Abstract
Anguibactin, the siderophore produced by Vibrio anguillarum 775, is synthesized via a nonribosomal peptide synthetase (NRPS) mechanism. Most of the genes required for anguibactin biosynthesis are harbored by the pJM1 plasmid. Complete sequencing of this plasmid identified an orf encoding a 108 kDa predicted protein, AngN. In this work we show that AngN is essential for anguibactin biosynthesis and possesses two domains with homology to cyclization (Cy) domains of NRPSs. Substitution by alanine of the aspartic acid residues within a conserved motif of either Cy1 or Cy2 domain demonstrated the importance of these two domains in AngN function during siderophore biosynthesis. Site-directed mutations in both domains (D133A/D575A and D138A/D580A) resulted in anguibactin-deficient phenotypes while mutations in each domain did not abolish siderophore production but caused a reduction in the amounts produced. The mutations D133A/D575A and D138A/D580A also resulted as expected in a dramatic attenuation of the virulence of V. anguillarum 775 highlighting the importance of this gene for the biosynthesis of anguibactin within the vertebrate host. Regulation of the angN gene follows the patterns observed at the iron transport-biosynthesis promoter with angN transcription repressed in the presence of iron and enhanced by AngR and trans-acting factor (TAF) under iron limitation.
INTRODUCTION
The possession of specialized iron transport systems is crucial for bacteria to override the iron limitation imposed by the host or the environment (Braun and Killmann, 1999; Wandersman and Delepelaire, 2004). Pathogenic bacteria have evolved systems, such as siderophores, to scavenge ferric iron from the iron-binding proteins of the host (Ratledge, 2007). Peptide siderophores are in general low molecular-weight iron chelators that are synthesized by proteins belonging to the nonribosomal peptide synthetase (NRPS) family (Crosa and Walsh, 2002; Miethke and Marahiel, 2007). NRPSs catalyze the formation of a wide variety of peptides, such as antibiotics and siderophores, in the absence of an RNA template (Finking and Marahiel, 2004; von Dohren, et al., 1999; Walsh, 2004). These multimodular enzymes work as an enzymatic assembly line in which the order of the modules determines the order of the amino acids in the peptide (Fischbach and Walsh, 2006; Marahiel, et al., 1997). Each module contains the complete information for an elongation step combining the catalytic functions for the activation of the substrate amino acid (adenylation domain, A), the tethering of the corresponding adenylate to the enzyme-bound 4′-phosphopantetheinyl (4′-PP) cofactor (peptidyl carrier protein domain, PCP) and the formation of the peptide bond by the condensation domain, C (Keating and Walsh, 1999; Marahiel, et al., 1997; von Dohren, et al., 1999). In some cases the condensation steps can also be catalyzed by a specialized condensation domain, the cyclization domain (Cy) that converts specific amino acids such as cysteine and threonine to their cyclic derivatives, thiazoline and oxazoline respectively, in the process of peptide bond formation (Marshall, et al., 2001; Miller and Walsh, 2001; Quadri, et al., 1999; Walsh, et al., 2001).
The bacterial fish pathogen Vibrio anguillarum is the causative agent of vibriosis, a highly fatal hemorrhagic septicemic disease in salmonids and other fish including eels (Actis, et al., 1999). Many pathogenic strains of V. anguillarum possess a virulence plasmid that encodes an iron-sequestering system that includes a 348 Da siderophore, anguibactin (ω-N-hydroxy-ωN((2′-(2″,3″-dihydroxyphenyl)thiazolin-4′-yl)carboxy)histamine), and a transport protein system for the binding and transport of iron as a complex with the siderophore anguibactin into the cell cytosol (Actis, et al., 1986; Actis, et al., 1988). The sequence of the 65 kilobase (kb) virulence plasmid pJM1 of V. anguillarum strain 775 has been completed (Di Lorenzo, et al., 2003) and revealed that most of the proteins proposed to be involved in anguibactin biosynthesis are encoded by genes on the plasmid. Interestingly, the chromosome harbors redundant copies of genes encoding proteins for the biosynthesis of the anguibactin precursor 2,3-dihydroxybenzoic acid (DHBA) and anguibactin itself (Alice, et al., 2005; Naka, et al., 2008).
Several of the anguibactin biosynthetic proteins are part of the NRPS family (Di Lorenzo, et al., 2004; Welch, et al., 2000; Wertheimer, et al., 1999) and one of these proteins, AngR, has also regulatory properties. AngR acts as a positive regulator of the fatDCBAangRT operon (Wertheimer, et al., 1999); expression from this operon is also enhanced by TAF, an additional regulator encoded in a region of the virulence plasmid noncontiguous to the fatDCBAangRT operon (Tolmasky, et al., 1988).
In this work we describe one of the genes harbored by the virulence plasmid pJM1, that encodes a putative NRPS, AngN. AngN shows an unusual domain organization for an NRPS with only two cyclization domains in tandem. Transposon insertions in the angN gene resulted in anguibactin-deficient mutants (Tolmasky, et al., 1988). Our results demonstrate the essential role played by AngN and its cyclization domains in anguibactin biosynthesis.
MATERIALS AND METHODS
Bacterial strains and plasmids
Bacterial strains and plasmids used in this study are described in Table 1. V. anguillarum was grown at 25°C in either trypticase soy broth or agar supplemented with 1% NaCl, TSBS and TSAS respectively. To determine iron uptake characteristics, the strains were grown in M9 minimal medium (Sambrook and Russell, 2001) supplemented with 0.2% Casamino acid, 5% NaCl, the appropriate antibiotics and either various concentrations of ethylenediamine-di-(o-hydroxyphenyl acetic acid) (EDDA) for iron-limiting conditions or 4 μg/ml ferric ammonium citrate for iron-rich conditions. Antibiotic concentrations used for V. anguillarum were ampicillin (Ap) 1 mg/ml, tetracycline (Tc) 2.5 μg/ml, rifampicin (Rif) 100 μg/ml, chloramphenicol (Cm) 10 to 15 μg/ml and gentamicin (Gm) 10 μg/ml.
Table 1.
Strains and plasmids used in this study
| Bacterial strains | Genotype and relevant characteristics | Source or reference |
|---|---|---|
| Vibrio anguillarum 775 (pJM1) | Wild type | (Crosa, et al., 1980) |
| Vibrio anguillarum CC9-16(pJHC9-16) | Anguibactin-deficient, iron transport-proficient | (Walter, et al., 1983) |
| Vibrio anguillarum CC9-8(pJHC9-8) | Anguibactin-deficient, iron transport-deficient | (Walter, et al., 1983) |
| Vibrio anguillarum 775(pJM1-120) | 775 carrying pJM1 with Tn3::Ho-Ho1 insertion in angN | This study |
| Vibrio anguillarum 775(pJM1-4) | 775 carrying pJM1 with Tn3::Ho-Ho1 insertion in angR | Laboratory strain |
| Vibrio anguillarum H775-3 | Plasmidless derivative of 775 | (Crosa, et al., 1980) |
| Vibrio anguillarum H775-3(pJHC-T2612) | Plasmidless derivative of 775 carrying pJHC-T2612 (TAF−) | (Tolmasky, et al., 1988) |
| Escherichia coli XL1 blue | recA1, endA1, gyrA46, thi, hsdR17, supE44, relA, lacF′ [proAB+, lacIq, lacZM15 Tn10 (Tetr)] | Stratagene |
| Escherichia coli HB101 | supE44 hsd20 (r−B m−B) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | (Boyer and Roulland- Dussoix, 1969) |
|
| ||
| Plasmids | Relevant characteristics | Source or reference |
|
| ||
| pJM1 | Indigenous plasmid in strain 775 | (Crosa, et al., 1980) |
| pJHC-T2612 | Recombinant clone carrying a 24 kb region of pJM1cloned in pVK102, Tcr | (Tolmasky, et al., 1988) |
| pJHC-T2612#120 | pJHC-T2612 with a Tn3::Ho-Ho1 insertion in angN, Tcr, Apr | (Tolmasky, et al., 1988) |
| pJHC9-8 | pJM1 derivative carrying only the TAF region | (Tolmasky, et al., 1988) |
| pJHC9-16 | pJM1 derivative carrying TAF and transport genes, | (Tolmasky, et al., 1988) |
| pPH1JI | Plasmid with RP4 ori, incompatible with pJHC-T2612, Gmr | (Hirsch and Beringer, 1984) |
| pRK2073 | Helper plasmid for conjugation, Tpr, Tra+ | (Figurski and Helinski, 1979) |
| pBluescript SK+ | Cloning vector, Apr | Stratagene |
| pCR®-BluntII-TOPO® | Cloning vector, Kmr | Invitrogen |
| pBR325 | Cloning vector, Tcr, Cmr, Apr | (Bolivar, 1978) |
| pBR325-M200 | Cloning vector derived from pBR325, Tcr, Cmr, Aps | (Di Lorenzo, et al., 2004) |
| pTL61T | lacZ reporter vector, Apr | (Linn and St Pierre, 1990) |
| pMDL6 | 3.2 kb PCR fragment from pJM1 containing the angN gene cloned in pCR®- BluntII-TOPO® | This study |
| pMDL6-D133A | pMDL6 with mutation D133A in Cy1 domain | This study |
| pMDL6-D138A | pMDL6 with mutation D138A in Cy1 domain | This study |
| pMDL6-D575A | pMDL6 with mutation D575A in Cy2 domain | This study |
| pMDL6-D580A | pMDL6 with mutation D580A in Cy2 domain | This study |
| pMDL6-D133A/D575A | pMDL6 with mutation D133A in Cy1 domain and D575A in Cy2 domain | This study |
| pMDL6-D138A/D580A | pMDL6 with mutation D138A in Cy1 domain and D580A in Cy2 domain | This study |
| pMDL30 | ClaI-BamHI fragment from pMDL6 cloned in pBR325-M200 | This study |
| pCy1D1-D133A | ClaI-BamHI fragment from pMDL6-D133A cloned in pBR325-M200 | This study |
| pCy1D2-D138A | ClaI-BamHI fragment from pMDL6-D138A cloned in pBR325-M200 | This study |
| pCy2D1-D575A | ClaI-BamHI fragment from pMDL6-D575A cloned in pBR325-M200 | This study |
| pCy2D2-D580A | ClaI-BamHI fragment from pMDL6-D580A cloned in pBR325-M200 | This study |
| pCy1/2D1-D133A/D575A | ClaI-BamHI fragment from pMDL6-D133A/D575A cloned in pBR325- M200 | This study |
| pCy1/2D2-D138A/D580A | ClaI-BamHI fragment from pMDL6-D138A/D580A cloned in pBR325- M200 | This study |
| pCRII-PangN | 350 bp PCR fragment from pJM1 containing the upstream region of the angN gene cloned in pCR®-BluntII-TOPO® | This study |
| pHNN-2 | XhoI-PstI fragment from pCRII-PangN cloned in pTL61T | This study |
| pSC50 | 125 bp Sau3AI fragment of the fatB gene cloned in pBluescript SK+ | This study |
| pQSH6 | 415 bp SalI-ClaI fragment of the aroC gene cloned in pBluescript SK+ | (Di Lorenzo, et al., 2004) |
E. coli strains were grown in Luria-Bertani (LB) medium in the presence of the appropriate antibiotics. Antibiotic concentrations used for E. coli were Ap 100 μg/ml, Tc 10 μg/ml, Cm 30 μg/ml, Gm 10 μg/ml and trimethropim (Tp) 10 μg/ml.
General methods
Plasmid DNA preparations were performed using the alkaline lysis method (Birnboim and Doly, 1979). Restriction endonuclease digestion of DNA was performed under the conditions recommended by the supplier (Invitrogen, Roche, NEB). Transformations in the E. coli strains HB101 and XL1 blue and other cloning strategies were performed according to standard protocols (Sambrook and Russell, 2001). Plasmids were transferred from E. coli to V. anguillarum by conjugation as previously described (Tolmasky, et al., 1988).
The Wizard® Plus SV Minipreps (Promega) and Qiaprep® Spin Miniprep Kit (Qiagen) were used to generate sequence quality plasmid DNA. DNA sequencing reactions were carried out by the OHSU-MMI Research Core Facility (http://www.ohsu.edu/core) using a model 377 Applied Biosystems Inc. automated fluorescence sequencer. Sequencing primers were designed using Oligo 6.8® primer analysis software and purchased from the OHSU-MMI Research Core Facility (http://www.ohsu.edu/core) and Invitrogen. DNA and protein sequence analysis were carried out at the NCBI using the BLAST network service (Altschul, et al., 1990).
Mobilization of the angN mutation #120 from pJHC-T2612 to the pJM1 plasmid
To generate strain 775(pJM1-120), plasmid pJHC-T2612#120 was transferred to V. anguillarum 775 by conjugation. In a second conjugation, plasmid pPH1JI, whose origin of replication is incompatible with the replicon of pJHC-T2612#120, was transferred to the strain obtained from the first conjugation. By plating in the presence of Gm (the resistance marker of pPH1JI) and Ap (the resistance gene harbored by the Tn3::HoHo1 transposon), it was possible to select for those cells in which the angN gene with the Tn3::HoHo1 insertion had replaced the wild type gene on the pJM1 plasmid. The loss of pJHC-T2612 was confirmed by Southern blot hybridization using the pVK102 vector sequence for probing (data not shown). The Tn3::HoHo1 transposon harbors a mutation in the transposase gene that prevents transposition to occur to other locations on the plasmid or in the chromosome.
Construction of the complementing clone
A 3.2 kb fragment containing the angN gene was amplified by PCR using the angN-F and the angN-BamHIR primers (Table 2) and pJM1 as a template. Reactions consisted of 2 minutes at 95°C followed by 30 cycles of 1 minute at 95°C, 1 minute at 53°C, and 4 minutes at 72°C followed by a single cycle at 72°C for 10 minutes. The PCR product was cloned in the pCR®-BluntII-TOPO® vector using the Zero Blunt® TOPO® PCR Cloning Kit (Invitrogen) resulting in the pMDL6 plasmid. A ClaI-BamHI fragment was subcloned from pMDL6 into the ClaI-BamHI sites of pBR325-M200 to generate the pMDL30 plasmid carrying the angN gene. After cloning in pBR325-M200 the entire angN gene was sequenced to verify that no mutation was generated in the angN gene during amplification or cloning. The pBR325-M200 cloning vector was derived from pBR325 by digestion with PstI, blunting of the ends with T4 DNA polymerase (Gibco) and religation, resulting in a 4-bp deletion leading to the inactivation of the ampicillin resistance gene.
Table 2.
DNA primers used in this study.
| Primer name | Nucleotide sequencea |
|---|---|
| angN-F | 5′-ACGACGATTGATGGGTGTAGC-3′ |
| angN-BamHIR | 5′-TTGTATTCACTATGGATCCTTGC-3′ |
| D133A-F | 5′-CGCTTACATATTGATAGCGCTATGATTGCTATTGACCCAG-3′ |
| D133A-R | 5′-CTGGGTCAATAGCAATCATAGCGCTATCAATATGTAAGCG-3′ |
| D138A-F | 5′-CGATATGATTGCTATTGCCCCAGATAGTTGCCGAG-3′ |
| D138A-R | 5′-CTCGGCAACTATCTGGGGCAATAGCAATCATATCG-3′ |
| D575A-F | 5′-GTATTTTCTCGCTTTGCTGCATTAATTCTTGATGCTCGCTC-3′ |
| D575A-R | 5′-GAGCGAGCATCAAGAATTAATGCAGCAAAGCGAGAAAATAC-3′ |
| D580A-F | 5′-CTTTGATGCATTAATTCTTGCTGCTCGCTCCATTGCTTC-3′ |
| D580A-R | 5′-GAAGCAATGGAGCGAGCAGCAAGAATTAATGCATCAAAG-3′ |
| PangN-XhoI | 5′-CTCGAGGTTTCCTCATCAATCCAAAAGGTC-3′ |
| PangN-PstI | 5′-CTGCAGCAACATGCAGCCTGCATTGGTGTTAATTC-3′ |
| PEX-angN | 5′-TGCCCATTATCTTTTCTTCC-3′ |
| angN-T7L | 5′-CGAAAGTTCTATTATTGATGTT-3′ |
| angN-T7R | 5′-ggatcctaatacgactcactatagggaggAGGTAATAAACTAACGGAGAAT-3′ |
Restriction sites are underlined, base changed from the wt sequence are shown in italics, T7 promoter sequence in lower case
Site-directed mutagenesis
The plasmids pMDL6-D133A, pMDL6-D138A pMDL6-D575A and pMDL6-D580A were generated using the Quickchange™ site-directed mutagenesis kit (Stratagene), plasmid pMDL6 as a template and the primers listed in Table 2. The whole procedure was performed according to the manufacturer recommendations, with 16 cycles consisting of: 30 seconds at 95°C followed by 1 minute at 55°C and 16 minutes at 68°C. Plasmids pMDL6-D133A/D575A and pMDL6-D138A/D580A were generated with the same procedure but using plasmids pMDL6-D133A and pMDL6-D138A as templates, respectively, and the appropriate primers. Site-specific mutations were confirmed by DNA sequencing. Once mutated, a ClaI-BamHI fragment from each derivative was subcloned into the ClaI-BamHI sites of pBR325-M200 to generate plasmids carrying the angN derivatives with mutations in the Cy domains listed in Table 1. Once cloned in pBR325-M200 the entire angN mutant genes were sequenced to verify that no other region of angN was affected during mutagenesis or cloning.
Construction of promoter fusions in pTL61T
Primers PangN-XhoI and PangN-PstI (Table 2) were used to amplify from pJM1 DNA a 350 bp fragment containing the 5′-end of the angN gene and the upstream region. The PCR product was cloned in the pCR®-BluntII-TOPO® vector using the Zero Blunt® TOPO® PCR Cloning Kit (Invitrogen) and subsequently subcloned into the XhoI-PstI sites of pTL61T to generate the pHNN-2 plasmid. The insert in pTL61T was sequenced to verify that the sequence of the PCR product is identical to the pJM1 sequence.
Growth in iron-limiting conditions and detection of anguibactin
For each mutant and wild type strain we determined the minimal inhibitory concentration (MIC) for EDDA by using liquid cultures at increasing concentrations of EDDA in M9 minimal medium at 25°C. From these analyses we chose a range of concentrations of EDDA to determine the ability of all the strains to grow in iron-limiting conditions.
The siderophore anguibactin was detected by CAS assay and by bioassays using strains CC9-16 and CC9-8 as previously described (Welch, et al., 2000; Wertheimer, et al., 1999) with few modifications. For the bioassay experiment each strain was grown in M9 minimal medium supplemented with 0.25 μM EDDA and after 16 hours the culture volume corresponding to an OD600nm = 1 was collected to obtain the supernatant. The supernatants were lyophilized to dryness, resuspended in 500 μl of methanol and stored at −80°C. Before spotting on the bioassay plates 10 μl of each sample were dried and resuspended in 3 μl of water.
Presence of anguibactin in the supernatant of strains grown in iron limiting conditions was also determined by HPLC analysis. 100 μl aliquots from 1,000 fold concentrated supernatant were applied to HPLC (Beckmann Coulter System Gold) and the samples were analyzed on a ODS-AQ 120, 5 μm reversed-phase column C18, 150 mm length, 6 mm inner diameter (YMC, Waters). A binary gradient consisting of solvent A (0.01% trifluoracetic acid) and solvent B (100% acetonitrile and 0.01% trifluoracetic acid) was used: 1–26 min with 0–100% solvent B, 2 min 100% solvent B, 1 min 100-0% solvent B. Elution was carried out a room temperature with a flow rate of 1 ml/min−1. The column effluent was analyzed by monitoring the absorbance at 230 and 260 nm and fractions collected. Pools of 5 fractions (40 fractions were collected per sample, resulting in 8 pools per sample) were tested in bioassays using strains CC9-16 and CC9-8.
Fish infectivity assays
Virulence tests were carried out on juvenile rainbow trout (Oncorhynchus mykiss) weighing ca. 2.5–3 g, which were anesthetized with tricaine methane sulfonate (0.1 g/liter). A total of 50 anesthetized fish were inoculated intramuscularly with 0.05 ml of each bacterial dilution, i.e., 50 fish per bacterial dilution. The dilutions were prepared with saline solution from 16 h cultures grown at 25°C in TSBS containing antibiotics for selection of the various plasmids harbored by the strains. The dilutions were prepared to test a range of cell concentrations from 101 to 107 cells/ml per strain. Therefore, 350 fish were tested per strain. After bacterial challenge, fish were maintained in fresh water at 13°C for 1 month and mortalities were checked daily. Virulence was quantified as the 50% lethal dose (LD50) as determined by the method of Reed and Muench (Reed and Muench, 1938).
β-galactosidase assay
β-galactosidase assays were performed as previously described (Miller, 1972) on sodium dodecyl sulfate-chloroform-permeabilized cells grown in M9 medium supplemented with 0.5 μM EDDA (iron-limiting) and 4 μg/ml ferric ammonium citrate (iron rich). Cells grown in the iron-limiting medium were harvested after 12 h of growth at 25 °C while for the iron rich medium the cells were harvested at 10 h. For all the strains, 1 ml of culture was used in the assay except for 775 pHNN-2 in iron-limiting conditions for which the assay was performed with 0.1 ml. The assays were repeated 4 times and each sample was tested in duplicate.
RNA isolation
A 1:100 inoculum from an overnight culture was grown in minimal medium with appropriate antibiotics. Cultures were grown with 2 μg/ml ferric ammonium citrate (iron-rich) or with EDDA (iron-limiting) supplemented to achieve similar levels of iron-limiting stress for each strain tested (see figure legend). Total RNA was prepared when the culture reached an OD600 of 0.3 to 0.5 using the RNAwiz™ (Ambion) isolation kit, as previously described (Di Lorenzo, et al., 2004).
Primer extension
Primer extension experiments were carried out with the synthetic primer PEX-angN (Table 2), which is complementary to the 5′-end region of the angN gene. The primer was end-labeled with T4-polynucleotide kinase (Life Technologies, Inc.) in the presence of [γ-32P]-ATP and annealed to V. anguillarum 775 total RNA (50 μg). Reverse transcription from the primer by avian myeloblastosis virus reverse transcriptase (Promega) and separation on an urea-PAGE (6%) were carried out as previously described (Sambrook and Russell, 2001). Manual sequencing was performed by the dideoxy chain-termination method using the Sequenase Version 2.0 DNA SequencingKit (USB), plasmid pMDL6 as template and the same primer used in the primer extension experiment.
Ribonuclease protection assays
Labeled riboprobes were generated by in vitro transcription of 1 μg of the linearized DNA with T7 or T3 RNA polymerase (MAXIscript® by Ambion) in the presence of [α-32P]-UTP using as a template pSC50 (linearized with BamHI) for fatB and pQSH6 (linearized with RsaI) for aroC. The template DNA for the angN probe was generated by PCR using the two primers angN-T7L and angN-T7R in which the T7 promoter sequence was added at the 5′-end of the angN-T7R primer (Table 2). The probes were purified on a 6% polyacrylamide gel. For each probe, the amount corresponding to 4×105 cpm was mixed with each RNA sample (20 μg). Ribonuclease protection assays were performed using the RPA III™ (Ambion) kit following the supplier’s instructions. The aroC riboprobe was used in each reaction as an internal control for the amount and quality of RNA.
RESULTS
Analysis of the AngN sequence
We have reported the complete sequence of plasmid pJM1 and identified several orfs that encode predicted proteins that are part of the iron uptake system (Di Lorenzo, et al., 2003). orf10, named angN, is predicted to encode a polypeptide (AngN) of 956 amino acids, with a calculated molecular mass of about 108 kDa (GenBank accession number NP943556 and Di Lorenzo, et al., 2003). The predicted AngN amino acid sequence showed significant matches to members of the family of NRPSs such as VibF of V. cholerae, PchE and PchF of Pseudomonas aeruginosa and HMWP2 of Yersinia pestis (Butterton, et al., 2000; Guilvout, et al., 1993; Quadri, et al., 1999). The similarity that AngN shares with these NRPSs is limited only to the cyclization (Cy) domain of these proteins. Like VibF but differently from the other proteins, the AngN protein possesses two tandem cyclization domains with 54% similarity and 35% identity with 927 amino acids at the N-terminal end of VibF.
Disruption of the angN gene and complementation with a wild type angN gene
To establish the role of this putative NRPS in anguibactin production, we selected from a collection of transposon insertions generated with Tn3::HoHo1 on a cloned region of pJM1, pJHC-T2612, two insertions, #120 and #68. These insertions occurred downstream of the iron transport-biosynthesis operon (Fig. 1) and each mutant was affected in anguibactin biosynthesis (Tolmasky, et al., 1988). DNA sequencing of the #120 and #68 mutants proved that these insertions were in angN at 417 bp and 1,913 bp respectively from the 5′-end. Since the insertions were on pJHC-T2612 containing only a partial sequence of pJM1 (Fig. 1) we had to determine the effect of angN mutations on the iron-uptake system encoded by the whole pJM1 plasmid. Therefore, the Tn3::HoHo1 insertion in mutant #120 was integrated by allelic-exchange onto the pJM1 plasmid resulting in V. anguillarum 775(pJM1-120). As expected strain 775(pJM1-120) was unable to grow in iron-limiting conditions at increasing concentrations of the iron chelator ethylenediamine-di-(O-hydroxyphenylacetic acid) (EDDA) since it did not produce anguibactin as detected by CAS (data not shown) and bioassay (Fig. 2A and 2B).
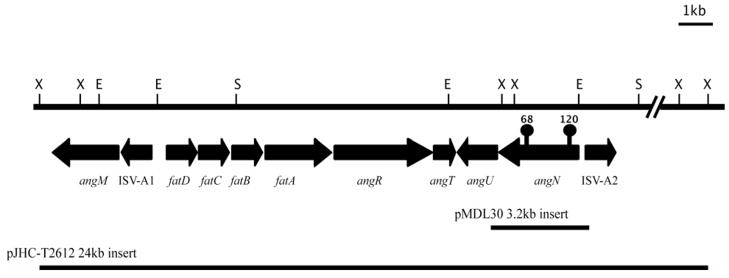
Fig. 1.
Schematic of a 24 kb DNA region from the pJM1 plasmid encoding the angN gene and the ITB operon showing the site of Tn3::HoHo1 insertion in mutant #68 and #120. Restriction endonucleases: X, XhoI; E, EcoRI; S, SalI.
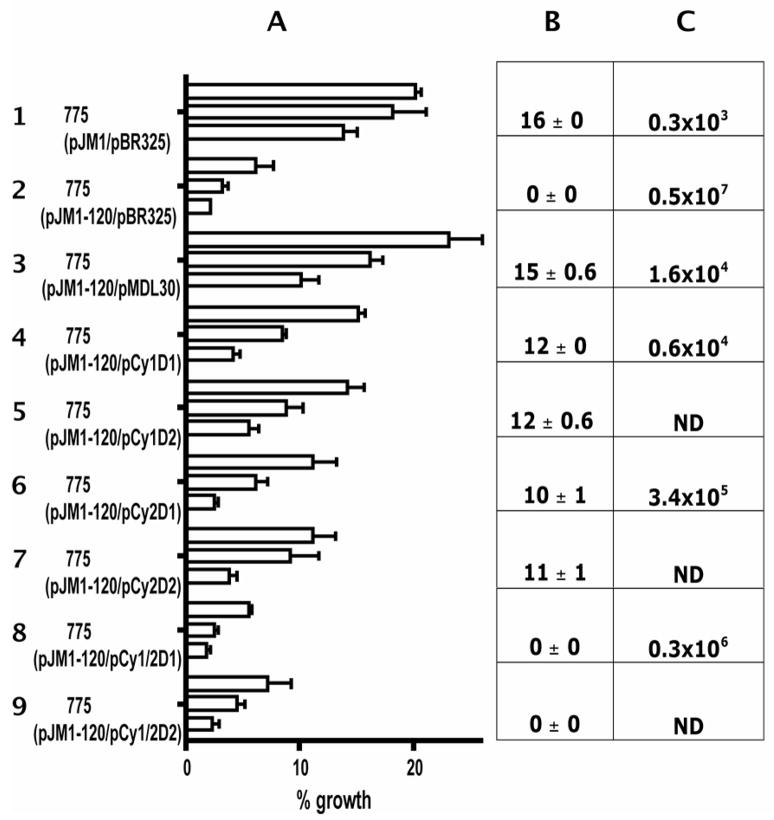
Fig. 2.
Growth in iron-limiting conditions, anguibactin production and virulence of the angN mutant strains. A. The ability of V. anguillarum strains to grow under-iron limitation is expressed as a percentage of the growth in EDDA normalized to the growth in iron-rich conditions (4 μg/ml of ferric ammonium citrate) for each strain. Each column for each strain corresponds to concentrations of EDDA, from top to bottom, of 0.5, 0.75 and 1μM, respectively. Results are the mean of three independent experiments with the error bars showing the standard error of the mean. B. Measurement of anguibactin production by bioassay in mm of growth. The results shown are the average of three independent experiments with corresponding error. C. Virulence experiments were carried out as described in Methods and the LD50 values were calculated by the method of Reed and Muench (Reed and Muench, 1938).
For complementation studies we constructed a clone, pMDL30, containing the complete angN gene (Fig. 1) expressed under the control of the tetracycline resistance gene promoter of the pBR325 vector and tested its ability to restore the growth of the V. anguillarum mutant 775(pJM1-120) under iron limitation. Plasmid pMDL30 was introduced into V. anguillarum 775(pJM1-120) and growth of the complemented mutant was determined in the presence of increasing concentrations of EDDA. The complemented strain grew as well as the wild type strain 775 under these conditions as seen by comparing growth of strains 1 and 3 in Fig. 2A. The fact that the angN gene was sufficient to restore growth of 775(pJM1-120) indicates that the insertion in mutant #120 was not polar to downstream genes.
The presence of anguibactin in the supernatant of strains 775, 775(pJM1-120) and 775(pJM1-120/pMDL30) grown in iron limiting conditions was also determined by HPLC analysis using a reversed-phase C18 column and a binary gradient as described in the Methods section. A peak identified as anguibactin by its retention time (12.06 min) and the UV spectra could be observed only in the wild type strain and in the 775(pJM1-120) mutant complemented with a wild type copy of the angN gene (Fig. 3). No peak corresponding to anguibactin was detected in strain 775(pJM1-120) confirming that a mutation affecting angN results in an anguibactin deficient phenotype (Fig. 3, Panel A). Pools of fractions collected from the HPLC column were tested in bioassay plates with strains CC9-16 and CC9-8 (the latter strain is used as a control for any iron contamination since it does not possess the anguibactin specific transport complex while strain CC9-16 can specifically transport anguibactin). Only the pools from strain 775 and 775(pJM1-120/pMDL30) containing the fraction of anguibactin allowed the growth of strain CC9-16 and no growth was observed in the plates seeded with strain CC9-8 (data not shown).
Fig. 3.
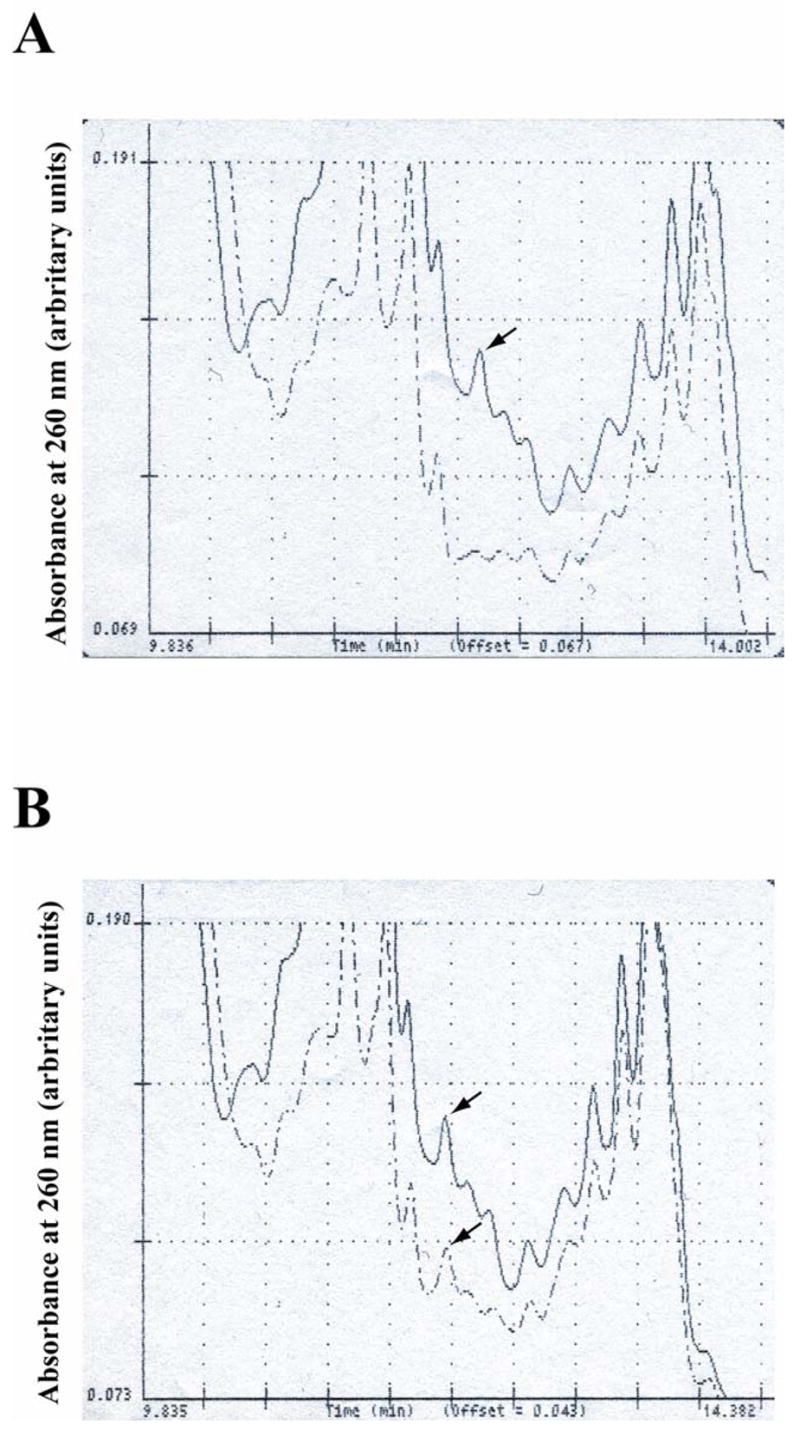
HPLC profiles of supernatants from cultures of 775 and mutant derivatives. Panel A. Detail (from min 9.836 to min 14.002) of the HPLC profiles of supernatants from 775 (continuous line) and 775(pJM1#120), dashed line. The arrow points to the anguibactin peak. Panel B. Detail (from min 9.835 to min 14.382) of the HPLC profiles of supernatants from 775 (continuous line) and 775(pJM1#120/pMDL30), dashed line. The arrows point to the anguibactin peak. Shown are representative HPLC profiles.
Effect of site-directed modification of the angN gene on anguibactin production
It has been demonstrated that cyclization domains catalyze peptide bond formation and cyclization of amino acids such as threonine and cysteine (Duerfahrt, et al., 2004; Marshall, et al., 2002; Quadri, et al., 1999). In the case of anguibactin it is likely that the thiazoline ring in anguibactin results from the incorporation of the amino acid cysteine in the siderophore. We wanted to determine the functionality of the two Cy domains of AngN in anguibactin biosynthesis. We performed site-directed mutagenesis of the first and second aspartic acid in the conserved Cy domain motif (DxxxxDxxS). Mutations in each of the aspartic acid residues within this motif have been shown to affect the activity of the Cy domains of VibF (Marshall, et al., 2002). As shown in Fig. 4, both Cy domains of AngN possess the two aspartic acid residues in the Cy motif (DMIAIDPDS in Cy1 and DALILDARS in Cy2). Each aspartic acid residue (shown as bold above) was mutated by site-directed mutagenesis to an alanine in the complementing construct pMDL30 generating four mutant constructs, pCy1D1-D133A, pCy1D2-D138A, pCy2D1-D575A and pCy2D2-D580A. Each plasmid was conjugated to the angN-deficient mutant 775(pJM1-120) and the resulting strains tested for their ability to grow in iron-limiting conditions. As shown in Fig. 2A, the insertion mutant #120 can still be complemented by the constructs harboring each of the mutated Cy domains (strains 4 to 7) although not as efficiently as with the construct harboring the wild type angN gene (strain 3). Furthermore, D to A mutations of each aspartic acid of the second cyclization domain seems to have a greater effect on the ability to complement the knock-out mutant. The growth in iron-limited medium of each strain correlates to their ability to produce anguibactin as determined by bioassays (Fig. 2B).
Fig. 4.

Alignment of the conserved Cy motifs of AngN with the Cy motif found in other NRPSs. The conserved residues are shown in bold.
Since a mutation in each of the aspartic acids had only a minor effect in anguibactin biosynthesis and growth, we decided to generate double mutations of the two Cy domains. Two plasmid derivatives of pMDL30 were constructed in which the first or the second aspartic acid of each domain, were mutated to an alanine (pCy1/2D1-D133A/D575A and pCy1/2D2-D138A/D580A). These two constructs were no longer able to complement the AngN-deficient mutant on pJM1 and no anguibactin could be detected by bioassay (Fig. 2A and B, strains 8 and 9).
AngN cyclization domains and anguibactin production in vivo
Our laboratory has shown that a clear correlation exists between anguibactin production and the multiplication of the bacterium in the fish host (Crosa, et al., 1980; Di Lorenzo, et al., 2004; Wertheimer, et al., 1999). Since the mutations in angN affected anguibactin production (Fig. 2B), we wanted to determine how this would be reflected in the virulence phenotype of each strain. Experimental infections of rainbow trout were performed with several dilutions of the wild type and the 775(pJM1-120) mutant and this mutant complemented by the constructs harboring wild type or mutant angN to calculate the LD50 (Reed and Muench, 1938). As expected mutations that resulted in an anguibactin-deficient phenotype (#120 insertion and #120 insertion complemented with the double aspartic acid mutant) had a reduced LD50 as compared to any strain that still produced anguibactin (Fig. 2C). Interestingly the LD50 for the mutant in the first Cy domain was of the same order of magnitude as the LD50 obtained with strain 775(pJM1-120) complemented with the wild type gene (Fig. 2C, strains 3 and 4), while the LD50 for the Cy2 mutant strain was reduced by one order of magnitude as compared with the two strains above (Fig. 2C, strains 3, 4 and 6).
Transcription and regulation of the angN gene
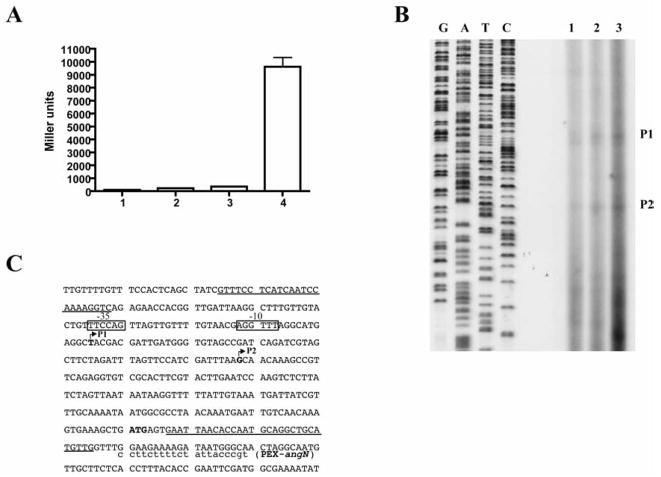
The location of the promoter for the angN gene was determined by transcriptional fusion of the region upstream of angN with a promoterless lacZ gene. As described in the Materials and Methods section, a 350 bp fragment encompassing the 3′-end of the gene was cloned upstream of the lacZ gene in pTL61T, obtaining plasmid pHNN2. This construct was conjugated in strain 775 by triparental mating and the resulting strain was tested for β-galactosidase activity. The construct showed high activity in iron-limiting conditions (0.5μM EDDA) while in iron rich conditions the Miller units measured were comparable to those of the empty vector control strain (Fig. 5A).
Fig. 5.
Mapping of angN promoter and the transcription start sites of angN mRNA. Panel A. β-galactosidase assays with V. anguillarum strains (lanes 1 and 2, 775 pTL61T; lanes 3 and 4 775 pHNN-2) grown in iron rich (4 μg/ml ferric ammonium citrate) or in iron-limiting (0.5 μM EDDA) conditions. Panel B. Primer extension analysis using a primer complementary to the 5′-end of the angN gene and RNA obtained from cultures grown in iron rich (lane 1, 2 μg/ml ferric ammonium citrate) and in iron-limiting conditions (lane 2, 1.5 μM EDDA; lane 3, 2 μM EDDA). Lanes G, A, C and T represent the sequence of plasmid pMDL6 using primer PEX-angN. P1 and P2 are putative transcription start sites for the angN gene. Panel C. The nucleotide sequence of the region upstream of the angN gene is shown with the primer used in the primer extension experiment and the location of the putative transcription starts (arrowheads P1 and P2 and bold letters). The –35 and –10 sequences for the P1 product are shown as open boxes and the ATG for AngN is in bold. The sequences corresponding to the primers used to construct the lacZ-fusion are underlined.
To determine the transcription start site of the angN mRNA, primer extension analysis was performed using primer PEX-angN complementary to the 5′-end of the angN gene and RNAs isolated from cultures grown at increasing iron limitation. Two iron-regulated products can be identified upstream of the angN gene (Fig. 5B, P1 and P2) with the first putative transcription start site 143 nt upstream of the start codon. Both transcription start sites fall within the region identified by β-galactosidase assay as the promoter region.
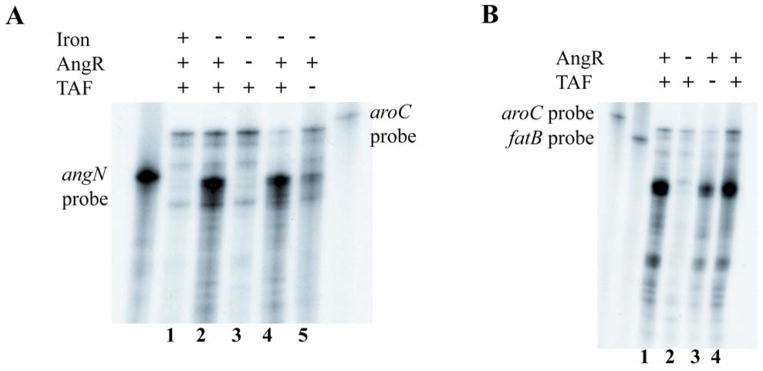
From these experiments it can be gathered that expression of the angN gene is repressed in iron rich conditions. To confirm this result we performed ribonuclease protection assays with total RNAs obtained from strain 775 cultures grown in iron-rich (minimal medium supplemented with 2 mg/ml of ferric ammonium citrate) and in iron-limiting conditions (minimal medium supplemented with 0.5 μM EDDA and 1.5 μM EDDA). The ribonuclease protection results (Fig. 6 panel A, lanes 1 and 2) clearly show that angN mRNA is virtually undetected in iron-rich conditions, while angN is highly transcribed in iron-limiting conditions. A riboprobe specific for the aroC-mRNA was added to each reaction as a control for RNA quality and loading.
Fig. 6.
Effect of iron, AngR and TAF on transcription of angN and fatB. A. Ribonuclease protection assay using a probe specific for the angN gene and total RNA isolated from cultures grown under iron-rich and iron-limiting conditions. V. anguillarum wild type (lanes 1, 2 and 4); V. anguillarum AngR-deficient strain (lane 3); V. anguillarum TAF-deficient strain (lane 5). RNA obtained under: iron-rich conditions, lane 1 (2 μg/ml ferric ammonium citrate); iron-limiting conditions, lanes 2 and 4 (1.5 μM EDDA) and lanes 3 and 5 (0.5 μM EDDA). B. Ribonuclease protection assay with the same RNA used in panel A from the cultures grown under iron-limiting conditions using a fatB-specific riboprobe. V. anguillarum wild type (lanes 1 and 4); V. anguillarum AngR-deficient strain (lane 2); V. anguillarum TAF-deficient strain (lane 3). The aroC-specific riboprobe is included in both Ribonuclease protection assays to provide an internal control for the quality of the RNA and the amount of RNA loaded onto the gel, since the aroC gene is expressed independently of the iron concentration of the cell.
In the anguibactin-mediated iron uptake system two positive regulators, AngR and TAF, have been identified that regulate expression of genes included in the iron transport-biosynthesis operon, fatDCBAangRT (Chen, et al., 1996; Wertheimer, et al., 1999). Since the angN promoter is adjacent to an ISVA2-transposase gene and transcribed in the opposite orientation, mirroring the organization of the promoters of the fatDCBAangRT and the tnp-angM operons (Di Lorenzo, et al., 2004), we wished to determine whether AngR and/or TAF products also regulate the transcription of angN. RNA was extracted from cultures of angR−taf + and angR+taf − strains that were grown in iron-limiting conditions (minimal medium supplemented with 0.5 μM EDDA) and the Ribonuclease protection assay was performed using an angN specific probe. We compared the changes in expression of the angN-mRNA and the fatDCBAangRT mRNA using a fatB-specific probe. Fig. 6 shows that AngR and TAF have a similar effect on expression of angN (panel A, lanes 3 to 5) as of fatB (panel B, lanes 1 to 4) demonstrating that in addition to the iron transport-biosynthesis operon promoter they can also enhance transcription from the angN promoter.
DISCUSSION
Sequencing of the plasmid pJM1 of V. anguillarum resulted in the identification of several genes encoding NRPSs and tailoring enzymes (Di Lorenzo, et al., 2003). In this work we have characterized one of these genes, angN, and established its role in vivo in anguibactin production and in virulence.
As part of this analysis we found that the predicted AngN protein showed homology with the cyclization domain of other NRPSs. AngN consists of two Cy domains in tandem with no associated additional domains and it is the first NRPS so far identified that has two free-standing Cy domains. Each domain has the two aspartic acids residues in the highly conserved motif that have been shown in in vitro experiments with VibF to be essential for the function of Cy domains (Marshall, et al., 2002). In this work we have shown that single amino acid substitutions of the conserved aspartic acid in either one of the domains partially abolishes anguibactin production, suggesting the possibility that one wild type Cy domain of AngN is sufficient to catalyze the cyclization reaction. Furthermore, mutations in the aspartic acid residues of both Cy domains (D133A/D575A and D138A/D580A) resulted in cessation of anguibactin biosynthesis demonstrating that these Cy domains are essential for production of anguibactin in vivo. Using HPLC analysis we showed that anguibactin can be detected only in supernatants from strains in which an active AngN is present. We also confirmed that the bioassay with strain CC9-16 is specific for anguibactin, since only the pools containing the anguibactin fraction resulted in growth of this strain.
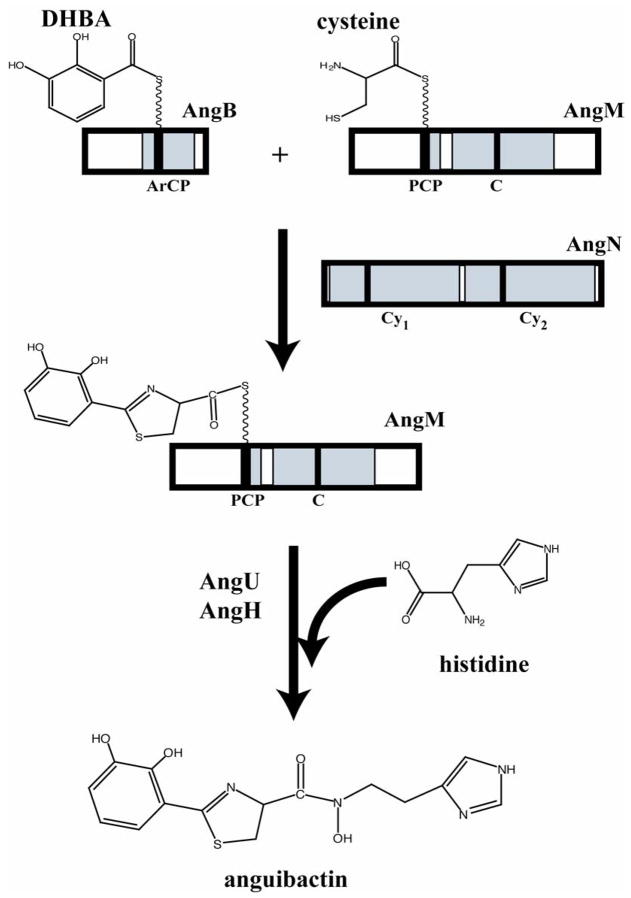
It is of interest that AngR, which plays a role in regulation of the expression of iron transport genes as well as in the production of anguibactin, possesses also a cyclization (Cy) domain (Wertheimer, et al., 1999). However the first aspartic acid residue of the conserved motif is replaced by an asparagine in the Cy domain of AngR. From the results presented in this work the Cy domain of AngR is unable to functionally replace the mutated Cy domains of AngN in anguibactin production. Therefore, an attractive possibility is that AngR provides the adenylation domain required to activate cysteine, prior to tethering it on the peptidyl carrier protein domain of AngM while either of the Cy domains of AngN is in charge of the condensation step between DHBA and cysteine (Fig. 7).
Fig. 7.
Model of the biosynthetic step catalyzed by AngN. The Cy domains of AngN catalyze peptide bond formation and cyclization between DHBA loaded on the ArCP domain of AngB and cysteine tethered onto the PCP domain of AngM. AngU and AngH are tailoring enzymes that convert histidine to hydroxyhistamine.
The unusual domain arrangement of AngN and the results obtained with the mutants affecting only one domain of AngN suggest that AngN is a protein with a redundant catalytic activity, in which each domain is functional independently of the other. However, the differences in the LD50 of the mutants in the virulence experiments suggest that only an AngN protein with two functional Cy domains results in the most efficient synthesis of anguibactin.
It is intriguing that a division of labor by two Cy domains was demonstrated in an in vitro experiment with VibF (Marshall, et al., 2002), an NRPS that in addition to two cyclization domains also has an adenylation, a peptidyl carrier protein and two condensation domains, and thus is not a free standing cyclization enzyme like AngN. From the experiments with VibF it was proposed that the first cyclization domain does the actual cyclization reaction while the second one does the condensation step (Marshall, et al., 2002). Sequence alignment of each Cy domain of AngN with other seven Cy domains (VibFCy1, VibFCy2, HMWP2Cy1, HMWP2Cy2, PchFCy, PchECy and the other Cy domain of AngN) revealed an average of 22.7% identity for Cy1 and 18.6% identity for Cy2. The percentage of identity of AngNCy2 to each one of the other Cy domains is relatively lower than the overall average of 25.4% of the other domains while the similarity shared with VibFCy2 is significantly higher (32%). Furthermore, the same sequence alignment showed that the amino acid residues that have been shown to be important in cyclization but not in condensation activity of Cy domains (Duerfahrt, et al., 2004) were not conserved in the Cy2 domain of AngN and in the Cy2 domain of VibF. However, we cannot conclude from our in vivo results with the AngN mutants that the division of labor observed in VibF Cy domains occurs also in AngN domains since site-directed mutations in either Cy1 or Cy2 are still proficient in anguibactin production.
In this work we also determined that expression of the angN gene is negatively regulated by the iron concentration of the culture medium. Analysis of the sequence at the angN promoter shows that it overlaps a putative transposase promoter that transcribes in the opposite direction, thus mirroring the situation described for the tnp-angM and iron transport-biosynthesis promoters (Di Lorenzo, et al., 2004). An exciting result was that the angN promoter is positively regulated by AngR and TAF, making it another member of the AngR and TAF regulon.
Acknowledgments
This project was supported by Grants from the National Institute of Health, AI19018 and GM64600, to J.H.C. We are grateful to Christopher T. Walsh for his insightful discussions.
References
- Actis LA, Fish W, Crosa JH, et al. Characterization of anguibactin, a novel siderophore from Vibrio anguillarum 775(pJM1) J Bacteriol. 1986;167:57–65. doi: 10.1128/jb.167.1.57-65.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Actis LA, Tolmasky ME, Farrell DH, et al. Genetic and molecular characterization of essential components of the Vibrio anguillarum plasmid-mediated iron-transport system. J Biol Chem. 1988;263:2853–2860. [PubMed] [Google Scholar]
- Actis LA, Tolmasky ME, Crosa JH. Vibriosis. In: Woo P, Bruno D, editors. Fish diseases and disorders Viral, bacterials, and fungal infections. Cab International Publishing; Wallingford: 1999. pp. 523–557. [Google Scholar]
- Alice AF, Lopez CS, Crosa JH. Plasmid- and chromosome-encoded redundant and specific functions are involved in biosynthesis of the siderophore anguibactin in Vibrio anguillarum 775: a case of chance and necessity? J Bacteriol. 2005;187:2209–2214. doi: 10.1128/JB.187.6.2209-2214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978;4:121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Boyer HW, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Braun V, Killmann H. Bacterial solutions to the iron-supply problem. Trends Biochem Sci. 1999;24:104–109. doi: 10.1016/s0968-0004(99)01359-6. [DOI] [PubMed] [Google Scholar]
- Butterton JR, Choi MH, Watnick PI, et al. Vibrio cholerae VibF is required for vibriobactin synthesis and is a member of the family of nonribosomal peptide synthetases. J Bacteriol. 2000;182:1731–1738. doi: 10.1128/jb.182.6.1731-1738.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Wertheimer AM, Tolmasky ME, et al. The AngR protein and the siderophore anguibactin positively regulate the expression of iron-transport genes in Vibrio anguillarum. Mol Microbiol. 1996;22:127–134. doi: 10.1111/j.1365-2958.1996.tb02662.x. [DOI] [PubMed] [Google Scholar]
- Crosa JH, Hodges LL, Schiewe MH. Curing of a plasmid is correlated with an attenuation of virulence in the marine fish pathogen Vibrio anguillarum. Infect Immun. 1980;27:897–902. doi: 10.1128/iai.27.3.897-902.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo M, Stork M, Tolmasky ME, et al. Complete sequence of virulence plasmid pJM1 from the marine fish pathogen Vibrio anguillarum strain 775. J Bacteriol. 2003;185:5822–5830. doi: 10.1128/JB.185.19.5822-5830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo M, Poppelaars S, Stork M, et al. A nonribosomal peptide synthetase with a novel domain organization is essential for siderophore biosynthesis in Vibrio anguillarum. J Bacteriol. 2004;186:7327–7336. doi: 10.1128/JB.186.21.7327-7336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerfahrt T, Eppelmann K, Muller R, et al. Rational design of a bimodular model system for the investigation of heterocyclization in nonribosomal peptide biosynthesis. Chem Biol. 2004;11:261–271. doi: 10.1016/j.chembiol.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides1. Annu Rev Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- Guilvout I, Mercereau-Puijalon O, Bonnefoy S, et al. High-molecular-weight protein 2 of Yersinia enterocolitica is homologous to AngR of Vibrio anguillarum and belongs to a family of proteins involved in nonribosomal peptide synthesis. J Bacteriol. 1993;175:5488–5504. doi: 10.1128/jb.175.17.5488-5504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch PR, Beringer JE. A physical map of pPH1JI and pJB4JI. Plasmid. 1984;12:139–141. doi: 10.1016/0147-619x(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Keating TA, Walsh CT. Initiation, elongation, and termination strategies in polyketide and polypeptide antibiotic biosynthesis. Curr Opin Chem Biol. 1999;3:598–606. doi: 10.1016/s1367-5931(99)00015-0. [DOI] [PubMed] [Google Scholar]
- Linn T, St Pierre R. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J Bacteriol. 1990;172:1077–1084. doi: 10.1128/jb.172.2.1077-1084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahiel MA, Stachelhaus T, Mootz HD. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- Marshall CG, Burkart MD, Keating TA, et al. Heterocycle formation in vibriobactin biosynthesis: alternative substrate utilization and identification of a condensed intermediate. Biochemistry. 2001;40:10655–10663. doi: 10.1021/bi010937s. [DOI] [PubMed] [Google Scholar]
- Marshall CG, Hillson NJ, Walsh CT. Catalytic mapping of the vibriobactin biosynthetic enzyme VibF. Biochemistry. 2002;41:244–250. doi: 10.1021/bi011852u. [DOI] [PubMed] [Google Scholar]
- Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DA, Walsh CT. Yersiniabactin synthetase: probing the recognition of carrier protein domains by the catalytic heterocyclization domains, Cy1 and Cy2, in the chain-initiating HWMP2 subunit. Biochemistry. 2001;40:5313–5321. doi: 10.1021/bi002905v. [DOI] [PubMed] [Google Scholar]
- Miller J. Experiments in molecular genetics. Cold Spring Harbor; NY: 1972. [Google Scholar]
- Naka H, Lopez CS, Crosa JH. Reactivation of the vanchrobactin siderophore system of Vibrio anguillarum by removal of a chromosomal insertion sequence originated in plasmid pJM1 encoding the anguibactin siderophore system. Environ Microbiol. 2008;10:265–277. doi: 10.1111/j.1462-2920.2007.01450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadri LE, Keating TA, Patel HM, et al. Assembly of the Pseudomonas aeruginosa nonribosomal peptide siderophore pyochelin: In vitro reconstitution of aryl-4, 2-bisthiazoline synthetase activity from PchD, PchE, and PchF. Biochemistry. 1999;38:14941–14954. doi: 10.1021/bi991787c. [DOI] [PubMed] [Google Scholar]
- Ratledge C. Iron metabolism and infection. Food Nutr Bull. 2007;28:S515–523. doi: 10.1177/15648265070284S405. [DOI] [PubMed] [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. The American Journal of Hygiene. 1938;27:493–497. [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Tolmasky ME, Actis LA, Crosa JH. Genetic analysis of the iron uptake region of the Vibrio anguillarum plasmid pJM1: molecular cloning of genetic determinants encoding a novel trans activator of siderophore biosynthesis. J Bacteriol. 1988;170:1913–1919. doi: 10.1128/jb.170.4.1913-1919.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dohren H, Dieckmann R, Pavela-Vrancic M. The nonribosomal code. Chem Biol. 1999;6:R273–279. doi: 10.1016/s1074-5521(00)80014-9. [DOI] [PubMed] [Google Scholar]
- Walsh CT, Chen H, Keating TA, et al. Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr Opin Chem Biol. 2001;5:525–534. doi: 10.1016/s1367-5931(00)00235-0. [DOI] [PubMed] [Google Scholar]
- Walsh CT. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science. 2004;303:1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- Walter MA, Potter SA, Crosa JH. Iron uptake system mediated by Vibrio anguillarum plasmid pJM1. J Bacteriol. 1983;156:880–887. doi: 10.1128/jb.156.2.880-887.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- Welch TJ, Chai S, Crosa JH. The overlapping angB and angG genes are encoded within the trans-acting factor region of the virulence plasmid in Vibrio anguillarum: essential role in siderophore biosynthesis. J Bacteriol. 2000;182:6762–6773. doi: 10.1128/jb.182.23.6762-6773.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheimer AM, Verweij W, Chen Q, et al. Characterization of the angR gene of Vibrio anguillarum: essential role in virulence. Infect Immun. 1999;67:6496–6509. doi: 10.1128/iai.67.12.6496-6509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]