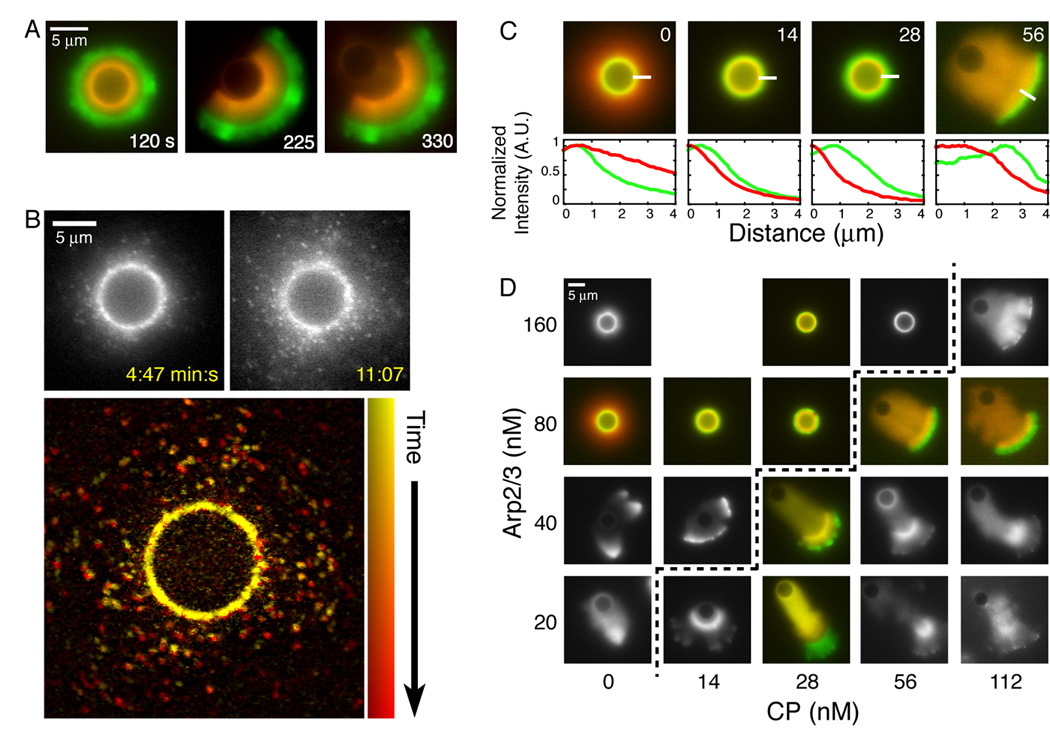
A. Actin assembly is restricted to the bead surface during shell growth. We mixed 5 µm ActA-coated beads with 2 µM actin (5% Alexa488-labeled), 50 nM Arp2/3, and 140 nM CP. After 30 seconds, the reaction was diluted ten-fold into a solution of identical protein composition containing 5% TMR-labeled actin.
B. Actin assembly is not restricted to the bead surface in the absence of CP.
Top: Selected frames from a time-lapse series of actin halo growth imaged using fluorescent speckle microscopy. Conditions: 5.8 µm ActA-coated beads mixed with 2 µM actin (1/16,000 TMR-labeled) and 80 nM Arp2/3.
Bottom: Actin speckles are stationary and new speckles appear farther away from the bead surface. 20 frames of the series shown above, representing 6:20 minutes of elapsed time, were used to generate this maximum intensity projection. Images were filtered to highlight the speckles. Time is represented by changing hues, starting with yellow at early time points, progressing through orange to red.
C. CP directs new actin assembly to the bead surface.
Top: New actin assembly was followed using fluorescence pulse-chase as in (A). 5 µm ActA-coated beads were mixed with 2 µM actin (first label 5% Alexa488, second label 5% TMR) , 80 nM Arp2/3, and CP as indicated in top right corner of each panel. Images were acquired 10–15 minutes after mixing.
Bottom: Intensity profiles of the green and red actin fluorescence along the 4 µm lines are plotted, starting from the endpoints closest to the beads. Each profile is normalized to its maximum value in the original image.
D. The concentration of CP required for symmetry breaking varies directly with the Arp2/3 concentration. Panel shows representative actin structures assembled on 5 µm ActA-coated beads in the presence of 2 µM actin (5% TMR-labeled), and Arp2/3 and CP as indicated. Dashed line marks the symmetry breaking threshold. Images were acquired 10–15 minutes after mixing.
CP is necessary for shell growth and symmetry breaking. Symmetry breaking requires the actin network to be built from the inside out, so that assembly on the bead surface continuously displaces older regions of the actin network (Bernheim-Groswasser et al., 2002; van der Gucht et al., 2005). Without CP, new actin assembly is no longer limited to the surface of the bead (Figures 3B and C). Long filaments can grow away from the bead and radial growth can occur by elongation of these filaments at the periphery of the network (Figure 3B and Movie S4). These conditions produce diffuse halos of actin which collapse into stellate structures when filament bundling is promoted by addition of crosslinking proteins (Haviv et al., 2006; Vignjevic et al., 2003) or methyl cellulose (Supplemental Figure 3). Vignjevic et al. generated similar stellate structures in brain extract and found that distal regions of the actin network lack the dendritic organization characteristic of Arp2/3 complex activity. These authors also found that addition of exogenous CP reorganizes the architecture of the network, producing dendritic arrays. Together, these results suggest that CP acts through the Arp2/3 complex to restrict actin assembly to the bead surface.
The concentration of CP required for symmetry breaking varies directly with the Arp2/3 concentration. With 2 µM actin and 80 nM Arp2/3, 14 nM CP is sufficient to produce actin networks that grow from the inside out, continuously displacing older material from the surface of the beads (Figure 3C, second panel). Although their final size increases with CP concentration, the shells grow slowly and do not break symmetry before running out of monomeric actin. We observed symmetry breaking only when CP exceeds a threshold concentration of 56 nM (Figure 3C, far-right panel). This threshold varies directly with the Arp2/3 concentration (Figure 3D). Increasing the CP concentration far above this threshold produces fast-growing, fragmented actin networks that do not form a continuous structure around the bead and frequently lose contact with the surface altogether (Movie S5, see Supplemental Discussion).
Biochemical and Kinetic Analysis of Shell Growth
To understand how changes in CP and Arp2/3 complex concentration translate into changes in actin network architecture, we developed a method to simultaneously measure the size and protein composition of actin shells. We initiated actin shell assembly and then, 60 seconds later, arrested actin dynamics with latrunculin B and phalloidin. We separated the arrested reactions into three fractions by differential centrifugation: (1) Beads and associated actin networks (low speed pellet), (2) filamentous actin and associated protein (high speed pellet), and (3) soluble protein (high speed supernatant). We separated proteins in each fraction by SDS-PAGE, and then stained and quantified them using a fluorescent dye (Supplemental Figure 4A). Actin shells survived fractionation and washing with little or no damage and, by fluorescence microscopy, were indistinguishable from untreated shells (Supplemental Figure 4B).
CP and Arp2/3 regulate shell size by controlling actin network density. We analyzed 16 combinations of CP and Arp2/3 concentrations (Figure 4A). Consistent with our observations of steady-state motility, shell size at 60 seconds increases with increasing CP concentration and decreases with increasing Arp2/3 concentration (Figure 4B). As with motility, shell size does not correlate with concentration of soluble actin, which actually decreases with increasing CP concentration (Figure 4C). This drop in soluble actin concentration is most likely due to in vitro filament nucleation by CP (Cooper and Pollard, 1985, Supplemental Figure 4C). Remarkably, the total amount of actin in the shells does not vary significantly with CP or Arp2/3 concentration (Figure 4D), suggesting that the overall rate of polymer assembly on the bead surface is insensitive to variations in the concentrations of either protein. This is also consistent with results obtained using motile beads (Figure 1E). CP and Arp2/3, therefore, control the spatial organization of the actin network but not the amount of actin contained in it. More CP decreases the network density while more Arp2/3 complex increases it (Figure 4E).
Figure 4. Effects of Arp2/3 and CP on shell growth.
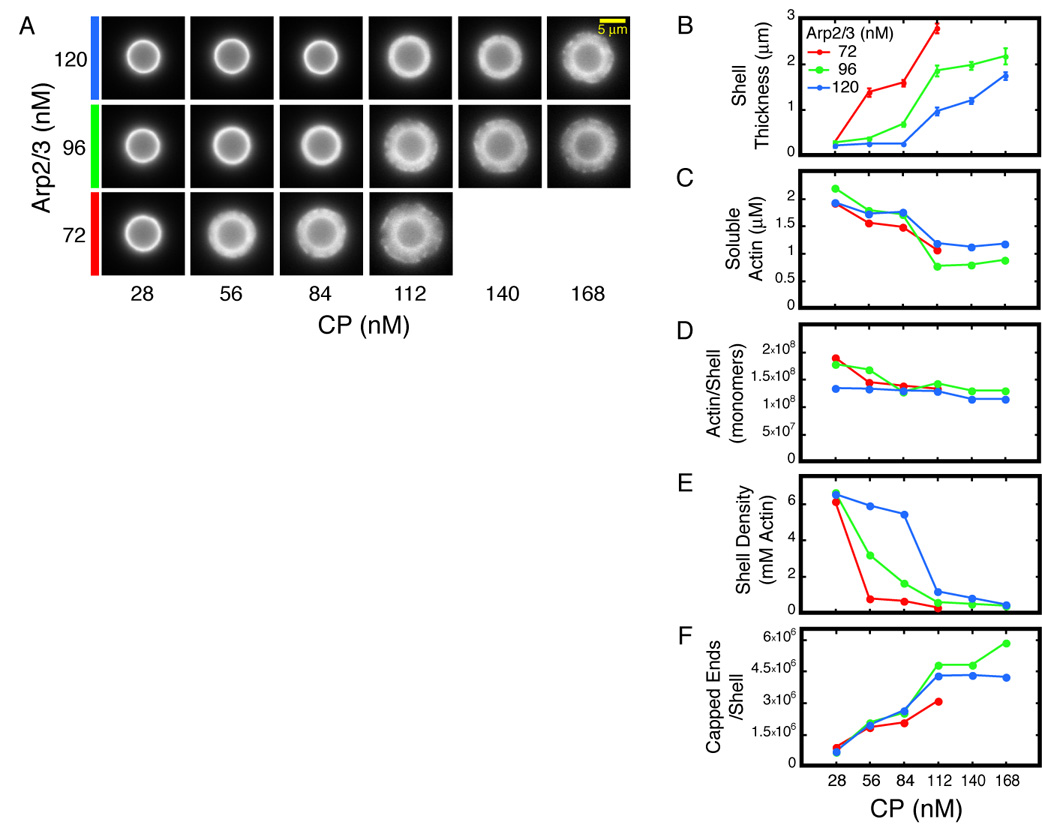
A–F. Reactions were Latrunculin B/phalloidin arrested 60 seconds after mixing, fractionated and analyzed as described. Conditions: 5.8 µm ActA-coated beads mixed with 3 µM actin (5% TMR-labeled), and Arp2/3 and CP as indicated.
A. Representative shells after arrest, fractionation, and washing.
B. Shell thickness decreases with [Arp2/3] and increases with [CP]. Error bars represent standard deviation (average n=17).
C. Soluble actin concentration at the time of arrest decreases with increasing [CP].
D. Total amount of actin in the shells does not vary with [Arp2/3] or [CP].
E. Average actin density in the shells increases with [Arp2/3] and decreases with [CP]. Density was calculated by dividing the actin mass in (D) by the volume of the shells as derived from (B).
F. Total number filaments in the shells is independent of [Arp2/3] but increases with [CP].
For technical reasons we could not use the Arp2/3 complex as a metric for filament number in symmetrical actin shells. After actin assembly is arrested, Arp2/3 continues to accumulate on the bead surface, where actin filaments and ActA, two complementary Arp2/3 binding partners, are juxtaposed (Supplemental Figure 2B). This strong targeting effect makes Arp2/3 an unreliable reporter for actin filament number; indeed, the amount of Arp2/3 recovered in arrested and washed shells is insensitive to changes in CP and Arp2/3 concentration (Supplemental Figure 4D).
We determined the number of actin filaments in each shell by quantifying the CP that remains stably associated with shells through fractionation and washing. The amount of CP in the shell accurately reflects the number of actin filament provided that: (1) CP does not associate non-specifically with actin shells; (2) the barbed ends that are free at the moment of arrest become capped after addition of latrunculin B/phalloidin; and (3) all filaments remain capped through fractionation and washing. To test the specificity of CP association, we incubated shells with additional CP after arrest and found that the amount of recovered CP changed only modestly (Supplemental Figure 5A). We also incubated arrested and washed shells with fluorescently labeled profilin:actin complexes and found that the new actin did not incorporate into the shells at free barbed ends, indicating that the latter two conditions were also satisfied (Supplemental Figure 5B). The mechanics of the assay (see Experimental Procedures) and the slow rate of CP dissociation (t1/2~30 minutes, Schafer et al., 1996) account for this result. Barbed ends that are free at the time of arrest become capped in the few minutes the shells spend in the presence of soluble CP before the first wash step. Very few filaments become uncapped during washing and final resuspension. Using CP as a reporter, we found that the total number of filaments per shell does not increase with Arp2/3 concentration (Figure 4F). Instead, the dominant trend is an increase in the number of filaments with increasing CP concentration.
The rate of nucleation is independent of Arp2/3 complex concentration but increases with the concentration of CP. By arresting shell growth at different time points, we characterized the kinetics of this reaction (Figure 5A). In agreement with the 60-second endpoint data (Figure 4), the shells grow faster with increasing CP concentration and more slowly with increasing Arp2/3 complex concentration (Figure 5B). The overall rate of actin assembly is insensitive to the concentration of either protein (Figure 5C). The total number of filaments in the actin shells increases linearly over time (Figure 5D). The slope, or the rate of filament nucleation, is the same for different Arp2/3 concentrations but increases with CP concentration.
Figure 5. The kinetics of shell growth is independent of the kinetics of actin assembly.
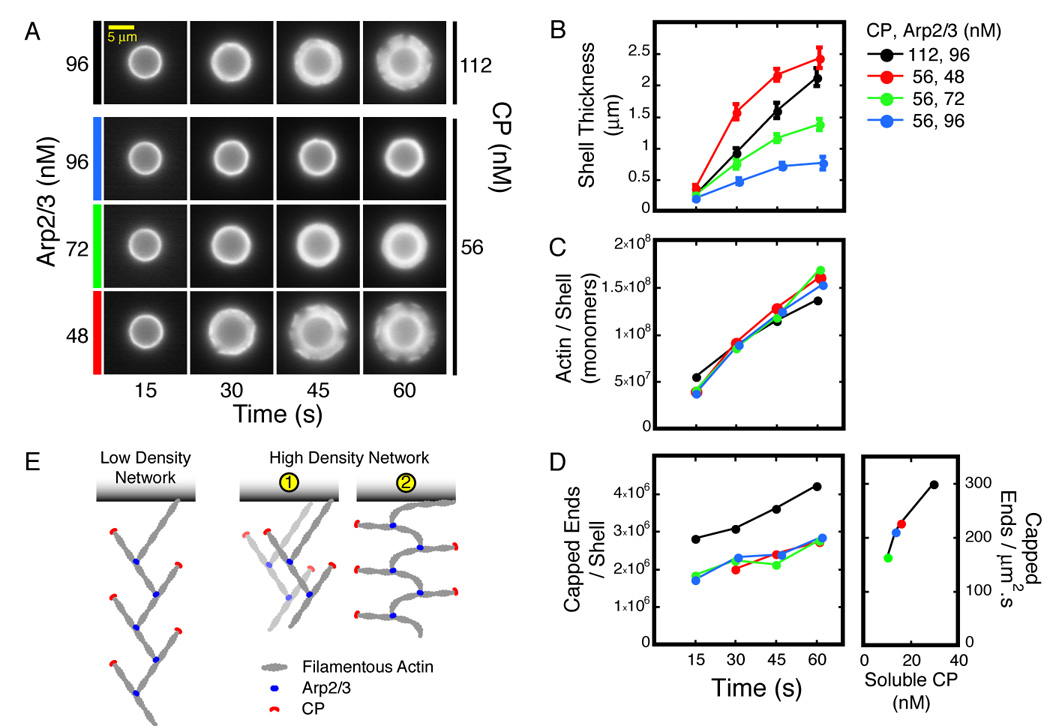
A–F. Aliquots from each reaction were Latrunculin B/phalloidin arrested 15, 30, 45, and 60 seconds after mixing, fractionated, and analyzed as described. Conditions: 5.8 µm ActA-coated beads mixed with 3 µM actin (5% TMR-labeled), and Arp2/3 and CP as indicated.
A. Representative shells after arrest, fractionation, and washing.
B. Rate of shell growth decreases with [Arp2/3] and increases with [CP]. Error bars represent standard deviation (average n=27).
C. Rate of actin assembly is independent of [Arp2/3] and [CP].
D. Rate of filament nucleation is independent of [Arp2/3] but increases with [CP]. Nucleation rate per square micron of bead surface was derived from the slopes of the time courses.
E. Two alternate mechanisms for increasing the density of a dendritic actin network imply different actin assembly kinetics. See text for details.
Effect of Arp2/3 on Actin Network Architecture
How does increasing the Arp2/3 concentration increase the density of the actin network without changing either the total amount of actin or the number of filaments? Denser packing can be achieved either by packing more filaments closer to the bead surface without altering their orientation (Figure 5E, Scheme 1), or by decreasing the glancing angle that each filament makes with the surface (Figure 5E, Scheme 2). These two mechanisms are distinguishable by the different actin assembly kinetics they imply. The first mechanism increases the density of barbed ends at the bead surface. As a result, the rate of filament elongation should decrease with increasing Arp2/3 concentration to maintain the invariant rate of actin assembly (Figure 5C). In contrast, the second mechanism preserves the barbed end density, so the rate of filament elongation should remain constant. We distinguished between these mechanisms by estimating the barbed end density and the elongation rate as a function of Arp2/3 concentration.
The surface density of free barbed ends is independent of the Arp2/3 concentration. We estimate the free barbed end density, [E], using the equation:
(Knuc: nucleation rate, kcap: filament capping rate constant, [CP]free: free CP concentration). When both Knuc and [CP]free are constant, [E] rapidly converges to the ratio Knuc/( kcap*[CP]free). The rate of nucleation is constant for the conditions studied (Figure 5D). We measured [CP]free from the soluble fractions of the arrested reactions and found the value to be approximately constant (Supplemental Figure 4D). Thus, the concentration of free barbed ends is constant during shell growth. Using the measured values, we found that the density of free barbed ends at the bead surface does not change with Arp2/3 concentration (2100±100 /µm2 for the three conditions studied).
The rate of filament elongation does not scale with the Arp2/3 concentration. With 56 nM CP, the kinetics of actin network assembly are identical at 48, 72, and 96 nM Arp2/3 (Figure 5C). We calculated the rates of filament elongation for these conditions (see Supplemental Experimental Procedures) and found the values to be very similar: The relative elongation rates at 48, 72, and 96 nM Arp2/3 are 1, 0.9, and 0.8, respectively (Supplemental Figure 6). Since neither the elongation rate nor the surface density of barbed ends changes with increasing Arp2/3 concentration, we conclude that the increase in network density is achieved through the second mechanism described above: More actin is packed into smaller volumes by changing the orientation of the filaments (Figure 5E, Scheme 2). This mechanism uncouples the kinetics of actin assembly and shell growth: The rate at which the actin network treadmills away from the bead surface varies by five-fold without significant changes in either the rate of nucleation or the rate of elongation.
Because increasing the concentration of the Arp2/3 complex slows shell growth without increasing nucleation rate, the effect of Arp2/3 on network architecture appears to be independent of its nucleation activity. To uncouple the nucleation activity of the Arp2/3 complex from its effect on shell growth, we replaced a portion of the active complex with inactive, dephosphorylated Arp2/3 complex. Dephosphorylation abolishes the actin nucleation activity of Arp2/3 without affecting actin filament or NPF binding (LeClaire et al., 2008). Dephosphorylated Arp2/3 complex is as effective as the nucleation-competent complex at slowing down shell growth (Figures 6A and B). We conclude that the excess Arp2/3 complex tethers actin filaments to surface-bound NPFs, and we propose that increased tethering increases network density and slows shell growth.
Figure 6. Arp2/3 slows down shell growth by tethering the actin network to the bead surface.
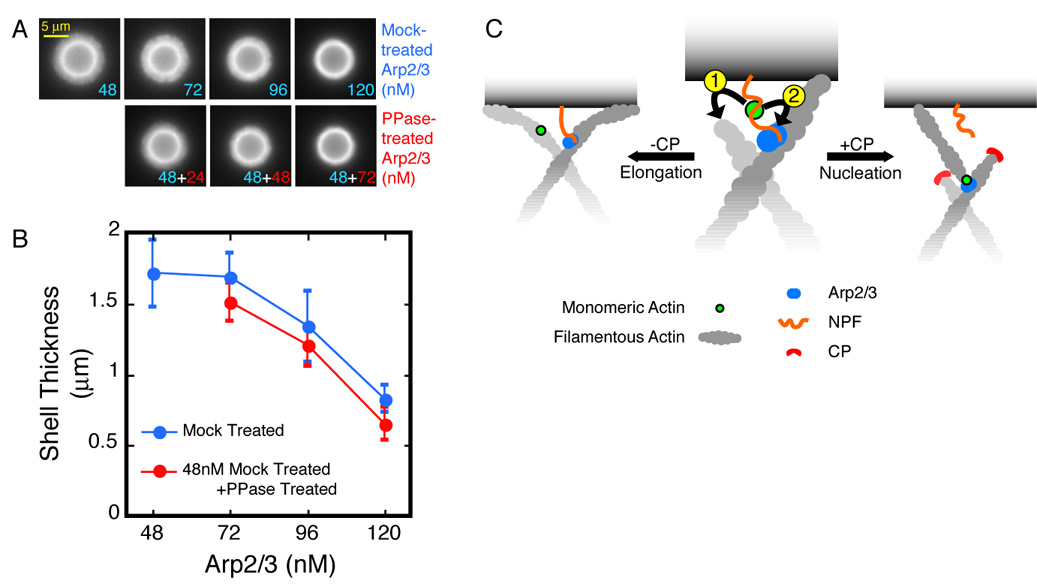
A. Dephosphorylated Arp2/3 complex slows down shell growth as effectively as the nucleation-competent complex. Conditions: 5.8 µm ActA-coated beads mixed with 3 µM actin (5% TMR-labeled), 112 nM CP, and Arp2/3 as indicated. Actin assembly was arrested 60 seconds after mixing.
B. Shell sizes for the conditions in (A). Error bars represent standard deviation (average n=20).
C. Monomer Gating: NPF-bound actin monomers can either associate with nearby barbed ends (1) and contribute to filament elongation, or they can participate in dendritic nucleation (2). Elongation is kinetically favored over nucleation, which requires the stable association of four protein partners—the mother filament, Arp/3, NPF, and the actin monomer—through a slow activation step. CP, by eliminating the competition from barbed ends, gates the biochemical path of actin monomers toward nucleation.
Discussion
The effect of CP on our reconstituted motility system is remarkably similar to its effect on actin organization and motility in living cells. Consistent with previous studies (Loisel et al., 1999; van der Gucht et al., 2005), we find that the rate at which Arp2/3-generated actin networks (both polarized comet tails and spherically symmetrical actin shells) move away from the surface of an ActA-coated bead increases with increasing CP concentration (Figure 1C and Figure 4B). Increased CP concentrations, however, do not increase the concentration of monomeric actin available during steady state motility (Figure 1F) or symmetrical shell growth (Figure 4C). Capping protein also has no effect on the average number of free barbed ends contributing to motility or the overall rate of polymer assembly. These results imply that CP also does not affect the average rate of filament elongation. All of these observations are incompatible with the major predictions of the Actin Funneling Hypothesis, namely that capping protein: (1) decreases the number of free barbed ends; (2) increases the soluble actin monomer concentration; and (3) increases the rate of filament elongation (Carlier and Pantaloni, 1997). The Funneling Hypothesis cannot, therefore, explain the effect of capping protein on actin network architecture and motility.
CP and Nucleation: The Monomer Gating Model
The most dramatic biochemical effect of capping protein in our system is to increase the rate of Arp2/3-dependent filament formation (Figure 2B and Figure 5D). How does capping filament barbed ends enhance the nucleation activity of the Arp2/3 complex? At the surface of an ActA-coated bead (or the leading edge of a motile cell) actin monomers can participate in one of two processes: elongating existing filaments or promoting dendritic nucleation by the Arp2/3 complex. Dendritic nucleation requires assembly of a four-member complex: the Arp2/3 complex itself, a pre-formed actin filament, an NPF, and an actin monomer (Figure 6C). The actin monomer required for nucleation must be bound to the WH2 domain of the NPF (Dayel and Mullins, 2004). An actin monomer bound to a WH2 domain, however, can interact with either the Arp2/3 complex or the barbed end of an actin filament (Co et al., 2007; Higgs et al., 1999). In other words, free barbed ends can compete with the Arp2/3 complex for WH2-bound actin monomers. Because the density of elongating barbed ends at the surface of our ActA-coated beads is extremely high (~2000 /µm2), we propose that CP promotes nucleation by capping the barbed ends near the surface that compete with the Arp2/3 complex for WH2-bound actin. When CP terminates elongation of a filament, it provides nearby WH2-bound actin monomers more time to participate in the nucleation reaction (Figure 6C). In this way CP controls the destiny of the actin monomer, selecting between nucleation and filament elongation. We refer to this switching mechanism as ‘Monomer Gating’. In vivo, we expect that monomer gating by CP will also shunt poly-proline-bound profilin:actin complexes away from filament elongation and toward dendritic nucleation.
Monomer Gating and Actin Funneling are fundamentally different explanations for the effect of CP on actin assembly. The Funneling Hypothesis proposes that CP forms a constriction, like the neck of a funnel, which concentrates the flow of monomeric actin onto a small subset of the total number of barbed ends. The Monomer Gating model proposes that CP acts more as a switch than a constriction and selects between two different processes, filament elongation and filament nucleation. Because of this switch, CP does not change the number of free barbed ends, the concentration of monomeric actin, or the overall rate of actin polymerization but dramatically changes the architecture and movement of the actin network.
Monomer Gating predicts that cellular factors that protect barbed ends from CP would suppress Arp2/3-dependent nucleation and lead to the reorganization of the actin cytoskeleton. One such factor is VASP, which has been shown to antagonize CP and promote the transformation from dendritic/lamellipodial to bundled/filopodial actin architecture in vivo (Mejillano et al., 2004). This is consistent with the effect of VASP on in vitro actin-based motility, where it decreases the density of Arp2/3-dependent branches in actin comet tails (Samarin et al., 2003), increases the average length of actin filaments, and promotes their alignment along the direction of motion (Plastino et al., 2004). As noted recently (Applewhite et al., 2007), the effects of VASP on actin network assembly can not be attributed solely to protecting barbed ends from CP. In order to promote filopodia formation in vivo and enhance motility in vitro, VASP must be bringing elongating barbed ends together and keeping them engaged with the load. Thus, in keeping with Monomer Gating, VASP suppresses dendritic nucleation by inhibiting CP, while promoting an alternate, Arp2/3-independent organization for force generation.
Nucleation, Network Architecture, and Motility
Given a constant rate of polymer assembly, how does changing the frequency of branching affect actin-based motility? First, frequent rounds of dendritic nucleation continually redirect actin assembly to the load surface, onto new filaments that are more efficient at generating force. A filament formed by the Arp2/3 complex begins life very short, mechanically rigid, and positioned to elongate against the load. According to the Elastic Brownian Ratchet Model of actin-based motility, shorter and stiffer filaments are mechanically better suited to harness the free energy of polymerization to perform mechanical work (Mogilner and Oster, 1996). Second, nucleation incorporates Arp2/3 into a network branchpoint and promotes release of the NPF (Dayel et al., 2001; Martin et al., 2006). Thus, one outcome of more frequent nucleation is faster removal of excess surface-bound Arp2/3, which reduces the drag force opposing network displacement. We note that this second mechanism may be specific to our reconstituted system. We propose that the first mechanism is the cause of faster motility with increasing CP concentration both in vitro and in vivo. This view is supported by an in vivo study which reported a correlation between increased capping activity and faster cell migration with higher density of Arp2/3-mediated branching in the lamellipod actin network (Bear et al., 2002).
Tethering by the Arp2/3 Complex
Increasing the concentration of the Arp2/3 complex in our system has no effect on the rate of filament nucleation (Figure 2B and Figure 5D), suggesting that soluble Arp2/3 is not limiting for the nucleation reaction. In contrast to CP, however, increasing the concentration of the complex actually decreases the rate of actin-based motility. This effect is mimicked by addition of a nucleation-dead complex (Figure 6B), indicating that the slowdown of motility is independent of the nucleation activity of the complex. These results argue strongly that the complex opposes motility by acting as a tether, increasing the drag force opposed to motility. This is not surprising, as the Arp2/3 complex binds to both the NPF (ActA) on the surface of the bead and to the sides of preformed filaments. At the bead surface, we estimate the concentration of filamentous actin to be in the millimolar range, three orders of magnitude higher than the KD for actin binding to the Arp2/3 complex (Mullins and Pollard, 1999). Under these conditions every ActA-bound Arp2/3 complex on the surface of the bead will be attached to an actin filament.
Force Response of Motile Networks
How do motile actin networks respond to increasing force? Increasing the Arp2/3 concentration increases the drag force opposing motility. While the velocity of motility varies significantly with changes in Arp2/3 concentration, the rate of actin network assembly is largely insensitive to these changes. This uncoupling of velocity from rate of network assembly highlights the importance of network architecture in motility. By changing the average orientation of filaments with respect to the surface of the load, dendritic actin networks can achieve dramatically different packing densities. Rather than slowing or stalling with increasing force, actin filaments take the path of least resistance and assume shallower glancing angles, becoming more parallel to the bead surface. The failure mode of this force adaptation is loss of contact between barbed ends and the surface, as observed with non-motile actin halos and asters. By continually resupplying the surface with new barbed ends, dendritic nucleation ensures that elongating filaments continue to engage the load. External forces can cause changes in the density (and, therefore, the mechanical properties) of the network without altering the flux of actin through the network. In this way, dendritic nucleation resembles an automobile transmission system. The power output of actin assembly remains constant and higher loads can be tolerated at the cost of reduced velocities.
Experimental Procedures
Proteins were purified using established methods. Arp2/3 was dephosphorylated by incubation with Antarctic Phosphatase (NEB) for 60 minutes at 30°C. The effect of phosphatase treatment on the nucleation activity of Arp2/3 was measured in pyrene actin polymerization assays. The activity of the dephosphorylated samples were typically reduced to 7–10% of the parental stock; mock-treated samples were unaffected. The residual activity of dephosphorylated Arp2/3 was taken into account when calculating the total concentration of nucleation-competent Arp2/3 in shell growth reactions.
Carboxylated polystyrene beads (BangsLabs) were covalently coated with ActA using EDC:sulfoNHS (Pierce) chemistry. The reconstituted motility mix contained 0.5 mM ATP, 1 mM MgCl2, 1 mM EGTA, 15 mM TCEP-HCl , 50 mM KOH (to neutralize TCEP-HCl), and 20 mM HEPES [pH 7.0]. For microscopic observation, we included 2.5 mg/mL BSA and 0.2% methylcellulose (M0262, Sigma) to minimize non-specific protein adsorption and drift. We used 100 mM β-ME to reduce photobleaching during low light intensity time-lapse imaging. We reserved the use of the oxygen scavenging system (glucose, glucose oxydase, catalase) for brief observations at high light intensity (e.g. FSM) to avoid the effects of medium acidification.
Brain cytosolic extract was prepared from frozen bovine tissue as described previously (Laurent et al., 1999). In addition to extract, which contributed 10% of the final volume, motility reactions contained 2 mM ATP, 2 mM MgCl2, 1 mM EGTA, 15 mM TCEP-HCl , 50 mM KOH (to neutralize TCEP-HCl), 20 mM HEPES [pH 7.0], 3 mg/mL BSA, 0.1% methylcellulose, and 2 µM actin (10% TMR-labeled).
All image processing and analysis was performed with Matlab (Mathworks) using custom routines.
See Supplemental Experimental Procedures for more details.
Supplementary Material
Acknowledgements
We thank the members of the Mullins laboratory, and especially Margot E. Quinlan, Alex E. Kelly, and J. Bradley Zuchero for their support and helpful discussions. We are grateful to Alex Mogilner for his insights and to Henry Bourne and Wallace Marshall for their careful reading of the manuscript. Special thanks are due to Lawrence L. LeClaire, III, for sharing his work on Arp2/3 dephosphorylation before publication. Finally, we thank John Cooper and Tom Pollard for generously providing bacterial expression constructs for CP, human Profilin I, and actophorin. O.A. dedicates this work to his father, Osman N. Akin. This work was supported by grants to R.D.M. from the National Institutes of Health (RO1 grant GM61010) and the Sandler Family Supporting Foundation and by funding from the NIH via the UCSF/UC Berkeley Nanomedicine Development Center (NDC) and a Howard Hughes Medical Institute Predoctoral Fellowship to O.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Applewhite DA, Barzik M, Kojima S, Svitkina TM, Gertler FB, Borisy GG. Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol Biol Cell. 2007;18:2579–2591. doi: 10.1091/mbc.E06-11-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B, Kunda P. Actin nucleation: spire - actin nucleator in a class of its own. Curr Biol. 2005;15:R305–R308. doi: 10.1016/j.cub.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, Gertler FB. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Bernheim-Groswasser A, Wiesner S, Golsteyn RM, Carlier MF, Sykes C. The dynamics of actin-based motility depend on surface parameters. Nature. 2002;417:308–311. doi: 10.1038/417308a. [DOI] [PubMed] [Google Scholar]
- Cameron LA, Footer MJ, van Oudenaarden A, Theriot JA. Motility of ActA protein-coated microspheres driven by actin polymerization. Proc Natl Acad Sci U S A. 1999;96:4908–4913. doi: 10.1073/pnas.96.9.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron LA, Robbins JR, Footer MJ, Theriot JA. Biophysical parameters influence actin-based movement, trajectory, and initiation in a cell-free system. Mol Biol Cell. 2004;15:2312–2323. doi: 10.1091/mbc.E03-12-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Pantaloni D. Control of actin dynamics in cell motility. J Mol Biol. 1997;269:459–467. doi: 10.1006/jmbi.1997.1062. [DOI] [PubMed] [Google Scholar]
- Co C, Wong DT, Gierke S, Chang V, Taunton J. Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell. 2007;128:901–913. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, Pollard TD. Effect of capping protein on the kinetics of actin polymerization. Biochemistry. 1985;24:793–799. doi: 10.1021/bi00324a039. [DOI] [PubMed] [Google Scholar]
- Dabiri GA, Sanger JM, Portnoy DA, Southwick FS. Listeria monocytogenes moves rapidly through the host-cell cytoplasm by inducing directional actin assembly. Proc Natl Acad Sci U S A. 1990;87:6068–6072. doi: 10.1073/pnas.87.16.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayel MJ, Holleran EA, Mullins RD. Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments. Proc Natl Acad Sci U S A. 2001;98:14871–14876. doi: 10.1073/pnas.261419298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayel MJ, Mullins RD. Activation of Arp2/3 complex: addition of the first subunit of the new filament by a WASP protein triggers rapid ATP hydrolysis on Arp2. PLoS Biol. 2004;2:E91. doi: 10.1371/journal.pbio.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy RJ, Han J, Condeelis JS. Capping protein terminates but does not initiate chemoattractant-induced actin assembly in Dictyostelium. J Cell Biol. 1997;139:1243–1253. doi: 10.1083/jcb.139.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Rodenbusch SE, Martin AC, Welch MD. Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol Cell. 2004;16:269–279. doi: 10.1016/j.molcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- Haviv L, Brill-Karniely Y, Mahaffy R, Backouche F, Ben-Shaul A, Pollard TD, Bernheim-Groswasser A. Reconstitution of the transition from lamellipodium to filopodium in a membrane-free system. Proc Natl Acad Sci U S A. 2006;103:4906–4911. doi: 10.1073/pnas.0508269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN, Blanchoin L, Pollard TD. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- Hug C, Jay PY, Reddy I, McNally JG, Bridgman PC, Elson EL, Cooper JA. Capping protein levels influence actin assembly and cell motility in dictyostelium. Cell. 1995;81:591–600. doi: 10.1016/0092-8674(95)90080-2. [DOI] [PubMed] [Google Scholar]
- Iwasa JH, Mullins RD. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr Biol. 2007;17:395–406. doi: 10.1016/j.cub.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kelleher JF, Atkinson SJ, Pollard TD. Sequences, structural models, and cellular localization of the actin-related proteins Arp2 and Arp3 from Acanthamoeba. J Cell Biol. 1995;131:385–397. doi: 10.1083/jcb.131.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AE, Kranitz H, Dotsch V, Mullins RD. Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J Biol Chem. 2006;281:10589–10597. doi: 10.1074/jbc.M507470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR. Molecular details of formin-mediated actin assembly. Curr Opin Cell Biol. 2006;18:11–17. doi: 10.1016/j.ceb.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Laurent V, Loisel TP, Harbeck B, Wehman A, Grobe L, Jockusch BM, Wehland J, Gertler FB, Carlier MF. Role of proteins of the Ena/VASP family in actin-based motility of Listeria monocytogenes. J Cell Biol. 1999;144:1245–1258. doi: 10.1083/jcb.144.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClaire LL, III, Baumgartner M, Iwasa JH, Mullins RD, Barber DL. Phosphorylation of the Arp2/3 Complex is Necessary for Actin Nucleation. Manuscript in review. 2008 doi: 10.1083/jcb.200802145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Reeves E, Wientjes F, Mattheyse FJ, Grogan A, Totty NF, Burlingame AL, Hsuan JJ, Segal AW. Mammalian actin-related protein 2/3 complex localizes to regions of lamellipodial protrusion and is composed of evolutionarily conserved proteins. Biochem J. 1997;328(Pt 1):105–112. doi: 10.1042/bj3280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- Martin AC, Welch MD, Drubin DG. Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nat Cell Biol. 2006;8:826–833. doi: 10.1038/ncb1443. [DOI] [PubMed] [Google Scholar]
- May RC, Caron E, Hall A, Machesky LM. Involvement of the Arp2/3 complex in phagocytosis mediated by FcgammaR or CR3. Nat Cell Biol. 2000;2:246–248. doi: 10.1038/35008673. [DOI] [PubMed] [Google Scholar]
- Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Qualmann B, Kessels MM, Almers W. Neural Wiskott Aldrich Syndrome Protein (N-WASP) and the Arp2/3 complex are recruited to sites of clathrin-mediated endocytosis in cultured fibroblasts. Eur J Cell Biol. 2004;83:13–18. doi: 10.1078/0171-9335-00356. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Tsuji T, Higashida C, Hertzog M, Fujita A, Narumiya S, Scita G, Watanabe N. Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. J Cell Biol. 2006;175:947–955. doi: 10.1083/jcb.200604176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A, Oster G. Cell motility driven by actin polymerization. Biophys J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Pollard TD. Rho-family GTPases require the Arp2/3 complex to stimulate actin polymerization in Acanthamoeba extracts. Curr Biol. 1999;9:405–415. doi: 10.1016/s0960-9822(99)80187-0. [DOI] [PubMed] [Google Scholar]
- Plastino J, Olivier S, Sykes C. Actin Filaments Align into Hollow Comets for Rapid VASP-Mediated Propulsion. Curr Biol. 2004;14:1766–1771. doi: 10.1016/j.cub.2004.09.054. [DOI] [PubMed] [Google Scholar]
- Samarin S, Romero S, Kocks C, Didry D, Pantaloni D, Carlier MF. How VASP enhances actin-based motility. J Cell Biol. 2003;163:131–142. doi: 10.1083/jcb.200303191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Jennings PB, Cooper JA. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J Cell Biol. 1996;135:169–179. doi: 10.1083/jcb.135.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JM, Galyov EE, Stevens MP. Actin-dependent movement of bacterial pathogens. Nat Rev Microbiol. 2006;4:91–101. doi: 10.1038/nrmicro1320. [DOI] [PubMed] [Google Scholar]
- van der Gucht J, Paluch E, Plastino J, Sykes C. Stress release drives symmetry breaking for actin-based movement. Proc Natl Acad Sci U S A. 2005;102:7847–7852. doi: 10.1073/pnas.0502121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D, Yarar D, Welch MD, Peloquin J, Svitkina T, Borisy GG. Formation of filopodia-like bundles in vitro from a dendritic network. J Cell Biol. 2003;160:951–962. doi: 10.1083/jcb.200208059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol. 1997;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner S, Helfer E, Didry D, Ducouret G, Lafuma F, Carlier MF, Pantaloni D. A biomimetic motility assay provides insight into the mechanism of actin-based motility. J Cell Biol. 2003;160:387–398. doi: 10.1083/jcb.200207148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D, To W, Abo A, Welch MD. The Wiskott-Aldrich syndrome protein directs actin-based motility by stimulating actin nucleation with the Arp2/3 complex. Curr Biol. 1999;9:555–558. doi: 10.1016/s0960-9822(99)80243-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


