Abstract
BLUF domains constitute a recently discovered class of photoreceptor proteins found in bacteria and eukaryotic algae. BLUF domains are blue-light sensitive through a FAD cofactor that is involved in an extensive hydrogen-bond network with nearby amino acid side chains, including a highly conserved tyrosine and glutamine. The participation of particular amino acid side chains in the ultrafast hydrogen-bond switching reaction with FAD that underlies photoactivation of BLUF domains is assessed by means of ultrafast infrared spectroscopy. Blue-light absorption by FAD results in formation of FAD•− and a bleach of the tyrosine ring vibrational mode on a picosecond timescale, showing that electron transfer from tyrosine to FAD constitutes the primary photochemistry. This interpretation is supported by the absence of a kinetic isotope effect on the fluorescence decay on H/D exchange. Subsequent protonation of FAD•− to result in FADH• on a picosecond timescale is evidenced by the appearance of a N-H bending mode at the FAD N5 protonation site and of a FADH• C=N stretch marker mode, with tyrosine as the likely proton donor. FADH• is reoxidized in 67 ps (180 ps in D2O) to result in a long-lived hydrogen-bond switched network around FAD. This hydrogen-bond switch shows infrared signatures from the C-OH stretch of tyrosine and the FAD C4=O and C=N stretches, which indicate increased hydrogen-bond strength at all these sites. The results support a previously hypothesized rotation of glutamine by ∼180° through a light-driven radical-pair mechanism as the determinant of the hydrogen-bond switch.
INTRODUCTION
The last decade has witnessed the discovery and characterization of a large number of novel photoreceptors, notably the LOV domains (1–4) and the BLUF domains (5–7). BLUF and LOV domains are of special interest as they bind a flavin rather than an isomerizing cofactor, making their photochemistry radically different from that of “traditional” photoreceptors such as the rhodopsins, phytochromes, and xanthopsins (8). The flavin photochemistry enables a strict separation regarding the roles of cofactor and protein: isomerizing cofactors show isomerization and twisting reactions in solution and even in vacuo (9), rendering the influence and catalyzing properties of the protein difficult to assess. In contrast, flavins need partner molecules to react. Thus, flavin-based photoreceptors pose us with new concepts and opportunities to understand how light absorption may be coupled to biological sensory function through efficient and selective photochemistry (4,10–19).
The vibrational spectrum of a protein or a protein-bound chromophore contains a wealth of information about its structure, the interaction with the environment and electronic properties. Time-resolved IR spectroscopy is a powerful tool that can show many of the dynamic structural and physical-chemical properties of chromophores involved in (photo)biological reactions (20,21). In addition, it can show the involvement of those parts of the protein that partake in the ongoing reactions. As the primary reactions in biological photoreceptors proceed on the ultrafast timescale, femtosecond IR spectroscopy is the method of choice to identify reaction mechanisms of biological photoreceptors (22–28).
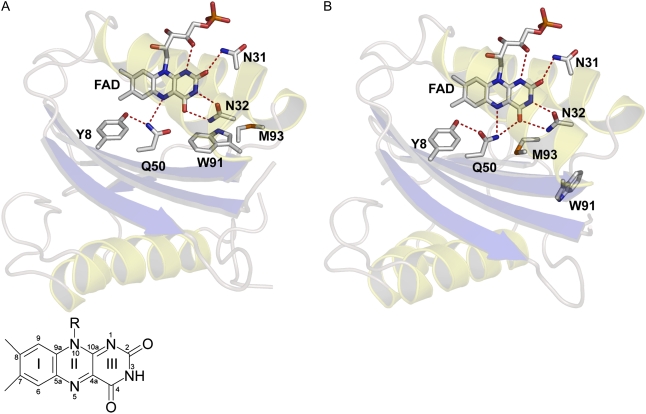
BLUF domains are found in bacteria and eukaryotic algae where they serve a number of physiological functions such as phototaxis, photophobia, and photosynthesis gene expression (5–7,29). The structure of the BLUF domain shows a ferredoxin-like fold consisting of a five-stranded β-sheet with two α-helices packed on one side (30–35). The noncovalently bound isoalloxazine ring of FAD is positioned between the two α-helices. Fig. 1 shows the x-ray structure of the Synechocystis Slr1694 BLUF protein (also referred to as PixD), with the FAD-binding pocket highlighted. FAD is involved in an extensive hydrogen-bond network with the side chains lining its binding pocket, including a highly conserved tyrosine (Tyr-8) and glutamine (Gln-50). On illumination with blue light, BLUF domains show a characteristic spectral red-shift of their absorption spectrum by ∼10 nm, which is thought to correspond to the signaling state (7). Vibrational spectroscopy has indicated that a light-induced hydrogen-bond rearrangement among FAD and nearby amino acid side chains underlies BLUF photoactivation (36–38). Light-induced rotation of the conserved glutamine would readily enable such putative hydrogen-bond switch (11,30,39,40), as indicated in Fig. 1, A and B, for the proposed dark and light conformations of the Slr1694 BLUF domain (35).
FIGURE 1.
X-ray structure of the Synechocystis Slr1694 BLUF domain (35) with the hydrogen bond patterns lining the FAD binding pocket in proposed dark state (A) and light state (B) configurations. The inset of A shows structure and numbering of the FAD isoalloxazine ring.
Femtosecond UV-vis spectroscopy has shown that in the Slr1694 BLUF domain from Synechocystis, blue-light absorption was followed by sequential formation of anionic FAD•− and neutral FADH• radicals on a picosecond timescale, showing that sequential electron and proton transfer constitute the BLUF photoactivation mechanism, which, on radical-pair recombination, results in the red-shifted product state in <200 ps (39). By comparing this result with the x-ray structure, a molecular mechanism was put forward by which light absorption by FAD drives sequential electron and proton transfer from the conserved Tyr to FAD, followed by a ∼180° rotation of the conserved Gln and ensuing hydrogen-bond switch. Yet, the catalytic role of particular amino acid side chains in the BLUF photoreaction remains to be shown. We present a femtosecond IR study of the Slr1694 BLUF domain from Synechocystis to experimentally assess the involvement of amino acid side chains in the vicinity of the FAD cofactor and their hydrogen-bond patterns with FAD in the ultrafast and efficient photo-induced hydrogen bond-switching reaction.
MATERIALS AND METHODS
Femtosecond IR spectroscopy was carried out with a setup described previously (22). Time-resolved fluorescence experiments were carried out by means of a synchroscan streak camera (41). The time-resolved data were analyzed using global and target analysis techniques as described in van Stokkum et al. (42). The sample preparation has been described in Zirak et al. (43). A detailed account of sample preparation, the experimental setups and data analysis techniques is given in the Supplementary Material, Data S1. A HPLC analysis of the Slr1694 sample indicates a heterogeneous flavin composition of 37% FAD, 38% FMN, and 25% riboflavin. Because all flavin vibrational bands observed in this study belong to the isoalloxazine ring and their frequencies are identical for FAD, FMN, and riboflavin within the 4 cm−1 spectral resolution of the ultrafast IR apparatus (38,44,45) no significant effect of the cofactor heterogeneity on the ultrafast vibrational spectra is expected. For convenience of notation, we will refer to the flavin cofactor as FAD throughout this article.
RESULTS AND DISCUSSION
Time-resolved fluorescence spectroscopy
The time-resolved fluorescence of the Slr1694 BLUF domain was recorded by means of a synchroscan streak camera with excitation at 400 nm. On excitation, the flavin is promoted to its singlet excited state, hereafter denoted FAD*. The fluorescence decay was highly nonexponential: five time constants were required for an adequate fit. Fig. 2 shows the resulting DAS. The first four DAS have a fluorescence maximum at ∼500 nm with a shoulder at ∼520 nm, as observed previously in the AppA BLUF domain (46). The lifetimes are 6 ps with 31% amplitude, 26 ps (37%), 92 ps (25%), and 335 ps (7%). In the Slr1694 BLUF domain in D2O, the lifetimes and amplitudes are essentially identical: 7 ps (29% amplitude), 28 ps (35%), 94 ps (25%), and 354 ps (11%). The fifth DAS has a lifetime of 4.5 ns, peaks at ∼520 nm and is associated with a minor fraction of unbound flavin in the sample. The concentrations and lifetimes of the resulting five DAS in H2O and D2O buffer are reported in Table 1. The streak images of Slr1694 in H2O and D2O buffer are shown in Fig. S1 in Data S1.
FIGURE 2.
DAS that follow from a global analysis of time-resolved fluorescence experiments on the Synechocystis Slr1694 BLUF domain in H2O (A) and D2O (B). The lifetime of each DAS in given in picoseconds with the relative contribution to the total fluorescence in parenthesis. The fifth DAS (bold line) is due to a minor fraction of unbound flavin and has a lifetime of 4.5 ns. The contribution of the free flavin was subtracted in the estimation of the relative contribution of the single DAS to the total fluorescence.
TABLE 1.
Lifetimes and fractional contributions in percentage for the fluorescence that result from fitting the picosecond time-resolved fluorescence data (panel A), the femtosecond time-resolved transient absorption data (global analysis (panel B); target analysis (panel C) for the Synechocystis Slr1694 BLUF domain in H2O and D2O
The absence of a KIE on the fluorescence decay on H/D exchange supports the idea that excited-state deactivation proceeds via electron transfer, presumably from the conserved Tyr-8 to FAD (11,39,47). The multiexponential decay of FAD* is very likely caused by the presence of conformational substates in the dark, as shown for the AppA BLUF domain by NMR spectroscopy. The slow timescale (μs-ms) exchange of the conserved Tyr between different conformational states leads to subpopulations having variations in the distance between FAD and Tyr, with an ensuing distribution of (fast) electron transfer rates as a result (31,46,48). In the AppA BLUF domain, electron transfer from the conserved Trp (Trp-104, AppA numbering) also contributes to the fluorescence quenching (48,49). It is important to note that the multiexponential decay of FAD* does not result from the heterogeneous flavin composition of the sample: in the heterologously expressed AppA BLUF domain reconstituted with FAD only, similar multiexponential FAD* decays were observed, in wild-type as well as W104 and Y21 mutants (46,48,49), Moreover, the fluorescence quantum yields of AppA samples reconstituted with riboflavin, FMN or FAD did not differ appreciably (50).
Infrared transient absorption spectroscopy on Slr1694
Femtosecond mid-IR spectra on excitation of the FAD cofactor were collected on Slr1694 dissolved in a D2O buffer (1780 to 1430 cm−1) and an H2O buffer (1300 to 1125 cm−1). These conditions were chosen to avoid the large absorption by O-H and O-D bending vibrations of H2O and D2O, respectively. The resulting data were first analyzed globally with a sequential kinetic scheme (see Fig. S2 B in Data S1). Five components were required for an adequate fit, with lifetimes 2.5 ps, 11 ps, 33 ps, 210 ps, and a long-living component in D2O. In H2O the lifetimes are estimated at 1.0 ps, 3 ps, 22 ps, 170 ps, and a long-living component (Table 1). These lifetimes are in close agreement with those found with femtosecond UV-visible transient absorption measurements on Slr1694 (39). Fig. 3 reports kinetic time traces at selected vibrational frequencies to illustrate the temporal evolution. The spectral evolution, represented by EADS, is reported in Fig. S3 in Data S1. To provide a complete overview of the ultrafast IR data, kinetic traces at all measured frequencies are shown in Data S1.
FIGURE 3.
Kinetic traces at indicated mid-IR vibrational frequencies of Synechocystis Slr1694 BLUF domain in D2O (A–F) and H2O (G–L) (circles). The excitation wavelength was 475 nm. The result of the target analysis is shown as a solid line. Note that the time axis is linear from −10 to 10 ps, and logarithmic thereafter.
It was shown before that from FAD*, formation of the long-lived red-shifted product state (SlrRED) proceeds via two distinct reaction intermediates called Q1 and Q2 (39). Due to the multiexponential decay of FAD*, the EADS that follow from the sequential analysis are made up from a mixture of these molecular species, i.e., FAD*, Q1, Q2, and SlrRED. To disentangle the contributions from these molecular species and obtain the infrared signature of the “pure” reaction intermediates, a specific kinetic model must be applied in a so-called target analysis procedure. In this way, the reaction mechanism can be assessed in terms of a number of discrete reaction intermediates (42). We applied the kinetic model used for the ultrafast UV-vis data (39) with the kinetic scheme described in Fig. 4. The observation of essentially identical time constants in the sequential analysis of UV-vis and this femtosecond IR data gives confidence that such approach will provide a realistic description of the photo-induced reaction dynamics. Moreover, as we showed for the UV-visible data, an evolution in terms of the two intermediates, Q1 and Q2, fits the ultrafast IR data significantly better than inclusion of only one intermediate Q1 (see Figs. S5 and S6 in Data S1). The multiexponential nature of FAD* decay is taken into account by four FAD* compartments, FAD*1–4 that are spectrally identical and for which the decay times from the time-resolved fluorescence experiments were used. We note that a stretched exponential function (51) could not adequately describe the decay of FAD*. FAD*1–3 evolve into the first intermediate Q1, whereas FAD*4 recombines to the ground state. Q1 evolves into the second intermediate Q2; it also features a loss channel of ∼50% to the ground state to account for the nonunity quantum yield of signaling-state formation in the Slr1694 BLUF domain (that amounts to ∼40% (39)). From Q2, the long-lived state SlrRED is formed. Fig. 5, A–C show the species-associated difference spectra (SADS) for Slr1694 in D2O and H2O, respectively. In Table 1, the concentrations and rate constants for each component forming the kinetic model are shown. The transient concentrations of FAD*, Q1, Q2, and SlrRED for Slr1694 in D2O and H2O are shown in Fig. S4 in Data S1.
FIGURE 4.
Kinetic scheme used for target analysis of the time-resolved data on the Slr1694 BLUF domain from Synechocystis. See text for details.
FIGURE 5.
(A) SADS that follow from a target analysis of the time-resolved mid-IR data of the Synechocystis Slr1694 BLUF domain in D2O on excitation at 475 nm; (B and C): same as A for the Slr1694 BLUF domain in H2O. See text for details.
Identification of FAD* vibrational signature
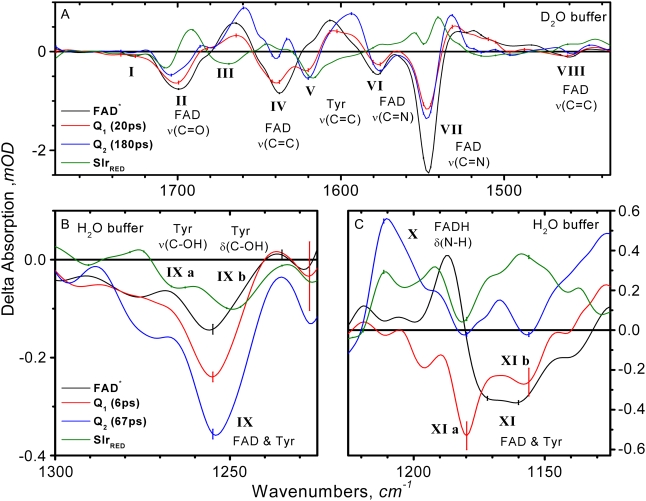
The vibrational signature of FAD* is represented by the black SADS in Fig. 5. Note that the FAD vibrational modes but not those of the protein bleach immediately after excitation and may thus readily be pinpointed. In this SADS, all the negative bands are thus attributed to the FAD cofactor ground state modes. Six bleaches are observed at 1728, 1699, 1638, 1578 and 1547, and 1460 cm−1, marked as I, II, IV, VI, VII, and VIII, respectively (note that because of band shifts in the various SADS, the band numbering refers to frequency regions rather than separate bands). These negative bands well resemble the vibrational frequencies of various flavin cofactors in solution (52,53).
Bands I and II at 1728 and 1699 cm−1 are assigned to carbonyl stretching modes (ν(C4=O), ν(C2=O)), which are coupled to N3-H bending (δ(N3H)), and having a major contribution from C4=O (54). These carbonyl modes vibrate at lower frequencies in the singlet excited state, showing an IA at 1665 cm−1 (Table 2). Such a downshift of the FAD C=O frequencies in the singlet excited state probably results from a decreased bond order associated with the π–π* transition. The bands at 1638 cm−1 (band IV) and 1460 cm−1 (band VIII) are predominant C=C stretching vibrations (ν(C=C)), associated with ring I of the isoalloxazine (38,45,53,54): the ring I vibration at 1638 cm−1 (bleach) downshifts to 1607 cm−1 (IA) on formation of FAD*. The negative bands at 1578 and 1547 cm−1 (bands VI and VII, respectively) are typical of the C=N double bond stretching in-phase and out-of-phase configuration [ν(C=N)in, ν(C=N)out] (38,45,52–56). These modes downshift on promotion to the singlet excited state with an IA at 1520 cm−1.
TABLE 2.
Spectral positions and assignments of the observed vibrational bands in the SADS that follow from a target analysis on femtosecond time-resolved transient absorption data of Slr1694 BLUF domain; see Fig. 5 and text for details
| SADS
|
||||||
|---|---|---|---|---|---|---|
| Bands | FAD* (black) | Q1 (red) | Q2 (blue) | SlrRED (green) | Assignment | |
| I | 1728 (−w) | ν(C4=O), ν(C2=O), δ(N3H) | ||||
| II | 1699 (−m) | 1701 (+2) | 1705 (+6) | 1707 (+8) | ν(C4=O), ν(C2=O), δ(N3H) | |
| 1665 (+m) | 1659 (−6) | 1692 | ||||
| 1685 (+vw) | ||||||
| III | 1670 (−) | Unknown, possibly amide I or ν(C=O) Gln-50 | ||||
| IV | 1638 (−m) | 1640 (+2) | 1640 | ν(C=C) I Ring | ||
| 1632 (+w) | ||||||
| V | 1621 (−m) | 1619 | 1617 (−4) | ν(C=C) Tyr-8 | ||
| 1607 (+m) | 1607 (−2) | |||||
| 1594 (+sh) | ||||||
| VI | 1578 (−w) | ν(C4a=N5), ν(C10a=N1) in phase | ||||
| VII | 1547 (−vs) | ν(C4a=N5), ν(C10a=N1) out phase | ||||
| 1528 (+m) | 1530 (+2) | 1532 (+4) | 1541 (+13) | |||
| 1519 (+m) | ||||||
| VIII | 1460 (−w) | 1462 (+2) | ν(C=C), ν(CMe), δ(CH) I Ring, δ(N3H) | |||
| 1276 (+) | ν(C-OH) Tyr-8 | |||||
| a | 1266 (−) | |||||
| IX | 1256 (−w) | 1256 | 1253 (−3) | ν(CC), ν(CN) III Ring, ν(C-OH)Tyr-8 | ||
| b | 1248 (−) | δ(C-OH) Tyr-8 | ||||
| 1235 (+) | ||||||
| X | 1211 (+m) | δ(N5-H) FADH | ||||
| a | 1179 (−m) | |||||
| XI | 1167 (−m) | ν(CC) III Ring, β-OH Tyr-8 | ||||
| b | 1157 (−m) | |||||
vw, very weak; w, weak; m, medium; s, strong; sh, strong shoulder; −, bleaching; +, induced absorption; −#, red shift of # wave numbers; +#, blue shift of # wavenumbers.
The observed bleach and IA bands of FAD* can be compared with those observed on flavin in solution by Kondo et al. (57). There, lower vibrational frequencies were observed for the carbonyl modes (1703 and 1646 cm−1) that probably resulted from the aqueous environment of flavin with ensuing extensive H-bonding to its carbonyls. Moreover, FAD in solution shows a lower frequency for the ring I vibration (1624 cm−1), a sign that the molecule may be less distorted in solution. The two C=N vibrations at 1578 and 1546 cm−1 reported by Kondo et al. (57) are in good agreement for both spectral position and relative intensity.
In our previous work on the AppA BLUF domain with ultrafast UV-vis spectroscopy, we observed a vibrational cooling process occurring in ∼2 ps for FAD* on excitation at 400 nm. On excitation at 475 nm, which is in the 0-0 vibronic transition of FAD, the spectroscopic signature for vibrational cooling was absent (46). Here, FAD is excited at 475 nm as well, so no significant contribution from vibrational cooling is expected for the FAD* IR spectra.
Q1: identification of a FAD•−-Tyr radical transient intermediate
Q1 was identified previously as a FAD•− radical anion by ultrafast UV/vis spectroscopy (39). Comparison of the Q1 SADS (Fig. 5, red lines) with the radical anionic IR spectrum of the flavin model compound RBTA in deutero-acetonitrile (CD3CN) (58) shows an overall good agreement. The principal C=O (band II) and C=N (bands VI and VII) stretch vibrations exhibit a downshift as a result of radical anion formation in both Q1 and RBTA. The C=O stretch downshift is significantly larger in RBTA as compared to Q1 (∼60 and 35 cm−1, respectively). However, this difference may only be apparent because of the large bleach contribution by ring I near 1640 cm−1 (band IV) in Q1. In RBTA, no such Ring I vibration was observed. Note that with its downshifted C=O and C=N stretch vibrations, the IR signature of the FAD anionic radical is quite similar to that of FAD*.
Strikingly, band V near 1620 cm−1 appears in the Q1 SADS. The rise of this bleach signal is clearly resolved in the kinetic trace of Fig. 3 D. The rise of negative features during the time evolution indicates the involvement of molecules other than those originally excited in the reaction. As a rule, only the amino acid side chains in close vicinity to FAD may reasonably be considered. The frequency of band V at ∼1620 cm−1 is typical for the C=C ring vibration of tyrosine (59) and it is accordingly assigned to Tyr-8. On selective labeling with [4-13C]Tyr, this band was downshifted in a FTIR experiment on the essentially identical PixD BLUF domain from Synechococcus (60), showing that this band belongs to tyrosine. Band IX (∼1256 cm−1) is present in the FAD* SADS and its intensity increases on population of Q1, as can also be seen in Fig. 3 G. A ∼ 1250 cm−1 frequency corresponds to the δ(C-OH) and ν(C-OH) modes of Tyr (59,61–63). As a bleach is already present immediately after excitation (cf. the FAD* SADS and Fig. 3 G), band IX likely represents a mixture of FAD and Tyr-8.
Band XI appears with two peaks at 1179 (XIa) and 1157 cm−1 (XIb). The time trace of XI is reported in Fig. 3 I. Evidently, the bands become more negative during the time evolution. The 1179 and 1157 cm−1 modes are typical for a Tyr ring vibration mixed with δ(CH) and δ(C-OH) (64–67). Similar 1179 and 1157 cm−1 bands were also observed in flavin in solution (45,54,55), and for FMN bound to LOV1 from Chlamydomonas reinhardtii (45). In this particular case, FAD* shows a large and sharp IA at 1188 cm−1 that distorts the FAD bleaches, precluding distinction between FAD and amino acid signals in this vibrational frequency region.
We conclude that Q1 represents a superposition of FAD•− vibrational features together with a bleach of Tyr-8 modes. The infrared spectral signature of Q1 is consistent with a photoactivation model where light absorption by FAD triggers electron transfer from Tyr-8 to FAD, resulting in formation of FAD•−-Tyr•+ (39).
As large electric fields may shift the vibrational frequencies of complex molecules (68), it is possible that signals from amino acids near FAD are affected by electric field effects arising from excited or charge-separated states. Such Stark effects would expected to be most prominent in the Q1 intermediate, with its short-range one-electron charge-separated state, and to a less extent FAD*, where the lowest excited state is known to exhibit a modest dipole change |Δμ| of 1.5 D with respect to the ground state (69). However, the FAD* and Q1 SADS reported in Fig. 5 are well understood in terms of a superposition of FAD excited and FAD-Tyr radical states, and no contributions from vibrational Stark signals need to be invoked. We note that for bacteriorhodopsin and PYP, where the protonated Schiff base of retinal and p-coumaric acid exhibit a very large |Δμ| of 13.5 and 26 D, respectively (70,71), no vibrational Stark effects were reported (22,23).
Q2: identification of a FADH•-Tyr radical transient intermediate
The Q2 SADS is given by the blue lines in Fig. 4. Our previous work indicated that Q2 represents the neutral flavin semiquinone radical (FADH•) (39). As we did for the FAD•− spectrum, we compare this SADS with that of the flavin model compound RBTAH• (58). In RBTAH• and Q2, the flavin C=O stretches are similarly downshifted by ∼40 cm−1. Most notably, as compared to Q1, the C=N stretch frequency (the IA associated with band VII near 1550 cm−1) shows a significant narrowing and a small upshift of the IA to 1532 cm−1. The temporal evolution of this IA feature is shown in Fig. 3 F. In RBTAH•, a similar upshift and narrowing is observed as compared to RBTA•−.
An interesting feature of the Q2 SADS is the positive band at 1211 cm−1 (Band X). This band is evidently formed in the Q2-intermediate time range and then decays (Fig. 3 H). A similar band at 1232 cm−1 was observed in the neutral semiquinone FMNH• of flavodoxin in H2O and assigned to the N5-H bending mode (56). In the kinetic trace at 1211 cm−1 (Fig. 3 H) the Q1 to Q2 transition becomes fully apparent in the trace where we observe the rise of the FAD N5-H bending vibration, which is likely due to transient protonation at N5 on formation of the neutral semiquinone (i.e., FAD•− to FADH•) and its subsequent decay in 65 ps by deprotonation on formation of SlrRED. There is little interference from the multi-exponential decay of FAD* because its SADS is almost zero in this frequency range, and we consequently have a relatively clean view of the Q1 to Q2 dynamics. We conclude that the IR signature of the Q2 SADS is consistent with the presence of a FADH• neutral radical intermediate.
The bands assigned to Tyr-8 (V, IX) are still present in the Q2 SADS, indicating that Tyr-8 remains involved at this stage of the photoreaction. Considering the positive IA on the low-frequency side of band V near 1600 cm−1, we observe a progressive downshift from 1607 to 1593 cm−1 on going from Q1 to Q2. Although changes in the flavin Ring I vibration may also contribute to the observed signals (that are difficult to assess because the RBTA model compound from Martin et al. (58) did not exhibit a Ring I vibration), such progressive red shifting may be expected from the vibrational ring modes of tyrosine involved in sequential electron transfer and proton transfer, passing from Tyr to Tyr•+ and Tyr•+ to Tyr•, respectively. Loss of an electron from Tyr leads to an overall weakening of C=C and C-C bonds, downshifting its frequency. For tyrosinate (Tyr-O−), the CC ring mode was shown to downshift to ∼1600 cm−1 (59,63), indicating that proton transfer also results in a weakening of the CC bonds of the ring. Thus, if the effects of electron and proton transfer on the Tyr ring vibration are additive, the progressive down shifts of the IA can be explained as a successive evolution Tyr → Tyr-OH•+ and Tyr-OH•+ → Tyr-O·. Moreover, on formation of the neutral radical Tyr-O·, the oxygen will be more electronegative than in the Tyr-OH state, dragging away electron density from the ring and downshifting its C=C bond frequency. We conclude that the IR signature of Q2 corresponds to a FADH•-Tyr radical pair, where Tyr-8 may reasonably be assigned to its neutral radical tyrosyl form, Tyr-O·.
SlrRED: the ultrafast hydrogen bond-switched network
The formation of SlrRED (Fig. 5, green curve) from Q2 is isotope-dependent, taking 180 ps in D2O and 67 ps in H2O. SlrRED represents the re-oxidized FAD experiencing a different H-bond network with nearby amino acid side chains. A most conspicuous feature is Band II, assigned mainly to ν(C4=O), which shows up as a bandshift from 1707 to 1692 cm−1. The kinetic trace at 1692 cm−1 is shown in Fig. 3 B. The shift of Band II is similar to that observed previously with FTIR and accordingly interpreted as a weakening of the C4=O carbonyl stretching vibration through formation of an additional hydrogen bond to C4=O (36). Band VII, associated with the FAD C=N stretch, downshifts to 1541 cm−1 (Fig. 3 E). This observation indicates that in the SlrRED state, hydrogen-bonding to FAD N5 is strengthened.
The SlrRED IR signature presents pronounced changes in amino acid absorbance, in particular from Tyr-8. Band V at 1617 cm−1 (ν(C=C) of Tyr-8) shows a bleach similar to that in Q1 and Q2 indicating that Tyr-8 is involved in the altered hydrogen-bond patterns. Band V lacks an associated IA and we cannot assign a frequency shift for this Tyr mode. Strikingly, the shifting pattern of bands IXa and IXb near 1250 cm−1 is essentially identical to that observed by Takahashi et al. in FTIR on the Synechococcus PixD (Tll0078) BLUF domain (60). Using selective labeling of Tyr with [4-13C]Tyr, these modes were unambiguously assigned to C-O stretch vibration ν(CO) and to the C-OH bending vibration δ(C-OH) of Tyr-8 in the latter work. Band IXa upshifts from 1266 to 1276 cm−1 whereas Band IXb downshifts from 1248 to 1235 cm−1.
DFT calculations indicated that the Tyr ν(CO) frequency depends on specific H-bonding interactions: Tyr donating a hydrogen bond to a carbonyl resulted in a higher ν(CO) frequency than a hydrogen bond between Tyr and an amino group, with Tyr acting either as donor or acceptor (60). Thus, the ν(CO) upshift from 1266 to 1276 cm−1 is consistent with the dark and light hydrogen-bond patterns as described in Fig. 1, A and B: in the dark, a hydrogen bond exists between Tyr-8 and the amino group of Gln-50. In the light, Tyr-8 donates a hydrogen bond to the carbonyl of Gln-50. More specifically for the dark, the DFT calculations were quantitatively more in line with Tyr-8 acting as a H-bond donor than an acceptor, with Tyr-8 donating a weak H-bond to the amino moiety of Gln-50. Considering that the Tyr-8 OH proton definitely is more acidic than that of the Gln-50 NH2 amino, such a dark conformation does not seem unreasonable. Note that such conformation bears little consequences for the reaction mechanism because the dark orientation of Gln-50 remains identical in these cases.
A striking observation concerns the appearance of Band III at 1670 cm−1 (bleach), which is formed in 180 ps during the Q2 to SlrRED evolution (cf. Fig. 3 C). Band III is not straightforward to assign: its frequency matches that of a carbonyl stretch vibration. It might result from a change in Amide I vibrations, which would imply that significant backbone motion takes place on a 100 ps timescale, presumably resulting from strains near Tyr-8 and Gln-50 induced by the hydrogen-bond switch. Such very rapid backbone motion has been observed before in designed photo switchable azobenzene-peptide constructs (72) and in proteorhodopsin (62,73). It should be noted, however, that in the latter systems, the chromophore is covalently attached to the protein, establishing strains on isomerization that may be significantly larger than those resulting from the hydrogen-bond switch in the BLUF domain. Another possible origin of Band III is the ν(C=O) carbonyl stretching of Gln-50 (59,63). If this assignment is correct, its frequency at 1670 cm−1 indicates that the Gln-50 carbonyl would be hydrogen-bonded in the dark, because in the absence of hydrogen-bonding Gln will have its carbonyl stretch vibration above 1700 cm−1 (28). In the AppA BLUF domain dark structure, the conserved Gln carbonyl indeed receives a hydrogen bond from the conserved Trp-104 (30). However, in the Synechocystis BLUF x-ray structure, the Gln-50 carbonyl is not hydrogen-bonded because Trp-91 has a different position that is out of reach for hydrogen-bonding (35). Possibly, in solution, Trp-91 in Synechocystis BLUF assumes a slightly different conformation similar to that of Trp-104 in AppA, allowing for a hydrogen bond between Gln-50 and Trp-91. Alternatively, hydrogen bonding to Gln-50 might be provided by water molecules in the FAD binding pocket that remained unresolved in the x-ray structure.
The light activation mechanism of BLUF domains
In this study, the application of femtosecond IR spectroscopy on the Synechocystis Slr1694 BLUF domain has given detailed insights of its light activation mechanism. In particular, the involvement of the amino acid side chains in the photo-induced reaction has been demonstrated. With this information at hand, we may test and refine the original reaction model of Gauden et al. shown in Fig. 6 (15,39).
FIGURE 6.
Reaction mechanism for the photo-induced hydrogen-bond network switch reaction in the Synechocystis Slr1694 BLUF domain. Hydrogen bonds are represented by dashed lines. See text for details.
First of all, considering the absence of a kinetic isotope effect on H/D exchange on the multiexponential fluorescence decay in 7–180 ps of FAD*, we show that the primary event in BLUF domains does not involve proton or hydrogen transfer. In analogy with previously studied flavoenzymes (74,75) it most likely corresponds to electron transfer from an aromatic side chain to FAD. The IR and UV-visible signatures of the intermediate species Q1 that immediately follows decay of FAD* is consistent with that of an FAD•− anionic flavin semiquinone radical. Clear bleach signals in the Q1 IR signature can unambiguously be assigned to Tyr-8 (band V, IX), proving beyond doubt that Tyr-8 acts as electron donor to FAD (Fig. 6 B).
The second intermediate Q2 is formed from Q1 in ∼6 ps (20 ps in D2O) and its IR signature corresponds to a neutral flavin semiquinone radical FADH• as evidenced by the appearance of a FAD N5-H bending mode at 1211 cm−1 and a FAD C=N stretch marker mode at 1532 cm−1. Here, too, Tyr-8 modes are bleached, indicating that a FADH•-Tyr radical pair constitutes an intermediate in the BLUF photoreaction. A progressive downshift of Tyr induced absorption in the evolution from Q1 to Q2 is consistent with formation of Tyr•, indicating the presence of a FADH•-Tyr• radical pair.
The question arises whether Gln-50 rotates on formation of the FADH•-Tyr• radical pair, corresponding to the two conformations shown in Fig. 6 C. A flipping of Gln-50 would become apparent as a downshift of the FAD C4=O vibration on H-bond formation to FAD C4=O. However, such putative downshift is overwhelmed by the C4=O downshift that arises from formation of FADH• itself. Interestingly, the flavin model compound RBTA shows a higher frequency for C4=O in its neutral semiquinone form RBTAH• (1660 cm−1) than its anionic semiquinone form RBTA•− (1636 cm−1) (58). Consequently, with an unchanged H-bond pattern, the FAD C4=O would be expected to upshift during the transition from Q1 to Q2. In contrast, Q2 shows a downshift of C4=O (1659 cm−1) as compared to Q1 (1665 cm−1). This observation suggests that C4=O has received a H-bond in the Q2 intermediate and we conclude that most likely, Gln-50 has flipped at this stage with its amino moiety donating a H-bond to C4=O, corresponding to the lower molecular configuration of Fig. 6 C.
From Q2, FAD is reoxidized to result in a long-lived hydrogen-bond switched network SlrRED in 65 ps (160 ps in D2O). The IR signature of the long-lived state is consistent with a flip of Gln-50 (Fig. 6, A and D): the FAD C4=O and C=N stretch vibrations downshift and accordingly FAD C4=O and N5 become more strongly hydrogen-bonded through shared hydrogen bonds from the Gln-50 amino moiety. The overall increase of hydrogen-bond strength to FAD is consistent with the red-shift of the FAD absorption in the UV/vis by ∼10 nm on illumination. Also, the ν(CO) vibration of Tyr-8 upshifts, consistent with a hydrogen bond from Tyr-8 to the carbonyl of Gln-50 in the light (60).
The hydrogen-bond network switch in the BLUF domain as determined in this study shows an interesting feature: essentially all functional hydrogen bonds between FAD, Gln-50, and Tyr-8 strengthen on photoactivation: the hydrogen bond from Gln-50 to FAD C4=O, the hydrogen bond from Gln-50 to FAD N5, the hydrogen bond from Tyr-8 to Gln-50. This observation is consistent with the notion that the FAD binding pocket becomes significantly more rigid and ordered on photoactivation (76). It raises question about the energetics of dark-state stabilization and signal generation in the BLUF domain and the role of entropy therein.
Comparison with previous FTIR work
It is instructive to compare the IR signature of SlrRED with the light-minus-dark FTIR results reported before by Masuda et al. (36). In SlrRED, the C4=O stretch (Band II) exhibits a downshift from 1707 to 1692 cm−1. In FTIR, a similar downshift is observed that is somewhat larger, from 1710 to 1689 cm−1. These observations suggest that as the protein relaxes on timescales longer than 1 ns, the hydrogen-bonding from Gln-50 to FAD C4=O further strengthens. Masuda et al. assigned a 1545(+)/1507(−) feature in FTIR as an upshift of the FAD C=N stretches on formation of the light state. This was interpreted as a loss of a hydrogen bond to the FAD N5. However, with our data at hand, we find that this assignment is incorrect. From the FAD* SADS (Fig. 5 A, black line), we confidently assign bands VI and VII (1578 and 1547 cm−1) to the C=N vibrations in the FAD ground state, not a 1507 cm−1 band.
Finally, we consider the origin of Band I. This band has a low amplitude and is located as a shoulder at ∼1735 cm−1 on the high-frequency side of Band II (Fig. 5, green line). Interestingly, a similar 1734 cm−1 feature was observed recently by step-scan FTIR spectroscopy in the AppA BLUF domain (77) and assigned to a C=O stretching vibration of the carboxylic acid of Asp-82 (Asp-69 in Slr1694). We note, however, that a similar 1735 cm−1 band is present in our IR data immediately after excitation, as can be seen in the SADS that belongs to FAD* (Fig. 5 A, black line). Accordingly, this band must be assigned to a flavin mode; its relatively high frequency can only reasonably be correlated with a C4=O stretch. This observation indicates that in the dark, a heterogeneity exists in C4=O frequencies, with a major conformer (>90%) vibrating at 1707 cm−1 and a minor one (<10%) at 1735 cm−1 as judged from the integrated intensities of the bleaches at these frequencies in the FAD* SADS. Possibly, the latter represents a minor fraction of FAD that lacks the hydrogen bond from Asn-32 to FAD C4=O (see Fig. 1, A and B), which latter mode therefore attains a higher frequency.
No evidence for electron transfer from Trp-91 in Slr1694
In the AppA BLUF domain, it was shown that light-driven electron transfer from a conserved tryptophan (Trp-104) to FAD competes with electron transfer from the conserved tyrosine and represents a nonproductive deactivation channel (48,49). The question arises whether in Slr1694, a similar electron transfer process from Trp-91 occurs. In the Q1 and Q2 intermediates, essentially all observed IR bands can be accounted for by FAD and Tyr radical bands. Thus, no evidence for electron transfer from Trp-91 to FAD is found within experimental accuracy. Inspection of the Slr1694 x-ray structure indicates that Trp-91 in the “in” conformation is further away from FAD than in the AppA x-ray structure (30,35), which will result in a slower electron transfer rate from Trp in the Slr1694 BLUF domain. Given that electron transfer from Tyr to FAD is much faster in Slr1694 than in AppA (∼10 ps vs. ∼600 ps dominant components), it is likely that electron transfer from Trp-91 cannot compete with electron transfer from Tyr-8 in the Slr1694 BLUF domain. The absence of electron transfer from Trp-91 to FAD also implies that the multiexponential decay of FAD* is not caused by a heterogeneity in Trp-91 conformation.
The orientation of the conserved glutamine and alternative reaction models
In the various BLUF x-ray crystal structures, the dark-state orientation of Gln-50 and the positions of Trp-91 and Met-93 remained ambiguous (30,32–35). In Fig. 1, the dark (Fig. 1 A) and the light state (Fig. 1 B) of Slr1694 are presented as proposed by Yuan et al. (35) and by Anderson et al. (30) for the AppA BLUF domain. In the dark, the amino group of Gln-50 is H-bonded to Tyr-8 and to FAD N5. No hydrogen bonding occurs to the flavin C4=O. In the light state, Gln-50 has rotated by ∼180°, Tyr-8 becomes a H-bond donor to the Gln-50 carbonyl, and the Gln-50 amino is H-bonded to both N5 and C4=O of the FAD cofactor. The latter orientation of Gln-50 was proposed to correspond to the dark state in the BlrB, Tll0078, and AppA C20S BLUF domains by Jung et al. (32,33) and Kita et al. (34) on the basis of the prevalence of this orientation in their BLUF crystal structures.
The interpretation of Gln orientation by Anderson et al. (30) and Yuan et al. (35) is supported by a number of spectroscopic studies: FTIR and Raman spectroscopy on a number of BLUF domains indicated that the FAD C4=O becomes more strongly hydrogen bonded in the light (36–38), which is consistent with dark and light orientations as shown in Fig. 1. Using resonant Raman spectroscopy on AppA, Unno et al. (40) showed that the hydrogen-bond state of the flavin C4=O in the dark did not change appreciably on mutation of the conserved Gln, consistent with a dark-state orientation as in Fig. 1 A. Importantly, using NMR spectroscopy on the AppA BLUF domain, Grinstead et al. (78) identified a strong H-bond from the conserved Tyr to the conserved Gln in the light, corresponding to the Gln orientation in Fig. 1 B. In the dark such strong hydrogen bond was absent. Moreover, Grinstead et al. (31,78) showed that in the dark, the conserved Trp (Trp-104 in AppA) is buried in core of Appa WT, as it is in the dark-state AppA crystal structure by Anderson et al. (30). Tryptophan fluorescence experiments on AppA and Slr1694 indeed indicated a buried conformation for the conserved Trp in the dark (35,76).
In a recent study by Domratcheva et al. (79), a number of spectroscopic observations on BLUF domains could be rationalized in terms of the dark- and light-state structures as proposed by Schlichting and co-workers (32, 33). From QM/MM calculations two distinct Gln conformations were found. The first conformation had the Gln carbonyl receiving a hydrogen bond from Tyr and the Gln amide donating a shared hydrogen bond to the FAD C4=O and N5, similar to that shown as the light state in Fig. 1 B and Fig. 6 D. The second conformation corresponded to rotation and tautomerization of the conserved glutamine to imidic acid (HN=CR-OH). Tautomerization of Gln was invoked in view of the short distance between the FAD and Gln carbonyls in the BLUF crystal structures (2.7 Å), considered too short a proximity for bare electronegative oxygens. In the tautomerized form, the Gln COH donates a hydrogen bond to FAD C4=O. The Gln HN=C donates a hydrogen bond to FAD N5, whereas the Gln HN=C receives a strong hydrogen bond from Tyr. Interestingly, the latter conformation corresponded to the lowest calculated FAD excited-state energy and to the highest calculated hydrogen-bond strength to FAD C4=O. It was accordingly proposed by the authors that the latter conformation represented the light state of the BLUF domain and the former the dark state, the reverse of models forwarded here and elsewhere (30,35,39,40,78). Light-induced Gln tautomerization was recently proposed on the basis of ultrafast IR experiments on the AppA BLUF domain. An induced absorption at 1667 cm−1 (that appeared within the instrument response of ∼200 fs) was assigned to a keto-enol tautomerization of the conserved Gln, proposed to result from a dipole change of nearby FAD on photon absorption (80). In our FAD* spectrum, a similar absorption is observed at 1665 cm−1 (Fig. 5, black line). However, we assign this band to a downshift of the FAD C=O modes on formation of FAD* (vide supra) rather than to Gln-50.
At present, no singular molecular model of the FAD binding pocket can account for all structural and spectroscopic data that pertain to the conformation of the conserved glutamine, or, for that matter, that of the conserved tryptophan. We consider the Gln-50 orientation and photoactivation model described in Figs. 1 and 6 as the most plausible given that it is consistent with most of the spectroscopic evidence, especially that by NMR from Grinstead et al. (78). However, more experimentation and high-level theoretical computation is certainly needed. In terms of IR spectroscopy, vibrational studies on BLUF domains with isotopically labeled Gln may help to resolve the critical issue of Gln conformation in the BLUF dark and light states.
CONCLUSIONS
The application of femtosecond IR spectroscopy to the BLUF domain, a novel flavin-binding photoreceptor, has shown IR signatures of key catalytic events among the light-absorbing cofactor and nearby amino acid side chains that efficiently transform the energy of a photon into a long-lived local structural perturbation that is required for biological signal generation. Electron transfer followed by proton transfer from the conserved tyrosine to FAD on a picosecond timescale constitute the primary photochemical events (Fig. 6). This leads to disruption of hydrogen bonds between the conserved glutamine and reduced FAD in its transient anionic and neutral radical states, allowing side chain isomerization of the glutamine. This, in turn, results in a switch of the hydrogen-bond network whereby, most likely, the glutamine amino hydrogen bonds to C4=O and N5 of the FAD chromophore. Radical-pair recombination then takes place in 65 ps whereby a hydrogen is shuttled back from FADH· to tyrosine, resulting in a new, strong hydrogen bond between tyrosine and the carbonyl of glutamine. Ambiguities persist on the IR signature of the conserved glutamine during the reaction; further experimentation involving isotopic substitutions of this side chain will be required to elucidate the precise hydrogen-bond dynamics of Gln-50.
We conclude that through strategic localization and mutual orientation of a flavin, a redox-active side chain (tyrosine) and a two-sided isomerizable amino acid with particular hydrogen-bonding capabilities (glutamine) as shown in Fig. 1 A, the hydrogen bond network connecting the three entities can be efficiently switched on the ultrafast timescale through a light-driven radical-pair mechanism. The challenges that lie ahead involve a detailed understanding of the BLUF reaction mechanism in quantum-chemical terms and the further relay of the locally generated signal to the molecular surface of the BLUF domain and ahead to its signaling partners.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
The authors thank Maxime Alexandre for stimulating discussions, Gabriele Reinke for excellent technical assistance, and Cathrin Lempfuhl for protein preparation.
This work was supported by the European Union through the LaserLab Europe Access Programme (T.M. and P.H.). C.B. was supported by the Life Sciences Council of the Netherlands Organization for Scientific Research (NWO-ALW). K.M.M by was supported by Computational Science grant 635.000.014 from the Netherlands Organization for Scientific Research. J.T.M.K. was supported by the Life Sciences Council of the Netherlands Organization for Scientific Research through a VIDI fellowship.
Abbreviations used: BLUF, blue-light sensing using FAD; DAS, decay-associated spectrum; EADS, evolution-associated difference spectrum; FAD, flavin adenine dinucleotide; FAD*, FAD singlet excited-state; FMN, flavin mononucleotide; IA, induced absorption; KIE, kinetic isotope effect; LOV, light, oxygen or voltage; RBTA, riboflavin tetraacetate; SADS, species-associated difference spectrum; IR, infrared.
Editor: Feng Gai.
References
- 1.Christie, J. M., and W. R. Briggs. 2005. Blue-light sensing and signaling by the phototropins. In Handbook of Photosensory Receptors. W. R. Briggs and J. L. Spudich, editors. Wiley-VCH Verlag GmbH & Company, Weinheim, Germany. 277–304.
- 2.Crosson, S., S. Rajagopal, and K. Moffat. 2003. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 42:2–10. [DOI] [PubMed] [Google Scholar]
- 3.Losi, A. 2006. Flavins: Photochemistry and Photobiology. E. Silvia and A. M. Edwards, editors. Royal Society of Chemistry, Cambridge, UK. 217–269.
- 4.Swartz, T. E., S. B. Corchnoy, J. M. Christie, J. W. Lewis, I. Szundi, W. R. Briggs, and R. A. Bogomolni. 2001. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem. 276:36493–36500. [DOI] [PubMed] [Google Scholar]
- 5.Gomelsky, M., and G. Klug. 2002. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 27:497–500. [DOI] [PubMed] [Google Scholar]
- 6.Iseki, M., S. Matsunaga, A. Murakami, K. Ohno, K. Shiga, K. Yoshida, M. Sugai, T. Takahashi, T. Hori, and M. Watanabe. 2002. A blue-light-activated adenylyl cyclase mediates photo avoidance in Euglena gracilis. Nature. 415:1047–1051. [DOI] [PubMed] [Google Scholar]
- 7.Masuda, S., and C. E. Bauer. 2002. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell. 110:613–623. [DOI] [PubMed] [Google Scholar]
- 8.van der Horst, M. A., and K. J. Hellingwerf. 2004. Photoreceptor proteins, “star actors of modern times”: a review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc. Chem. Res. 37:13–20. [DOI] [PubMed] [Google Scholar]
- 9.Lee, I. R., W. Lee, and A. H. Zewail. 2006. Primary steps of the photoactive yellow protein: Isolated chromophore dynamics and protein directed function. Proc. Natl. Acad. Sci. USA. 103:258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berndt, A., T. Kottke, H. Breitkreuz, R. Dvorsky, S. Hennig, M. Alexander, and E. Wolf. 2007. A novel photoreaction mechanism for the circadian blue light photoreceptor Drosophila cryptochrome. J. Biol. Chem. 282:13011–13021. [DOI] [PubMed] [Google Scholar]
- 11.Dragnea, V., M. Waegele, S. Balascuta, C. E. Bauer, and B. Dragnea. 2005. Time-resolved spectroscopic studies of the AppA blue-light receptor BLUF domain from Rhodobacter sphaeroides. Biochemistry. 44:15978–15985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giovani, B., M. Byrdin, M. Ahmad, and K. Brettel. 2003. Light-induced electron transfer in a cryptochrome blue-light photoreceptor. Nat. Struct. Biol. 10:489–490. [DOI] [PubMed] [Google Scholar]
- 13.Kennis, J. T. M., I. H. M. van Stokkum, S. Crosson, M. Gauden, K. Moffat, and R. van Grondelle. 2004. The LOV2 domain of phototropin: a reversible photochromic switch. J. Am. Chem. Soc. 126:4512–4513. [DOI] [PubMed] [Google Scholar]
- 14.Kennis, J. T. M., S. Crosson, M. Gauden, I. H. M. van Stokkum, K. Moffat, and R. van Grondelle. 2003. Primary reactions of the LOV2 domain of phototropin, a plant blue-light photoreceptor. Biochemistry. 42:3385–3392. [DOI] [PubMed] [Google Scholar]
- 15.Kennis, J. T. M., and M. L. Groot. 2007. Ultrafast spectroscopy of biological photoreceptors. Curr. Opin. Struct. Biol. 17:623–630. [DOI] [PubMed] [Google Scholar]
- 16.Kottke, T., J. Heberle, D. Hehn, B. Dick, and P. Hegemann. 2003. Phot-LOV1: photocycle of a blue-light receptor domain from the green alga Chlamydomonas reinhardtii. Biophys. J. 84:1192–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kottke, T., B. Dick, R. Fedorov, I. Schlichting, R. Deutzmann, and P. Hegemann. 2003. Irreversible photoreduction of flavin in a mutated Phot-LOV1 domain. Biochemistry. 42:9854–9862. [DOI] [PubMed] [Google Scholar]
- 18.Sato, Y., T. Iwata, S. Tokutomi, and H. Kandori. 2005. Reactive cysteine is protonated in the triplet excited state of the LOV2 domain in Adiantum phytochrome3. J. Am. Chem. Soc. 127:1088–1089. [DOI] [PubMed] [Google Scholar]
- 19.Schleicher, E., R. M. Kowalczyk, C. W. M. Kay, P. Hegemann, A. Bacher, M. Fischer, R. Bittl, G. Richter, and S. Weber. 2004. On the reaction mechanism of adduct formation in LOV domains of the plant blue-light receptor phototropin. J. Am. Chem. Soc. 126:11067–11076. [DOI] [PubMed] [Google Scholar]
- 20.Groot, M. L., L. J. G. W. van Wilderen, and M. Di Donato. 2007. Time-resolved methods in biophysics. 5. Femtosecond time-resolved and dispersed infrared spectroscopy on proteins. Photochem. Photobiol. Sci. 6:501–507. [DOI] [PubMed] [Google Scholar]
- 21.Kotting, C., and K. Gerwert. 2005. Proteins in action monitored by time-resolved FTIR spectroscopy. ChemPhysChem. 6:881–888. [DOI] [PubMed] [Google Scholar]
- 22.Groot, M. L., L. J. G. W. van Wilderen, D. S. Larsen, M. A. van der Horst, I. H. M. van Stokkum, K. J. Hellingwerf, and R. van Grondelle. 2003. Initial steps of signal generation in photoactive yellow protein revealed with femtosecond mid-infrared spectroscopy. Biochemistry. 42:10054–10059. [DOI] [PubMed] [Google Scholar]
- 23.Herbst, J., K. Heyne, and R. Diller. 2002. Femtosecond infrared spectroscopy of bacteriorhodopsin chromophore isomerization. Science. 297:822–825. [DOI] [PubMed] [Google Scholar]
- 24.Heyne, K., O. F. Mohammed, A. Usman, J. Dreyer, E. T. J. Nibbering, and M. A. Cusanovich. 2005. Structural evolution of the chromophore in the primary stages of trans/cis isomerization in photoactive yellow protein. J. Am. Chem. Soc. 127:18100–18106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schumann, C., R. Gross, M. M. N. Wolf, R. Diller, N. Michael, and T. Lamparter. 2008. Subpicosecond mid-infrared spectroscopy of the P-fr reaction of phytochrome Agp1 from Agrobacterium tumefaciens. Biophys. J. 94:3189–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoner-Ma, D., A. A. Jaye, P. Matousek, M. Towrie, S. R. Meech, and P. J. Tonge. 2005. Observation of excited-state proton transfer in green fluorescent protein using ultrafast vibrational spectroscopy. J. Am. Chem. Soc. 127:2864–2865. [DOI] [PubMed] [Google Scholar]
- 27.van Thor, J. J., K. L. Ronayne, and M. Towrie. 2007. Formation of the early photoproduct Lumi-R of cyanobacterial phytochrome Cph1 observed by ultrafast mid-infrared spectroscopy. J. Am. Chem. Soc. 129:126–132. [DOI] [PubMed] [Google Scholar]
- 28.van Wilderen, L. J. G. W., M. A. van der Horst, I. H. M. van Stokkum, K. J. Hellingwerf, R. van Grondelle, and M. L. Groot. 2006. Ultrafast infrared spectroscopy reveals a key step for successful entry into the photocycle for photoactive yellow protein. Proc. Natl. Acad. Sci. USA. 103:15050–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okajima, K., S. Yoshihara, Y. Fukushima, X. X. Geng, M. Katayama, S. Higashi, M. Watanabe, S. Sato, S. Tabata, Y. Shibata, S. Itoh, and M. Ikeuchi. 2005. Biochemical and functional characterization of BLUF-type flavin-binding proteins of two species of cyanobacteria. J. Biochem. (Tokyo). 137:741–750. [DOI] [PubMed] [Google Scholar]
- 30.Anderson, S., V. Dragnea, J. Y. S. Masuda, K. Moffat, and C. E. Bauer. 2005. Structure of a novel photoreceptor, the BLUF domain of AppA from Rhodobacter sphaeroides. Biochemistry. 44:7998–8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grinstead, J. S., S. T. D. Hsu, W. Laan, A. M. J. J. Bonvin, K. J. Hellingwerf, R. Boelens, and R. Kaptein. 2006. The solution structure of the AppA BLUF domain: insight into the mechanism of light-induced signaling. ChemBioChem. 7:187–193. [DOI] [PubMed] [Google Scholar]
- 32.Jung, A., J. Reinstein, T. Domratcheva, R. L. Shoeman, and I. Schlichting. 2006. Crystal structures of the AppA BLUF domain photoreceptor provide insights into blue light-mediated signal transduction. J. Mol. Biol. 362:717–732. [DOI] [PubMed] [Google Scholar]
- 33.Jung, A., T. Domratcheva, M. Tarutina, Q. Wu, W. H. Ko, R. L. Shoeman, M. Gomelsky, K. H. Gardner, and I. Schlichting. 2005. Structure of a bacterial BLUF photoreceptor: insights into blue light-mediated signal transduction. Proc. Natl. Acad. Sci. USA. 102:12350–12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kita, A., K. Okajima, Y. Morimoto, M. Ikeuchi, and K. Miki. 2005. Structure of a cyanobacterial BLUF protein, Tll0078, containing a novel FAD-binding blue light sensor domain. J. Mol. Biol. 349:1–9. [DOI] [PubMed] [Google Scholar]
- 35.Yuan, H., S. Anderson, S. Masuda, V. Dragnea, K. Moffat, and C. E. Bauer. 2006. Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling. Biochemistry. 45:12687–12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuda, S., K. Hasegawa, A. Ishii, and T. A. Ono. 2004. Light-induced structural changes in a putative blue-light receptor with a novel FAD binding fold sensor of blue-light using FAD (BLUF); Slr1694 of Synechocystis sp PCC6803. Biochemistry. 43:5304–5313. [DOI] [PubMed] [Google Scholar]
- 37.Masuda, S., K. Hasegawa, and T. A. Ono. 2005. Light-induced structural changes of apoprotein and chromophore in the sensor of blue light using FAD (BLUF) domain of AppA for a signaling state. Biochemistry. 44:1215–1224. [DOI] [PubMed] [Google Scholar]
- 38.Unno, M., R. Sano, S. Masuda, T. A. Ono, and S. Yamauchi. 2005. Light-induced structural changes in the active site of the BLUF domain in AppA by Raman spectroscopy. J. Phys. Chem. B. 109:12620–12626. [DOI] [PubMed] [Google Scholar]
- 39.Gauden, M., I. H. M. van Stokkum, J. M. Key, D. C. Lührs, R. Van Grondelle, P. Hegemann, and J. T. M. Kennis. 2006. Hydrogen-bond switching through a radical pair mechanism in a flavin-binding photoreceptor. Proc. Natl. Acad. Sci. USA. 103:10895–10900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unno, M., S. Masuda, T. A. Ono, and S. Yamauchi. 2006. Orientation of a key glutamine residue in the BLUF domain from AppA revealed by mutagenesis, spectroscopy, and quantum chemical calculations. J. Am. Chem. Soc. 128:5638–5639. [DOI] [PubMed] [Google Scholar]
- 41.Gobets, B., I. H. M. van Stokkum, M. Rögner, J. Kruip, E. Schlodder, N. V. Karapetyan, J. P. Dekker, and R. van Grondelle. 2001. Time-resolved fluorescence emission measurements of photosystem I particles of various cyanobacteria: a unified compartmental model. Biophys. J. 81:407–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Stokkum, I. H. M., D. S. Larsen, and R. van Grondelle. 2004. Global and target analysis of time-resolved spectra. Biochim. Biophys. Acta. 1657:82–104. [DOI] [PubMed] [Google Scholar]
- 43.Zirak, P., A. Penzkofer, C. Lehmpfuhl, T. Mathes, and P. Hegemann. 2007. Absorption and emission spectroscopic characterization of blue-light receptor Slr1694 from Synechocystis sp. PCC6803. J. Photochem. Photobiol. B. 86:22–34. [DOI] [PubMed] [Google Scholar]
- 44.Abe, M., Y. Kyogoku, T. Kitagawa, K. Kawano, N. Ohishi, A. Takaisuzuki, and K. Yagi. 1986. Infrared-spectra and molecular association of lumiflavin and riboflavin derivatives. Spectrochim. Acta. 42:1059–1068. [Google Scholar]
- 45.Ataka, K., P. Hegemann, and J. Heberle. 2003. Vibrational spectroscopy of an algal Phot-LOV1 domain probes the molecular changes associated with blue-light reception. Biophys. J. 84:466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gauden, M., S. Yeremenko, W. Laan, I. H. M. van Stokkum, J. A. Ihalainen, R. van Grondelle, K. J. Hellingwerf, and J. T. M. Kennis. 2005. Photocycle of the flavin-binding photoreceptor AppA, a bacterial transcriptional antirepressor of photosynthesis genes. Biochemistry. 44:3653–3662. [DOI] [PubMed] [Google Scholar]
- 47.Zirak, P., A. Penzkofer, T. Schiereis, P. Hegemann, A. Jung, and I. Schlichting. 2005. Absorption and fluorescence spectroscopic characterization of BLUF domain of AppA from Rhodobacter sphaeroides. Chem. Phys. 315:142–154. [Google Scholar]
- 48.Gauden, M., J. S. Grinstead, W. Laan, I. H. M. van Stokkum, M. Avila-Perez, K. C. Toh, R. Boelens, R. Kaptein, R. van Grondelle, K. J. Hellingwerf, and J. T. M. Kennis. 2007. On the role of aromatic side chains in the photoactivation of BLUF domains. Biochemistry. 46:7405–7415. [DOI] [PubMed] [Google Scholar]
- 49.Laan, W., M. Gauden, S. Yeremenko, R. van Grondelle, J. T. M. Kennis, and K. J. Hellingwerf. 2006. On the mechanism of activation of the BLUF domain of AppA. Biochemistry. 45:51–60. [DOI] [PubMed] [Google Scholar]
- 50.Laan, W., T. Bednarz, J. Heberle, and K. J. Hellingwerf. 2004. Chromophore composition of a heterologously expressed BLUF-domain. Photochem. Photobiol. Sci. 3:1011–1016. [DOI] [PubMed] [Google Scholar]
- 51.Berberan-Santos, M. N., E. N. Bodunov, and B. Valeur. 2005. Mathematical functions for the analysis of luminescence decays with underlying distributions 1. Kohlrausch decay function (stretched exponential). Chem. Phys. 315:171–182. [Google Scholar]
- 52.Benecky, M., T. Y. Li, J. Schmidt, F. Frerman, K. L. Watters, and J. McFarland. 1979. Resonance Raman study of flavins and the flavoprotein fatty acyl coenzyme-a dehydrogenase. Biochemistry. 18:3471–3476. [DOI] [PubMed] [Google Scholar]
- 53.Copeland, R. A., and T. G. Spiro. 1986. Ultraviolet resonance Raman-spectroscopy of flavin mononucleotide and flavin adenine-dinucleotide. J. Phys. Chem. 90:6648–6654. [Google Scholar]
- 54.Abe, M. and Y. Kyogoku. 1987. Vibrational analysis of flavin derivatives—normal coordinate treatments of lumiflavin. Spectrochim. Acta. 43:1027–1037. [Google Scholar]
- 55.Bowman, W. D., and T. G. Spiro. 1981. Normal mode analysis of lumiflavin and interpretation of resonance Raman spectra of flavoproteins. Biochemistry. 20:3313–3318. [DOI] [PubMed] [Google Scholar]
- 56.Dutta, P. K., and T. G. Spiro. 1980. Resonance coherent anti-stokes Raman-scattering spectra of oxidized and semi-quinone forms of clostridium Mp flavodoxin. Biochemistry. 19:1590–1593. [DOI] [PubMed] [Google Scholar]
- 57.Kondo, M., J. Nappa, K. L. Ronayne, A. L. Stelling, P. J. Tonge, and S. R. Meech. 2006. Ultrafast vibrational spectroscopy of the flavin chromophore. J. Phys. Chem. B. 110:20107–20110. [DOI] [PubMed] [Google Scholar]
- 58.Martin, C. B., M. L. Tsao, C. M. Hadad, and M. S. Platz. 2002. The reaction of triplet flavin with indole. A study of the cascade of reactive intermediates using density functional theory and time resolved infrared spectroscopy. J. Am. Chem. Soc. 124:7226–7234. [DOI] [PubMed] [Google Scholar]
- 59.Barth, A. 2000. The infrared absorption of amino acid side chains. Prog. Biophys. Mol. Biol. 74:141–173. [DOI] [PubMed] [Google Scholar]
- 60.Takahashi, R., K. Okajima, H. Suzuki, H. Nakamura, M. Ikeuchi, and T. Noguchi. 2007. FTIR study on the hydrogen bond structure of a key tyrosine residue in the flavin-binding blue light sensor TePixD from Thermosynechococcus elongatus. Biochemistry. 46:6459–6467. [DOI] [PubMed] [Google Scholar]
- 61.Gerothanassis, I. P., N. Birlirakis, C. Sakarellos, and M. Marraud. 1992. Solvation state of the Tyr side-chain in peptides—an Ft-Ir and O-17 NMR approach. J. Am. Chem. Soc. 114:9043–9047. [Google Scholar]
- 62.Rothschild, K. J., P. Roepe, P. L. Ahl, T. N. Earnest, R. A. Bogomolni, S. K. Dasgupta, C. M. Mulliken, and J. Herzfeld. 1986. Evidence for a tyrosine protonation change during the primary phototransition of bacteriorhodopsin at low-temperature. Proc. Natl. Acad. Sci. USA. 83:347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolpert, M. and P. Hellwig. 2006. Infrared spectra and molar absorption coefficients of the 20 alpha amino acids in aqueous solutions in the spectral range from 1800 to 500 cm(-1). Spectrochim. Acta. 64:987–1001. [DOI] [PubMed] [Google Scholar]
- 64.Unno, M., M. Kumauchi, J. Sasaki, F. Tokunaga, and S. Yamauchi. 2002. Resonance Raman spectroscopy and quantum chemical calculations reveal structural changes in the active site of photoactive yellow protein. Biochemistry. 41:5668–5674. [DOI] [PubMed] [Google Scholar]
- 65.Unno, M., M. Kumauchi, J. Sasaki, F. Tokunaga, and S. Yamauchi. 2003. Assignment of resonance Raman spectrum of photoactive yellow protein in its long-lived blue-shifted intermediate. J. Phys. Chem. B. 107:2837–2845. [Google Scholar]
- 66.Unno, M., M. Kumauchi, N. Hamada, F. Tokunaga, and S. Yamauchi. 2004. Resonance Raman evidence for two conformations involved in the L intermediate of photoactive yellow protein. J. Biol. Chem. 279:23855–23858. [DOI] [PubMed] [Google Scholar]
- 67.Unno, M., M. Kumauchi, J. Sasaki, F. Tokunaga, and S. Yamauchi. 2000. Evidence for a protonated and cis configuration chromophore in the photobleached intermediate of photoactive yellow protein. J. Am. Chem. Soc. 122:4233–4234. [Google Scholar]
- 68.Park, E. S., S. S. Andrews, R. B. Hu, and S. G. Boxer. 1999. Vibrational stark spectroscopy in proteins: a probe and calibration for electrostatic fields. J. Phys. Chem. B. 103:9813–9817. [Google Scholar]
- 69.Stanley, R. J., and M. S. Siddiqui. 2001. A stark spectroscopic study of N(3)-methyl, N(10)-isobutyl-7,8-dimethylisoalloxazine in nonpolar low-temperature glasses: experiment and comparison with calculations. J. Phys. Chem. A. 105:11001–11008. [Google Scholar]
- 70.Mathies, R., and L. Stryer. 1976. Retinal has a highly dipolar vertically excited singlet-state—implications for vision. Proc. Natl. Acad. Sci. USA. 73:2169–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Premvardhan, L. L., M. A. van der Horst, K. J. Hellingwerf, and R. van Grondelle. 2003. Stark spectroscopy on photoactive yellow protein, E46Q, and a nonisomerizing derivative, probes photo-induced charge motion. Biophys. J. 84:3226–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bredenbeck, J., J. Helbing, J. R. Kumita, G. A. Woolley, and P. Hamm. 2005. Alpha-helix formation in a photoswitchable peptide tracked from picoseconds to microseconds by time-resolved IR spectroscopy. Proc. Natl. Acad. Sci. USA. 102:2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rupenyan, A., I. H. M. van Stokkum, J. C. Arents, R. van Grondelle, K. Hellingwerf, and M. L. Groot. 2008. Characterization of the primary photochemistry of proteorhodopsin with femtosecond spectroscopy. Biophys. J. 94:4020–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mataga, N., H. Chosrowjan, Y. Shibata, F. Tanaka, Y. Nishina, and K. Shiga. 2000. Dynamics and mechanisms of ultrafast fluorescence quenching reactions of flavin chromophores in protein nanospace. J. Phys. Chem. B. 104:10667–10677. [Google Scholar]
- 75.Zhong, D., and A. H. Zewail. 2001. Femtosecond dynamics of flavoproteins: charge separation and recombination in riboflavine (vitamin B2)-binding protein and in glucose oxidase enzyme. Proc. Natl. Acad. Sci. USA. 98:11867–11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Toh, K. C., I. H. M. van Stokkum, J. Hendriks, M. T. A. Alexandre, J. C. Arents, M. A. Perez, R. van Grondelle, K. J. Hellingwerf, and J. T. M. Kennis. 2008. On the signaling mechanism and the absence of photo reversibility in the AppA BLUF domain. Biophys. J. 95:312–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Majerus, T., T. Kottke, W. Laan, K. Hellingwerf, and J. Heberle. 2007. Time-resolved FT-lR Spectroscopy traces signal relay within the blue-light receptor AppA. ChemPhysChem. 8:1787–1789. [DOI] [PubMed] [Google Scholar]
- 78.Grinstead, J. S., M. Avila-Perez, K. J. Hellingwerf, R. Boelens, and R. Kaptein. 2006. Light-induced flipping of a conserved glutamine sidechain and its orientation in the AppA BLUF domain. J. Am. Chem. Soc. 128:15066–15067. [DOI] [PubMed] [Google Scholar]
- 79.Domratcheva, T., B. L. Grigorenko, I. Schlichting, and A. V. Nemukhin. 2008. Molecular models predict light-induced glutamine tautomerization in BLUF photoreceptors. Biophys. J. 94:3872–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stelling, A. L., K. L. Ronayne, J. Nappa, P. J. Tonge, and S. R. Meech. 2007. Ultrafast structural dynamics in BLUF domains: transient infrared spectroscopy of AppA and its mutants. J. Am. Chem. Soc. 129:15556–15564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.