Abstract
We have examined the distribution of RNA transcription and processing factors in the amphibian oocyte nucleus or germinal vesicle. RNA polymerase I (pol I), pol II, and pol III occur in the Cajal bodies (coiled bodies) along with various components required for transcription and processing of the three classes of nuclear transcripts: mRNA, rRNA, and pol III transcripts. Among these components are transcription factor IIF (TFIIF), TFIIS, splicing factors, the U7 small nuclear ribonucleoprotein particle, the stem–loop binding protein, SR proteins, cleavage and polyadenylation factors, small nucleolar RNAs, nucleolar proteins that are probably involved in pre-rRNA processing, and TFIIIA. Earlier studies and data presented here show that several of these components are first targeted to Cajal bodies when injected into the oocyte and only subsequently appear in the chromosomes or nucleoli, where transcription itself occurs. We suggest that pol I, pol II, and pol III transcription and processing components are preassembled in Cajal bodies before transport to the chromosomes and nucleoli. Most components of the pol II transcription and processing pathway that occur in Cajal bodies are also found in the many hundreds of B-snurposomes in the germinal vesicle. Electron microscopic images show that B-snurposomes consist primarily, if not exclusively, of 20- to 30-nm particles, which closely resemble the interchromatin granules described from sections of somatic nuclei. We suggest the name pol II transcriptosome for these particles to emphasize their content of factors involved in synthesis and processing of mRNA transcripts. We present a model in which pol I, pol II, and pol III transcriptosomes are assembled in the Cajal bodies before export to the nucleolus (pol I), to the B-snurposomes and eventually to the chromosomes (pol II), and directly to the chromosomes (pol III). The key feature of this model is the preassembly of the transcription and processing machinery into unitary particles. An analogy can be made between ribosomes and transcriptosomes, ribosomes being unitary particles involved in translation and transcriptosomes being unitary particles for transcription and processing of RNA.
INTRODUCTION
In eukaryotic cells the cytoplasmic translation machinery consists of ribosomes, whose subunits are assembled in the nucleolus and are then exported to the cytoplasm, where they serve as the substrate for protein synthesis. The discovery of this pathway was facilitated by the abundance of ribosomes, by the ease with which they could be isolated from the cytoplasm, and by the physical separation of ribosome assembly in the nucleus from ribosome function in the cytoplasm. Furthermore, the nucleolus was a well-known and prominent structure in the nucleus, and assembly of the ribosomes was coincident with synthesis of the most abundant RNA in the cell (Vincent and Miller, 1965). By contrast, an understanding of the cellular organization of the transcription and processing machinery has lagged behind that of the translation machinery. Indeed, whether a transcription and processing complex comparable with the ribosome even exists remains speculative. If it does exist, what are the constituents of the complex? Is it assembled on the chromatin template, or is it preassembled? If preassembled, where does assembly and/or storage take place? Is there a common pathway for the assembly of polymerase I (pol I), pol II, and pol III transcription and processing complexes?
Remarkable progress has been made in understanding the individual constituents of the transcription and processing machinery, primarily from biochemical studies on purified or partially purified complexes. Some of these complexes, referred to as holoenzymes, contain the core polymerase along with general transcription factors and other proteins (Greenblatt, 1997; Wang et al., 1997; Myer and Young, 1998; Parvin and Young, 1998; Roeder, 1998; Seither et al., 1998). Unfortunately, the physical properties of most interphase nuclei make the separation of morphologically well-defined subnuclear fractions difficult, and it is therefore not clear from what part of the nucleus these large complexes arise. Nucleoli can be isolated as a reasonably clean fraction, because they are large and relatively stable structures (Busch and Smetana, 1970; Rothblum, 1998), and recently Mintz et al. (1999) described a protocol by which morphologically well-defined interchromatin granules can be prepared. As yet, however, no general method for subnuclear fractionation exists, so that much of the most valuable information on the distribution of transcription and processing components comes from microscopical techniques, such as electron microscopy, immunofluorescence, and in situ hybridization (de Jong et al., 1996; Lamond and Earnshaw, 1998; Matera, 1998; Misteli and Spector, 1998). However, because most nuclei are small, even high-resolution microscopical analysis may fail to distinguish those regions involved in assembly and storage of components from those in which transcription and processing of RNA take place.
The amphibian germinal vesicle (GV) provides a way around the problem of size. The GV from a midsized Xenopus oocyte is ∼400 μm in diameter, and its giant lampbrush chromosomes transcribe RNA at a rate well above that in typical somatic nuclei (Callan, 1986; Davidson, 1986). When the GV contents are spread on a microscope slide, the transcriptionally active chromosomes and other nuclear organelles are well separated from each other. Thus, by a combination of immunofluorescent staining and in situ hybridization, it is possible to determine unambiguously the molecular composition of each organelle (Lacroix et al., 1985; Roth and Gall, 1987; Wu et al., 1991). Furthermore, injection experiments allow one to follow the targeting of protein and RNA molecules to specific structures within the nucleus (Roth and Gall, 1989; Kay, 1991; Wu et al., 1996; Lange et al., 1998a–c; Narayanan et al., 1999) and to make inferences about the sites of assembly of macromolecular complexes.
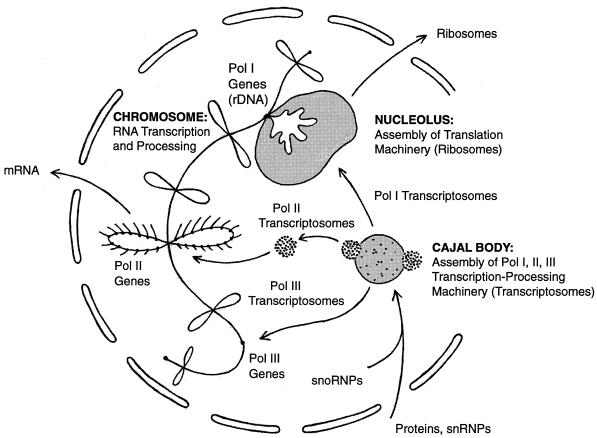
From experiments on the GV we suggest that the pol II transcription and processing machinery is assembled as a unitary particle in the Cajal bodies (coiled bodies; see note on terminology at end of DISCUSSION), is stored in the B-snurposomes, and functions on the loops of the lampbrush chromosomes (Figure 1). According to this model, the complex of pol II and its associated factors is visible by electron microscopy as a 20-to 30-nm particle, which we call a pol II transcriptosome. Pol II transcriptosomes are the major, if not the only structural component of B-snurposomes, and they probably correspond to the interchromatin granules described from somatic nuclei.
Figure 1.
A model of Cajal body function. The essential feature of the model is the preassembly of RNA transcription and processing complexes in the Cajal body. It is suggested that complexes involving pol I, pol II, and pol III are assembled in the Cajal body before transport to the appropriate genes on the chromosomes. The pol II complexes are stored in the B-snurposomes, where they are visible in electron micrographs as 20- to 30-nm particles, which we call pol II transcriptosomes. It is postulated that similar pol I and pol III transcriptosomes are assembled in the matrix of the Cajal body.
We postulate that pol I and pol III transcription and processing complexes are similarly assembled in the Cajal bodies. We suggest that pol I and associated processing factors are then transported to the nucleoli, where they function in rRNA transcription and processing. Pol III and its associated factors could be targeted from the Cajal bodies to specific pol III loci on the chromosomes.
In summary, we propose that pol I, pol II, and pol III transcription and processing complexes are assembled in the Cajal bodies, from which they are ultimately transported to the appropriate genes on the chromosomes (Figure 1).
MATERIALS AND METHODS
Oocytes and GV Spreads
A female Xenopus laevis or Notophthalmus viridescens was anesthetized in 0.1% methanesulfonate salt of 3-aminobenzoic acid ethyl ether (tricaine methane sulfonate or MS222; A5040, Sigma, St. Louis, MO). A sample of ovary was removed surgically and held in a small Petri dish of OR2 saline (Wallace et al., 1973) at 18–20°C for ≥12 h before use. General instructions for preparation of GV spreads were published earlier (Gall et al., 1991), but a few critical modifications dramatically improve their quality (Gall, 1998). Injected oocytes were transferred one at a time to isolation medium (83 mM KCl, 17 mM NaCl, 6.5 mM Na2HPO4, 3.5 mM KH2PO4, 1 mM MgCl2, 1 mM dithiothreitol, pH 7.0). The GV was removed with forceps and transferred within 20 s to spreading medium (21 mM KCl, 4 mM NaCl, 1.6 mM Na2HPO4, 0.9 mM KH2PO4, 1 mM MgCl2, 1 mM dithiothreitol, 0.1% paraformaldehyde, pH 7.0). The nuclear envelope was removed with jeweler’s forceps, and the nuclear gel was transferred with a pipette to a spreading chamber. If the procedure was carried out quickly, dispersal of the nuclear gel occurred within minutes, and centrifugation could begin almost immediately. Slides were centrifuged in a special holder in the Sorvall HS4 rotor (DuPont, Wilmington, DE) at 5000 rpm (4800 × g) at 4°C for 30 min. GV spreads were fixed with 2% paraformaldehyde in PBS plus 1 mM Mg2+ for ≥1 h.
Oocyte Injections
Oocytes were injected into the cytoplasm (23 nl) or the nucleus (4.6 or 9.2 nl), depending on the experiment. Before nuclear injection, oocytes were centrifuged at 1800 rpm (630 × g) for 20–30 min to bring the GV to the surface, where its position can be detected as a depigmented area. Glass needles were made from capillary tubing (0.5-mm inner diameter, 1.2-mm outer diameter) using a Vertical Pipette Puller (David Kopf Instruments, Tujunga, CA). Injections were made under a dissecting microscope with a Drummond Nanoject microinjection apparatus (Drummond Scientific, Broomall, PA), which uses a plunger to displace liquid in the needle.
Immunofluorescent Staining and Microscopy
GV spreads were rinsed in PBS and blocked with 10% horse serum in PBS for 5 min. Spreads were incubated with primary antibody for 1 h at room temperature, rinsed briefly in PBS, incubated 1 h at room temperature in secondary antibody, and rinsed again in PBS. mAbs were used at 0.5–5 μg/ml, when the concentration was known; otherwise, culture supernate was used undiluted or diluted 1:2–1:20 in 10% horse serum. Rabbit polyclonal sera were diluted 1:200–1:5000 with 10% horse serum. Secondary antibodies were Alexa 488- or Alexa 594-labeled goat anti-mouse immunoglobulin G (IgG) or goat anti-rabbit IgG (Molecular Probes, Eugene, OR). In some cases the secondary antibody was fluorescein-labeled goat anti-mouse IgM (Cappel/Organon Teknika, Durham, NC). In double-label experiments, the primary antibodies and their corresponding secondaries were applied successively. Spreads were sometimes stained for a few minutes in 0.01 μg/ml 4′,6-diamidino-2-phenylindole. Preparations were mounted in 50% glycerol containing 1 mg/ml phenylenediamine to prevent fading. Confocal laser scanning microscopy was carried out with the Leica (Heidelberg, Germany) TCS NT system.
Antibodies
The following mAbs were used: H1 against Xenopus coilin or SPH-1 (Tuma et al., 1993), H5 and H14 against the carboxyl-terminal domain (CTD) of human RNA pol II (Bregman et al., 1995), 8WG16 against the CTD of wheat germ pol II (Thompson et al., 1989), 9E10 against the c-myc epitope (Evan et al., 1985), Y12 against the Sm epitope (Lerner et al., 1981), SC35 against human SR protein SC35 (Fu and Maniatis, 1990), 3F10 (rat) against the hemagglutinin (HA) epitope (Boehringer Mannheim, Indianapolis, IN), BU-33 against bromodeoxyuridine (Sigma), 17C12 against fibrillarin (Pollard et al., 1997), No114 against Xenopus Nopp140 (Schmidt-Zachmann et al., 1984), No185 against Xenopus NO38 or B23 (Schmidt-Zachmann et al., 1987), and P7 1A4 against Xenopus nucleolin (Messmer and Dreyer, 1993).
Rabbit polyclonal sera against the following proteins were used: RPA194 and RPA127 (Hannan et al., 1998); transcription factor IIF (TFIIF) (RAP74), CstF77, and CPSF100 (from D. Bentley, University of Colorado Health Sciences Center); CPSF100 (Jenny et al., 1994); RPC19, RPC53, and RPC62 (Wang et al., 1997); and TFIIIA (from R.G. Roeder, Rockefeller University).
In Vitro RNA Transcription
RNA transcripts for in situ hybridization or microinjection were synthesized by T3 or T7 RNA polymerase with ∼1 μg of plasmid DNA as template in reaction volumes of 20–30 μl. The transcription reaction for capped, fluorescein-labeled RNAs for microinjection included 250 μM GTP; 2.5 mM m7G(5′)ppp(5′)G cap (New England Biolabs, Beverly, MA); 500 μM ATP and CTP; 250 μM UTP; 166 μM fluorescein-12-UTP (Boehringer Mannheim); and trace amounts of [α-32P]UTP. Fluorescein-labeled probes for in situ hybridization were transcribed in a similar reaction in which the GTP concentration was 500 μM and the cap was omitted. Probes for injection were precipitated twice at −20°C with 70% ethanol and 1 μg of glycogen as carrier and redissolved in 20 μl of H2O to give a concentration of 1–3 pmol/μl. Probes for in situ hybridization were similarly precipitated and redissolved in 50 μl of hybridization buffer (40% formamide, 4× SSC, 60 mM Na2HPO4, 40 mM KH2PO4, 300 μg/ml Escherichia coli DNA, and 300 μg/ml E. coli RNA). Hybridization probes were diluted 5- to 40-fold before use. Probes for microinjection were used without further dilution or diluted up to 10-fold (0.1–0.3 pmol/μl).
Capped, fluorescein-labeled U3 small nucleolar RNA (snoRNA) for injection was transcribed from a PCR product of a cloned wild-type Xenopus U3 gene (Lange et al., 1998c). Injections of 2–15 fmol (∼0.1–1 ng) were made into the GV. Capped, fluorescein-labeled U1 and U2 small nuclear RNAs (snRNAs) for injection were transcribed from cloned full-length Xenopus U1 and U2 genes as described (Bellini and Gall, 1998). Injections of 8–20 fmol (∼0.5–1.5 ng) were made into the cytoplasm. A synthetic, full-length U5 snRNA gene was constructed by annealing overlapping 5′ and 3′ oligodeoxynucleotides derived from the published sequence of the Xenopus U5 gene (Kazmaier et al., 1987). After annealing, both strands were extended with Klenow polymerase, digested with EcoRI and XbaI, and cloned into the plasmid pUC 19. A PCR product that contains the T3 RNA polymerase promoter was synthesized from the U5 clone as template with the following primers (T3 promoter underlined): 5′-CGGAATTCAATTAACCCTCACTAAAGGG-3′ and 5′-ATACCTGGTGTGAACCAGGCTTC-3′ Capped, fluorescein-labeled U5 snRNA was transcribed from the PCR product. Injection of 27 fmoles (∼1.0 ng) was made into the GV. Fluorescein-labeled antisense U1–U6 snRNA probes for in situ hybridization were transcribed from cloned fragments of human and mouse genes as described (Wu et al., 1991).
In Situ Hybridization
After fixation for ≥1 h in 2% paraformaldehyde, GV preparations were dehydrated in an ethanol series, deparaffinized in several changes of xylene, and rehydrated through an ethanol series to PBS. The area around the preparation was wiped dry, leaving a minimal drop of PBS over the specimen. Fluorescein-labeled antisense RNA probe (5 μl) was placed on the specimen area, and an 18-mm2 coverslip was added. The edges were sealed with rubber cement, and the slide was held at 42°C for 4–18 h. The coverslip was removed under PBS, and the slide was washed in three changes of 50% formamide and 2× SSC at 45°C for a total of 45 min (SSC is 0.15 M NaCl, 0.015 M Na citrate, pH 7). Some preparations were sufficiently bright to be observed directly by fluorescence microscopy. A much brighter and more stable preparation was obtained by amplifying the fluorescein signal with an antibody. In some cases we used an unlabeled mouse anti-fluorescein antibody followed by Alexa 488-labeled goat anti-mouse IgG (Molecular Probes). We also prepared Alexa 488-labeled mouse anti-fluorescein using the manufacturer’s labeling kit. In this case the amplification required only a single antibody step.
Electron Microscopy
Spread GV preparations on plastic microscope slides were fixed in 2% paraformaldehyde and PBS for ≥1 h. They were further fixed in 4% glutaraldehyde overnight, rinsed in PBS, and treated with 1% OsO4 plus 0.5% K3Fe(CN)6 in PBS for 45 min. After a further rinse in PBS, they were contrasted in 0.5% UO2(C2H3O2)2 for 45 min. The slide was dehydrated in an ethanol series, and the preparation was “flat embedded” in Epon resin plus 2% benzyldimethylamine. Silver sections were cut from the end of the resin block with a Reichert (Vienna, Austria) Ultracut S microtome and examined on parlodion-coated or unsupported copper grids in a JEOL (Tokyo, Japan) 100S electron microscope.
RESULTS
A single GV of a midsized Xenopus oocyte (Dumont stage IV–V; Dumont, 1972) contains 18 actively transcribing lampbrush chromosomes plus hundreds to thousands of granules of various sizes. Despite this seeming complexity, there are only three major types of extrachromosomal granules: 1) ∼1500 nucleoli, 2) 50–100 Cajal bodies, and 3) a highly variable number of B-snurposomes (Figure 2, A and E). The origin of the extrachromosomal nucleoli from amplified copies of the rDNA and their role in pol I transcription of rRNA have been extensively documented (Miller, 1966; Brown and Dawid, 1968; Gall, 1968; Macgregor, 1972). The two remaining structures, the B-snurposomes and the Cajal bodies, both contain pol II and a number of factors involved in pre-mRNA processing. Because pol II transcripts are made and probably processed on the loops of the lampbrush chromosomes, B-snurposomes and Cajal bodies must have other functions, such as assembly, storage, or modification of the pol II transcription and processing machinery.
Figure 2.
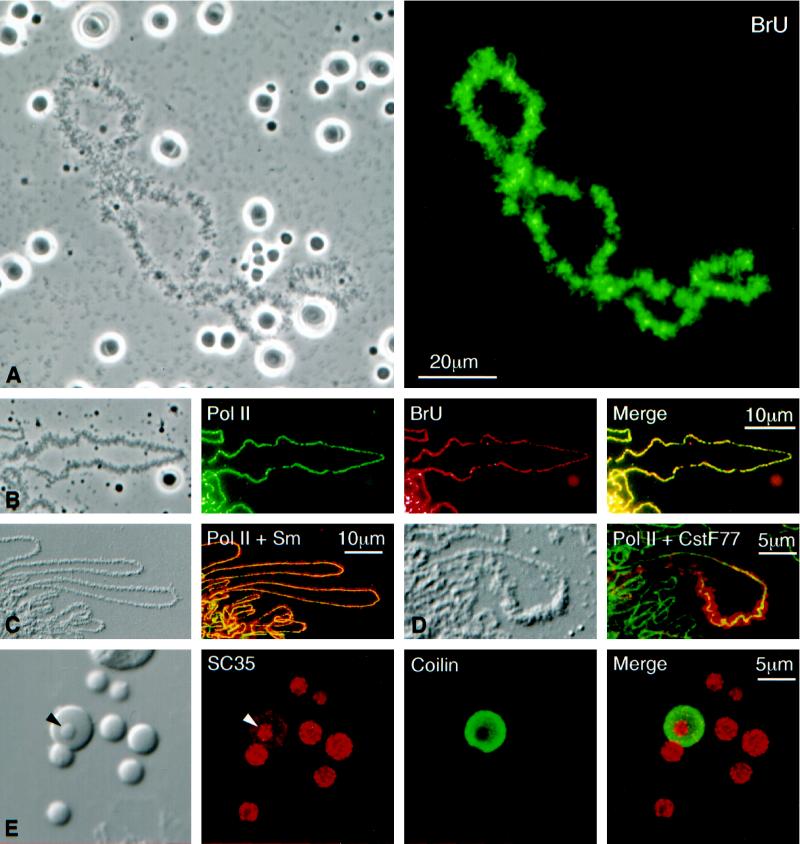
(A) Phase-contrast (left) and fluorescent (right) images of a single lampbrush chromosome with scattered nucleoli, Cajal bodies, and B-snurposomes from a spread preparation of a Xenopus GV. The oocyte had been injected 4.5 h earlier with 20 mM BrUTP (253 ng in 23 nl). Strong BrU label in the lampbrush chromosome U loops is revealed by immunostaining with an anti-BrU antibody (right). Nucleoli are weakly stained, but Cajal bodies and B-snurposomes are completely negative. (B) Single loop from a lampbrush chromosome of the newt Notophthalmus. The oocyte had been injected 1 h earlier with 20 mM BrUTP (253 ng in 23 nl). The left panel shows a phase-contrast image of the loop. The following three panels show staining of the loop axis with an antibody against pol II (mAb H5, green), BrU incorporation (red), and a merge of the two images. Pol II and newly transcribed RNA are precisely colocalized. (C) Differential interference contrast (DIC) and immunofluorescence images of loops from a Notophthalmus lampbrush chromosome after double staining for pol II (mAb H14, green) and the Sm epitope (mAb Y12, red). Pol II occurs only along the axis, whereas splicing snRNPs are present throughout the RNP matrix of the loop. (D) Loop from a Notophthalmus lampbrush chromosome after double staining for pol II (mAb H14, green) and the 77-kDa subunitof the cleavage stimulation factor CstF (red). CstF77 occurs through the matrix of a single “thin-to-thick” transcription unit. (E) The left panel shows the DIC image of a single Cajal body with an attached B-snurposome and an internal B-like inclusion (arrowhead). Nearby are six free B-snurposomes. The following three panels show the same field after staining for the SR protein SC35 (red), the Cajal body marker coilin (green), and a merge of the two images. The B-snurposomes and the inclusion stain strongly for SC35; coilin is limited to the matrix of the Cajal body, which also stains weakly for SC35.
Lampbrush Chromosomes
RNA synthesis on the loops of lampbrush chromosomes was first demonstrated by autoradiography after incorporation of radioactive nucleotide precursors (Gall, 1958; Gall and Callan, 1962; Izawa et al., 1963; Callan, 1986). This transcription is sensitive to low concentrations of α-amanitin (0.5 μg/ml), demonstrating that pol II is involved, whereas pol I transcription in the extrachromosomal nucleoli is resistant to much higher concentrations of the inhibitor (Bucci et al., 1971; Schultz et al., 1981). Antibodies against pol II provide a direct and striking demonstration of the enzyme as it moves along the DNA axis of the lampbrush loops. Both mAb H5 and mAb H14 (Bregman et al., 1995), which recognize different phosphorylated epitopes on the CTD of pol II (Patturajan et al., 1998), display a more or less continuous line of stain down the middle of each loop. Details of loop organization are particularly clear in the very large loops of newt lampbrush chromosomes. As shown in Figure 2, B–D, pol II stain appears as an essentially diffraction-limited line ∼0.3–0.4 μm wide, despite wide variations in overall loop length and thickness of the RNP matrix. The line tends to be strongest at the start or thin end of a transcription unit and fades out somewhat toward the thick end.
Some interesting features of transcription are revealed by labeling chromosomes with BrUTP, which can be monitored with a fluorescent antibody to provide high-resolution details of newly synthesized RNA (Jackson et al., 1993; Wansink et al., 1993). After a 1-h pulse of BrUTP, newly transcribed RNA occurs in a pattern overlapping that of pol II on the lampbrush loops (Figure 2, A and B). That is, a more or less uniform line of stain occurs down the middle of each loop, and the line is remarkably similar from loop to loop. Such a pattern is consistent with earlier electron micrographs by Miller and colleagues (Miller et al., 1970; Miller and Hamkalo, 1972; Beyer et al., 1979), which show closely spaced RNA transcripts extending laterally from the DNA axis of the loop. In a relatively brief pulse, each pol II complex will move a short distance along the axis, and the 3′ end of all the nascent transcripts will be increased in length by a constant amount, regardless of their initial length. Thus, newly incorporated BrU will be adjacent to the pol II and uniform in amount along the length of a transcription unit.
BrU incorporation is weak but detectable in the extrachromosomal nucleoli (Figure 2A). By contrast, when 3H-labeled nucleotides are used as precursors, nucleoli are as intensely labeled as the chromosomes (Macgregor, 1967; Schultz et al., 1981), suggesting that in the oocyte, as in cultured mammalian cells, pol I uses BrUTP somewhat inefficiently (Jackson et al., 1993; Wansink et al., 1993). Of particular importance is the complete absence of BrUTP incorporation in the B-snurposomes and Cajal bodies, although both structures contain easily detectable concentrations of pol II, as discussed in the next section.
Earlier studies showed that the matrix of lampbrush chromosome loops stains strongly with antibodies against splicing snRNPs and is positive after in situ hybridization for all five splicing snRNAs (Wu et al., 1991). Figure 2C shows the staining pattern with mAb Y12, which recognizes the Sm epitope common to several snRNP proteins (Lerner et al., 1981). Similarly, lampbrush loops are stained by antibodies such as mAb SC35 (Fu and Maniatis, 1990) and mAb 104, which recognize members of the SR group of non-snRNP essential splicing factors (Roth et al., 1990; Wu et al., 1991). Last, loops stain with antibodies against the RNA 3′-end processing factors CstF77 (Takagaki and Manley, 1994) and CPSF100 (Jenny et al., 1994), as shown in Figure 2D for CstF77. An important feature of the staining pattern with all these antibodies is that transcription units are stained along their entire length. This pattern is consistent with a model in which the various processing factors go onto the chromatin coincident with the polymerase and subsequently move with the nascent transcripts along the template.
Snurposomes
In our earlier immunofluorescent studies of the GV, we found splicing factors not only in the matrix of lampbrush chromosome loops but also in numerous extrachromosomal granules. Most of these granules have diameters of ∼1–4 μm and stain strongly with mAb Y12 against the Sm epitope and mAb K121 against the trimethylguanosine (TMG) cap of splicing snRNAs. Moreover, they are positive after in situ hybridization for all five splicing snRNAs, U1, U2, U4, U5, and U6 (Figure 3). Because they contain high concentrations of snRNPs (pronounced snurps in laboratory jargon), we called these bodies B-snurposomes (Wu et al., 1991). (Less abundant particles, called A-snurposomes, contain only U1 snRNA and are found in newt but not Xenopus GVs. The main body of the sphere or Cajal body was named the C-snurposome, a term we no longer use.) As later studies showed, the term B-snurposome is misleading, because these bodies also contain other important proteins of the transcription and processing machinery.
Figure 3.
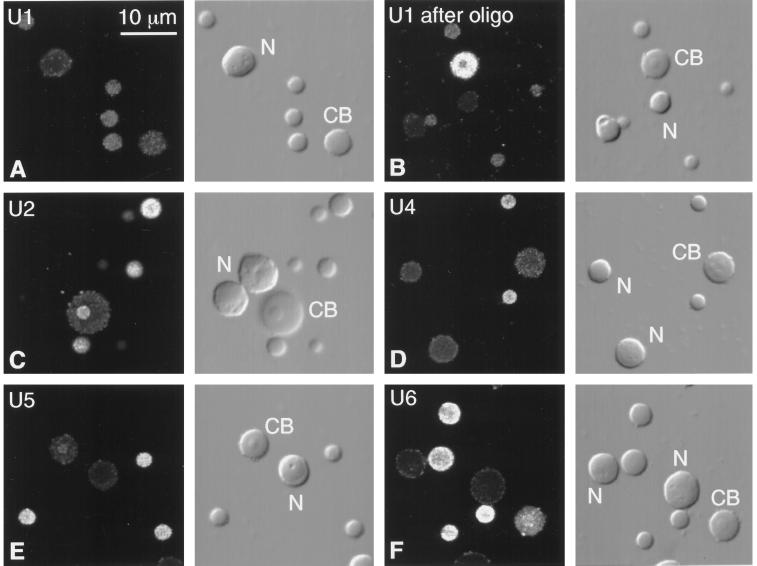
Fluorescent in situ hybridization of splicing snRNAs (U1, U2, U4, U5, and U6) in Cajal bodies, B-snurposomes, and nucleoli from spread GV preparations of Xenopus. Each pair of panels shows the fluorescent image on the left and the corresponding DIC image on the right. CB, Cajal body; N, nucleolus. (A) U1 is weakly detectable in B-snurposomes, but the Cajal body and nucleoli are barely above the background level of unhybridized controls. (B) Truncated U1 accumulates in Cajal bodies of oocytes treated with a deoxyoligonucleotide that removes the cap and first 20 nucleotides from the 5′ end. B-snurposomes and nucleoli are similar to those in untreated oocytes. (C) U2 is strong in B-snurposomes and easily detectable in the matrix of the Cajal body. (D) U4 is strong in B-snurposomes, but the Cajal body and nucleoli are at background level. (E) U5 is strong in B-snurposomes and detectably above background in the matrix of the Cajal bodies. (F) U6 is strong in B-snurposomes and gives the strongest reaction of all the splicing snRNAs in the matrix of the Cajal bodies.
The first non-snRNP components to be recognized in B-snurposomes were the SR proteins. Both mAb SC35 (Figure 2E) and mAb 104 stain B-snurposomes strongly. In fact, mAb 104 was described as an antibody that reacted with a phosphorylated epitope in B-snurposomes and lampbrush chromosome loops before it was realized that it defined the family of SR proteins (Roth et al., 1990, 1991; Zahler et al., 1992).
B-snurposomes are stained intensely by mAb H5, which reacts with the CTD of pol II when serine 2 of the seven-amino-acid repeat, YSPTSPS, is phosphorylated (Patturajan et al., 1998; Figure 4B). By contrast, mAb H14, which recognizes the CTD when serine 5 is phosphorylated, gives a very weak reaction, only slightly above the background level of staining in nucleoli (Figure 4C). There is no evidence that B-snurposomes contain DNA, and they do not label in short-term experiments with either [3H-]NTPs (Callan and Gall, 1991) or BrUTP (Figure 2, A and B). Therefore, the pol II in B-snurposomes is not transcriptionally active. A similar conclusion was reached by Bregman et al. (1995) regarding the clearly demonstrable pol II in the speckled regions of cultured Madin–Darby canine kidney cells.
Figure 4.
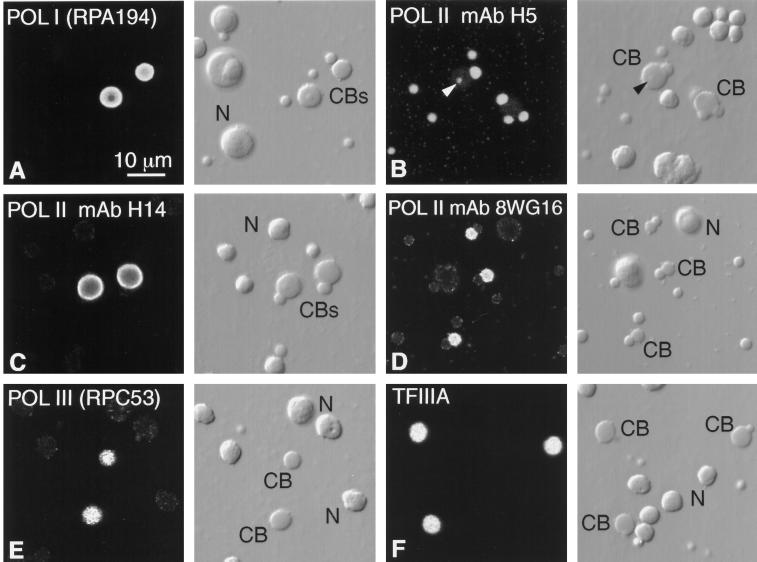
Detection of RNA polymerases and TFIIIA by immunofluorescent staining. Each pair of panels shows the fluorescent image on the left and the corresponding DIC image on the right. CB, Cajal body; N, nucleolus. (A) Stain is brilliant in the matrix of Cajal bodies with a polyclonal antibody against the largest subunit of pol I, RPA194. For unknown reasons the nucleoli are very much weaker, although this antibody stains nucleoli well in HeLa cells. (B) Pol II stains strongly in B-snurposomes with mAb H5, which detects the CTD when serine 2 is phosphorylated (Patturajan et al., 1998), whereas the matrix of the Cajal bodies is only weakly stained. The arrowhead points to stained B-like inclusion in one Cajal body. (C) Pol II stains strongly in the matrix of Cajal bodies with mAb H14, which detects the CTD when serine 5 is phosphorylated (Patturajan et al., 1998), whereas the B-snurposomes are essentially unstained. (D) Pol II stains strongly in the matrix of Cajal bodies with mAb 8WG16, which recognizes the nonphosphorylated CTD (Patturajan et al., 1998). (E) Pol III stains strongly in the matrix of the Cajal bodies with a polyclonal antibody against the 53-kDa subunit, RPC53. (F) Pol III transcription factor TFIIIA is readily demonstrable in Cajal bodies with a polyclonal antibody.
We have used antibodies to examine the distribution of two factors involved in the release of transcripts from the chromatin template: the 100-kDa subunit of the cleavage and polyadenylation specificity factor, CPSF100 (Jenny et al., 1994), and the 77-kDa subunit of the cleavage stimulation factor, CstF77 (Takagaki and Manley, 1994). Affinity-purified antipeptide antibodies against each of these proteins gave a positive reaction in B-snurposomes (Figure 5, C and D) and in the loops of the lampbrush chromosomes (Figure 2D), as did an independently produced polyclonal serum against CPSF100. Each of these antibodies gave an even stronger reaction in the matrix of the Cajal bodies, as discussed in a later paragraph.
Figure 5.
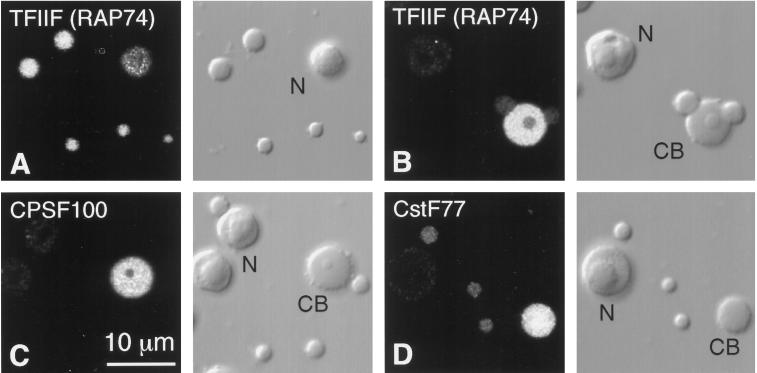
Each pair of panels shows the fluorescent image on the left and the corresponding DIC image on the right. CB, Cajal body; N, nucleolus. (A) Five B-snurposomes and a nucleolus stained with an antipeptide antibody against TFIIF (RAP74). (B) A Cajal body from the same preparation as A, printed at lower contrast. The Cajal body stains much more intensely than its associated B-snurposomes. (C) A Cajal body, nucleoli, and B-snurposomes stained with an antipeptide antibody against the 100-kDa subunit of the cleavage/polyadenylation specificity factor CPSF. The Cajal body stains intensely. (D) A Cajal body, three B-snurposomes, and a nucleolus stained with an antipeptide antibody against the 77-kDa subunit of the cleavage stimulation factor CstF. The Cajal body is the most intensely stained structure.
We have begun to examine the distribution of basal transcription factors using antibodies against rat and human proteins. We report here that a peptide antibody against TFIIF(RAP74) gives strong staining of B-snurposomes and the matrix of Cajal bodies (Figure 5, A and B).
In summary, B-snurposomes contain phosphorylated but inactive pol II, TFIIF(RAP74), all five splicing snRNPs, SR proteins, and at least two cleavage and polyadenylation factors (CstF77 and CPSF100).
Fine Structure of B-Snurposomes
Electron micrographs provide an idea of the type of complex(es) in which these pol II transcription and processing components exist. Earlier studies of thin sections of intact GVs showed that B-snurposomes contain many thousands of particles with diameters in the range of 20–30 nm (Callan and Gall, 1991; Gall et al., 1995). More recently we have examined sections of flat-embedded GV spreads, which give an even clearer picture of the constituent particles (Figure 6, A–C and E). The general appearance is of a uniform population of 20- to 30-nm particles. Further study of isolated whole particles will be necessary before their precise size and shape can be determined.
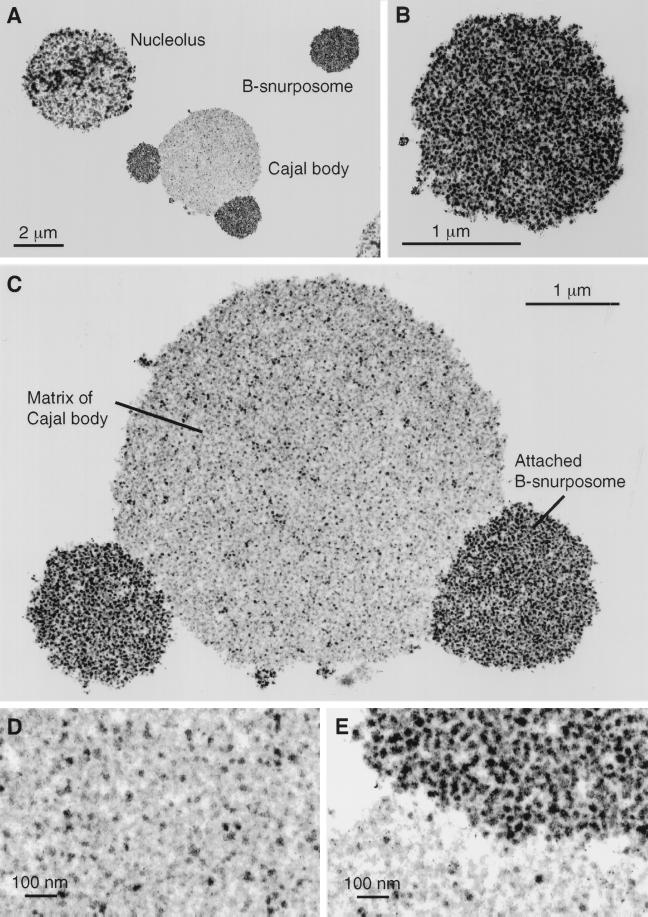
Figure 6.
Electron micrographs of thin sections through nuclear organelles, from a spread preparation of a Xenopus GV fixed in 4% glutaraldehyde and contrasted with OsO4, uranyl acetate, and lead citrate. (A) A nucleolus, a B-snurposome, and a Cajal body with two attached B-snurposomes. (B) A single B-snurposome composed of a relatively uniform aggregation of high-contrast 20- to 30-nm particles, which we call pol II transcriptosomes. (C) Cajal body from A at higher magnification showing that the matrix is heterogeneous in composition. The attached B-snurposomes are identical in appearance to free B-snurposomes. (D) At higher magnification the Cajal body matrix displays scattered 20- to 30-nm particles that look like pol II transcriptosomes, interspersed with numerous larger particles of lower contrast. (E) Boundary between the Cajal body matrix (below) and an attached B-snurposome (above) showing similar high-contrast particles in both compartments.
We postulate that each of these particles contains all of the pol II transcription and processing components detected in B-snurposomes by immunostaining and in situ hybridization. To emphasize their composition, we will borrow a term previously used in a different context (Halle and Meisterernst, 1996), and call these particles transcriptosomes or, more specifically, pol II transcriptosomes. The implication of the term is that many or most of the components required to produce an RNA transcript occur in the particle. An attractive hypothesis is that each transcriptosome represents a complex of pol II with its general transcription factors as well as splicing, cleavage, and polyadenylation machinery ready for transport to the chromosomes. Transcriptosomes might also represent a stored form of pol II transcription and processing factors that will be delivered to the egg cytoplasm at the time of GV breakdown. These two roles are not mutually incompatible.
Cajal Bodies
Our overall hypothesis is that Cajal bodies are the sites of assembly for pol I, pol II, and pol III transcription and processing complexes. We will first present evidence that pol II transcriptosomes might be assembled in the Cajal bodies. The evidence is essentially of three kinds: morphological, molecular, and kinetic.
1) Morphologically, Cajal bodies usually have B-snurposomes on their surface or embedded within their matrix (Figures 2E, 3C, and 6, A, C, and E). Hence, there are transcriptosomes on and in Cajal bodies.
2) Cajal bodies contain many if not all the same molecular components as B-snurposomes. (They also contain components not found in B-snurposomes.)
3) When components shared by Cajal bodies and B-snurposomes enter the GV, they appear first in the Cajal bodies and only later in the B-snurposomes and chromosomes.
Morphology of Cajal Bodies
In earlier publications we described the intimate association between Cajal bodies and B-snurposomes (Callan and Gall, 1991; Wu et al., 1991, 1994; Gall et al., 1995). In stage IV–V Xenopus oocytes, typical Cajal bodies have one or more B-snurposomes firmly embedded on their surface and one or more smaller B-snurposomes within their matrix (Figures 2E, 3C, and 6, A, C, and E). The numbers and sizes of the attached and included B-snurposomes are extremely variable. Cajal bodies from smaller oocytes tend to have more associated B-snurposomes, whereas those from fully mature oocytes often lack them entirely (Figures 3A, 4E, and 5D). In the newt Notophthalmus some giant Cajal bodies have diameters of 15–20 μm; they may have several dozen full-sized B-snurposomes on their surface and hundreds of smaller B-like inclusions (see Wu et al., 1991, their Figure 4A).
Electron micrographs of sectioned Cajal bodies show that the attached and included B-snurposomes are identical in structure to free B-snurposomes; that is, they consist of tightly packed 20- to 30-nm pol II transcriptosomes (Figure 6, A, C, and E). In addition, smaller clusters of what appear to be pol II transcriptosomes as well as single 20- to 30-nm particles occur throughout the matrix (Figure 6, C–E). If the free particles and smaller clusters represent scattered pol II transcriptosomes, the matrix of the Cajal body might be expected to contain molecular components found in B-snurposomes. The concentration of a given component might vary, depending on whether it is found only in the particles or in the particles and elsewhere in the matrix.
Composition of the Cajal Body Matrix
In discussing the composition of Cajal bodies, it is necessary to make a somewhat arbitrary distinction between the matrix and the included B-snurposomes. Because B-snurposomes do not contain the Cajal body marker p80-coilin (Andrade et al., 1991), the matrix can be defined as that part of the Cajal body that stains with antibodies against p80-coilin (Figure 2E). This definition is easy to apply when the Cajal body has no inclusions, or only a few large ones but becomes difficult when there are many small inclusions.
Every antibody or fluorescent in situ hybridization probe that gives a positive reaction in B-snurposomes also stains the matrix of the Cajal bodies, even when there are no inclusions resolvable by light microscopy. Signals from some probes are significantly weaker in the Cajal body matrix than in the attached and included B-snurposomes. In this category are antisense probes against all five of the splicing snRNAs (Figure 3) and mAb anti-SC35 against SR proteins (Figure 2E). A reasonable assumption is that these probes detect scattered transcriptosomes or unresolvable clusters of transcriptosomes in the matrix.
Because splicing snRNAs are at a lower concentration in the Cajal body matrix than in B-snurposomes, one would expect mAb Y12 and mAb K121 (against the Sm proteins and TMG, respectively) to stain the matrix weakly. Just the opposite is true, but in this case the explanation is known. The Cajal body matrix contains a high concentration of the U7 snRNP, not found in B-snurposomes (Wu and Gall, 1993; Wu et al., 1996). U7 snRNA has a TMG cap and is associated with the same Sm proteins as the splicing snRNAs (Birnstiel and Schaufele, 1988; Smith et al., 1991). When U7 in the GV is destroyed by injecting an antisense deoxyoligonucleotide complementary to its 5′ end, the TMG staining of the Cajal body matrix drops to ∼10% of its original value, well below that of the associated B-snurposomes (Bellini and Gall, 1998).
Several other antibodies that stain B-snurposomes give stronger reactions in the Cajal body matrix. Among these are anti-CstF77, anti-CPSF100, and anti-TFIIF(RAP73) (Figure 5). The relatively strong staining intensities suggest that these proteins may not be limited to pol II transcriptosomes in the Cajal bodies.
Pol II is a special case. mAb H14 gives an intense reaction in the matrix of Cajal bodies but negligible stain in B-snurposomes, just the opposite of mAb H5 (Figure 4, B and C). One possibility is that pol II transcriptosomes in the matrix contain pol II that is phosphorylated on serine 5. Alternatively, the weak staining of the matrix by mAb H5 may reflect the low abundance of pol II transcriptosomes in this compartment, and H14-reactive pol II may exist elsewhere in the matrix, that is, not in the pol II transcriptosomes. A third antibody that reacts with pol II is mAb 8WG16, which recognizes primarily the unphosphorylated CTD (Patturajan et al., 1998). Like mAb H14, this antibody stains the matrix of Cajal bodies but does not stain B-snurposomes (Figure 4D).
Uptake of Cajal Body Components
If pol II transcriptosomes are assembled in the Cajal bodies, we might expect their component molecules to pass through the Cajal bodies before reaching the B-snurposomes or the chromosomes. Although only a few such molecules have been examined, their kinetics are consistent with such a pathway. These molecules include three snRNAs (U1, U2, and U5), one snRNP protein (the U1-specific snRNP C protein), one cleavage and polyadenylation factor (CstF77), the elongation factor TFIIS, and a subunit of the core polymerase (RPB9).
Capped, fluorescein-labeled U1, U2, and U5 snRNA were transcribed in vitro and injected into the cytoplasm of Xenopus oocytes. There the transcripts associate with Sm proteins, the cap is hypermethylated, and the assembled snRNP particle is imported into the GV (Mattaj, 1988). Spread preparations of GV contents were made at various times after injection and examined by fluorescence microscopy. In each case, fluorescence was first detectable in the matrix of the Cajal bodies, despite the relatively low concentration of snRNAs in this compartment (Figure 7, A and C). Only later was fluorescence seen in the B-snurposomes and the loops of the lampbrush chromosomes (Figure 7B). The intensity of stain remained highest in the Cajal bodies throughout the duration of the experiment (∼24 h). The sustained labeling of the Cajal bodies could result from continued import of fluorescent snRNA from the cytoplasm or from recycling within the nucleus. Detailed kinetic experiments will have to be performed to determine the precise intranuclear movement of the snRNAs, but these data are consistent with the passage of splicing snRNPs through the Cajal bodies on their way to the B-snurposomes and chromosomes.
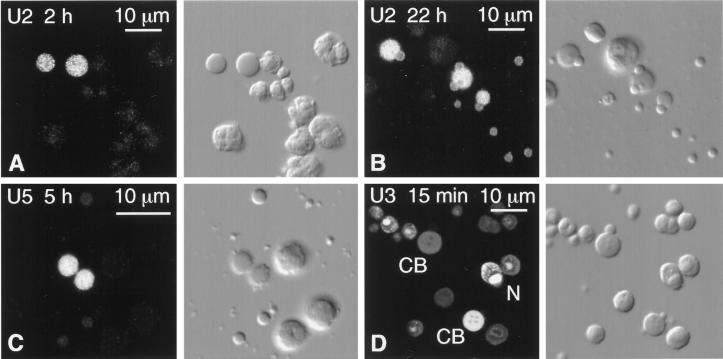
Figure 7.
Spread GV contents from oocytes injected with capped, fluorescein-labeled RNAs. The fluorescent signal was enhanced with goat anti-fluorescein labeled with Alexa 488. Each pair of panels shows the fluorescent image on the left and the corresponding DIC image on the right. CB, Cajal body; N, nucleolus. (A) Two hours after cytoplasmic injection of U2 snRNA, only the matrix of the Cajal bodies is labeled. (B) Twenty-two hours after injection of U2 snRNA, the matrix of the Cajal bodies is still labeled, but now the B-snurposomes are also labeled. (C) Five hours after nuclear injection of U5 snRNA, only the matrix of the Cajal bodies is significantly labeled. (D) Fifteen minutes after nuclear injection of U3 snoRNA, the matrix of Cajal bodies and the dense fibrillar region of the nucleoli are labeled. Individual Cajal bodies and nucleoli vary greatly in intensity.
We have not yet examined targeting of mutated versions of the U1, U2, and U5 snRNAs. It should be possible to define sequences in these RNAs that modify their intranuclear movements, as has been done for U7 snRNA (Wu et al., 1996) and several snoRNAs (Lange et al., 1998a–c; Samarsky et al., 1998; Narayanan et al., 1999). A suggestion that the 5′ end of U1 snRNA is required for exit from the Cajal bodies is provided by an observation on oocytes treated with an antisense oligodeoxynucleotide against U1. Earlier experiments showed that an oligo complementary to nucleotides 1–20 of Xenopus U1 caused loss of the 5′ end within minutes, presumably by RNase H digestion, leaving a truncated molecule that was stable for at least 48 h (Pan and Prives, 1988; Tsvetkov et al., 1992). In situ hybridization of GV spreads from treated oocytes shows a striking accumulation of truncated U1 snRNA in the Cajal bodies (Figure 3B), suggesting that the normal pathway of U1 through the Cajal body has been interrupted.
The intranuclear targeting of CstF77 was demonstrated by injecting HA-tagged transcripts into the cytoplasm of Xenopus oocytes and following the appearance of newly translated protein in the GV at various times after injection. HA-tagged protein appeared rapidly in the Cajal bodies and by 24 h showed a distribution similar to that of the endogenous protein (Figure 8). A similar pattern of labeling was previously seen with the U1-specific snRNP C protein (Jantsch and Gall, 1992). Recent experiments show that the transcription elongation factor TFIIS and a subunit of the core polymerase (RPB9) are targeted rapidly and specifically to oocyte Cajal bodies in the newt Triturus vulgaris (Morgan, personal communication).
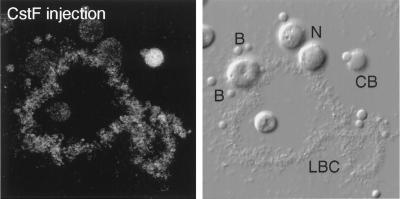
Figure 8.
Distribution of HA-tagged CstF77 in a GV spread, detected with an antibody against the HA tag. The fluorescent image is on the left; the corresponding DIC image is on the right. CB, Cajal body; N, nucleolus; B, B-snurposomes. The oocyte was injected 24 h previously with a transcript synthesized in vitro from a full-length cDNA clone of CstF77. The Cajal body is the most intensely stained structure, but label is detectable in the chromosome and the B-snurposomes.
Pol I Transcription and Processing Components in Cajal Bodies
It is well known that pol I occurs in the fibrillar centers of nucleoli (reviewed in Paule, 1998). Raška et al. (1991) reported that a pol I autoimmune serum did not stain Cajal bodies in HeLa nuclei, and other studies dealing with pol I fail to mention an association with Cajal bodies (Matera et al., 1994; Jordan et al., 1996). Nevertheless, polyclonal sera specific for the two largest subunits of rat pol I, RPA194 and RPA127 (Hannan et al., 1998), gave a positive reaction in the matrix of oocyte Cajal bodies, the serum against RPA194 being particularly strong (Figure 4A). Unexpectedly, both antibodies stained nucleoli only weakly, although rRNA transcription is active in nucleoli from oocytes of the size used for these studies (Dumont stages IV–V).
Processing of pre-rRNA involves a large number of snoRNAs (reviewed in Maxwell and Fournier, 1995; Bachellerie and Cavaille, 1997; Smith and Steitz, 1997). U3, U8, and U14 snoRNAs are required for cleavage of the 45S precursor into the 18S, 28S, and 5.8S molecules found in the mature ribosome. Approximately 200 additional snoRNAs act as guides to direct the conversion of specific uridines to pseudouridine and the insertion of 2′-O-methyl groups on specific riboses in the mature rRNA. Although both U3 and U8 have been reported as components of somatic Cajal bodies (Bauer et al., 1994; Jiménez-García et al., 1994), we have been unable to detect these molecules by in situ hybridization of Cajal bodies from the Xenopus GV, either with tritiated (Gall et al., 1995) or fluorescein-labeled probes (our unpublished experiments). Nevertheless, targeting of U3 and U8 to Cajal bodies has been seen by Narayanan et al. (1999) within minutes after fluorescein-labeled U3 or U8 was injected into the GV. During the first hour after injection, the intensity of label in the Cajal bodies decreased, whereas the intensity in the nucleoli continued to rise. We have repeated some of their experiments and have confirmed that Cajal bodies are rapidly labeled with either U3 or U8 probes (Figure 7D). Significantly, Narayanan et al. (1999) found that mutated U3 or U8 molecules that were not targeted to nucleoli remained in the Cajal bodies for several hours. In recent studies, Samarsky et al. (1998) saw targeting of U14 snoRNA to both Cajal bodies and nucleoli in monkey COS-1 cells, and Shaw et al. (1998) demonstrated a precursor to U14 snoRNA in Cajal bodies in maize. Thus, there is presumptive evidence that some snoRNAs involved in rRNA processing pass through the Cajal bodies on their way to the nucleoli.
Three important nucleolar proteins are also found in oocyte Cajal bodies: fibrillarin, Nopp140, and NO38(B23). Our original attempts to demonstrate these proteins in oocytes by immunofluorescence were unsuccessful, leading us to suggest that oocyte and somatic Cajal bodies differed in composition (Gall et al., 1995). Recently, however, good staining was obtained for each of these proteins after a change in fixation conditions (Figure 9, A–C). Another prominent nucleolar protein, nucleolin, is not detectable in oocyte Cajal bodies (Figure 9D). In earlier experiments we showed that myc-tagged NO38 was targeted to nucleoli and Cajal bodies in Notophthalmus oocytes (Peculis and Gall, 1992). When sequences were deleted from the carboxyl terminus, the truncated NO38 protein localized strongly in Cajal bodies but not in nucleoli. This result suggests that NO38 may normally pass through the Cajal body on its way to the nucleolus, and that truncation prevents the protein from leaving the Cajal body.
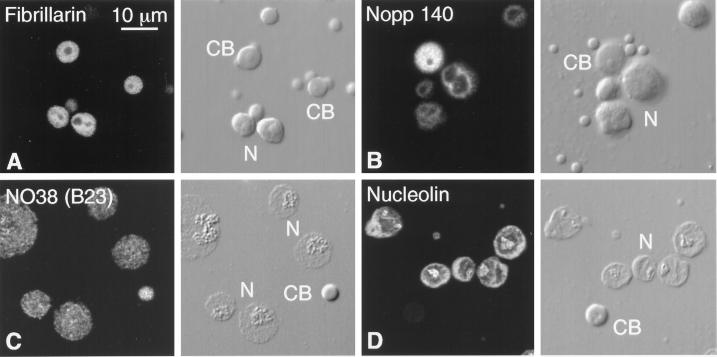
Figure 9.
Nucleolar proteins in the Cajal body. Each pair of panels shows the fluorescent image on the left and the corresponding DIC image on the right. CB, Cajal body; N, nucleolus. (A) Fibrillarin detected with mAb 17C12. The matrices of the Cajal bodies and the nucleoli are equally stained. (B) Nopp140 detected with mAb No114. The matrix of the Cajal body stains more intensely than the dense fibrillar zone of the nucleoli. (C) NO38 (B23) detected with mAb No185. This GV was isolated and spread in the absence of Mg2+. Under these conditions the nucleoli lose much of their granular zone, only the matrix of the Cajal bodies remains intact, and the B-snurposomes disappear. Such partially solubilized Cajal bodies retain their NO38, as shown here, as well as their fibrillarin and Nopp140. (D) Nucleolin detected with mAb P71A4, after Mg2+-free spreading. Nucleolin is readily detectable in the nucleoli but is absent from Cajal bodies.
The existence of pol I in Cajal bodies and the likelihood that some snoRNAs and some nucleolar proteins move through the Cajal bodies on their way to the nucleolus raise the interesting possibility that pol I and associated rRNA-processing components may be assembled in Cajal bodies as a unitary pol I transcriptosome. As already discussed, the matrix of Cajal bodies contains a scattering of 20- to 30-nm particles similar to the pol II transcriptosomes in the B-snurposomes. However, most of the matrix is composed of somewhat larger, less densely contrasted particles ∼40–50 nm in diameter (Figure 6, D and E). Some of these larger particles could be pol I transcriptosomes.
Pol III Transcription and Processing Components in Cajal Bodies
Little attention has been paid to the possibility that pol III and associated factors may occur in Cajal bodies. We examined the intranuclear staining pattern of antibodies against two unique subunits of human pol III, RPC62 and RPC53, and one subunit shared by pol I and pol III, RPC19 (Wang et al., 1997). These antibodies gave strong and specific Western blots with proteins of the appropriate size in GV extracts. In each case the Cajal bodies were strongly stained (Figure 4E). In addition, the antibody against RPC53 stained some 50–60 scattered sites on the lampbrush chromosomes, including 15 terminal granules that were previously identified as the loci of the oocyte-type 5S RNA genes (Callan et al., 1987). Because 5S RNA genes are transcribed by pol III, this staining pattern provides evidence for the specificity of the antibody. Many of the remaining stained regions could be tRNA loci, which, like the 5S RNA genes, are transcribed by pol III.
We also examined the distribution of TFIIIA, a protein that serves two related functions: first, as a specific transcription factor for the 5S RNA gene, and second, as part of the complex in which 5S RNA is stored (Engelke et al., 1980; Pelham and Brown, 1980). A polyclonal antibody against TFIIIA gave bright and highly specific staining of Cajal bodies (Figure 4F). Because 5S rRNA is synthesized primarily in very young oocytes (Dumont stages I and II), we thought that TFIIIA might be more prominent in Cajal bodies from early stages. However, the intensity of stain was approximately the same in GVs from small (0.4-mm-diameter) and large (1.3-mm-diameter) oocytes.
DISCUSSION
Molecular and cytological studies on the Xenopus GV suggest a model for the assembly of the RNA transcription and processing machinery of the nucleus in which the Cajal bodies and B-snurposomes play a central role (Figure 1). This model has three related but somewhat independent postulates:
1) the pol II transcription and processing machinery is preassembled as a unitary particle, for which the term pol II transcriptosome is suggested;
2) pol II transcriptosomes are assembled in the Cajal bodies and stored in the B-snurposomes before transport to the loops of the lampbrush chromosomes;
3) pol I and pol III transcriptosomes are similarly assembled in the Cajal bodies. Pol I transcriptosomes travel to the nucleoli, where they engage in transcription of the rRNA genes. Pol III transcriptosomes are targeted to a relatively small number of specific sites on the chromosomes, including the 5S rRNA and tRNA genes.
The assembly of the transcription machinery for both pol I and pol II at a single site provides a basis for understanding one of the more puzzling features of Cajal bodies: their clear relationship to the chromosomes and pre-mRNA processing on the one hand and to the nucleolus and pre-rRNA processing on the other (see discussions in Bohmann et al., 1995; Gall et al., 1995; Lamond and Earnshaw, 1998; Matera, 1999). Furthermore, this model predicts an unanticipated relationship between Cajal bodies and pol III transcription, which our data begin to demonstrate.
Pol II Transcriptosomes
The evidence for a unitary pol II transcriptosome comes from the composition of B-snurposomes and their fine structure. B-snurposomes contain phosphorylated but inactive pol II, TFIIF(RAP74), all five splicing snRNPs, SR proteins, and at least two cleavage and polyadenylation factors (CstF77 and CPSF100). Although this list contains only a small sampling of the factors known to be involved in transcription and processing, inclusion of both early- and late-acting factors as well as the polymerase itself is significant. Electron micrographs reveal that B-snurposomes consist of a relatively homogeneous population of 20- to 30-nm particles. We suggest that each particle contains polymerase II along with many or most of its associated transcription and processing components.
It has been known for many years that somatic nuclei contain clusters of similar particles, referred to in the electron microscopical literature as interchromatin granules (Monneron and Bernhard, 1969; Fakan and Puvion, 1980). Interchromatin granule clusters (ICGC) correspond to the speckled regions visible by light microscopy after immunofluorescent staining (reviewed in Spector, 1993). Like the B-snurposomes they contain pol II and a wide variety of transcription and processing factors. Recently, interchromatin granule clusters have been isolated as a biochemical fraction, and their molecular components have begun to be identified (Mintz et al., 1999). Included are the largest subunit of pol II and >30 previously identified proteins, many of which are known to be involved in pre-mRNA splicing.
The idea that the three RNA polymerases are preassembled with other proteins as a large “holoenzyme” complex before recruitment to promoters has received considerable support from recent biochemical studies in both yeast and mammalian systems (Greenblatt, 1997; Wang et al., 1997; Myer and Young, 1998; Parvin and Young, 1998; Roeder, 1998; Seither et al., 1998). In the case of pol II, the holoenzyme consists of the core pol II, itself a complex of 12 subunits, along with associated general transcription factors, the SRB–mediator complex, and additional proteins. In addition, the largest subunit of pol II can bind various RNA processing factors by means of its CTD, including those involved in capping, splicing, cleavage, and polyadenylation (reviewed in Greenleaf, 1993; Steinmetz, 1997; Bentley, 1999; Minvielle-Sebastia and Keller, 1999). It has been suggested that the CTD may regulate RNA processing by recruiting these components to the nascent transcripts as the polymerase moves along the template. Our data, which show that pol II is colocalized with several of these factors in the B-snurposomes and Cajal bodies, suggest that recruitment may begin well before the polymerase engages the template.
Polymerases in Cajal Bodies
A crucial feature of the model presented here is the occurrence of pol I, pol II, and pol III in Cajal bodies. Although in somatic nuclei pol II is primarily localized in the speckles and regions of active chromatin (Bregman et al., 1995; Grande et al., 1997; Kim et al., 1997), Schul et al. (1998) recently demonstrated a subset of pol II in Cajal bodies. Specifically, they showed that mAb 8WG16 against the unphosphorylated CTD labeled Cajal bodies in cultured human cells, whereas mAb H5, which detects the CTD when serine 2 is phosphorylated, did not. Both antibodies labeled numerous smaller foci throughout the nucleus. Similarly, 8WG16 stains the matrix of oocyte Cajal bodies, but H5 does not. In addition, mAb H14, which detects the CTD when serine 5 is phosphorylated, gives strong staining of the Cajal body matrix. Important additional evidence for pol II comes from unpublished experiments that show targeting of a myc-tagged subunit of pol II (RPB9) to oocyte Cajal bodies in the newt Triturus vulgaris (Morgan, personal communication).
Pol I is generally thought to be limited to nucleoli (Scheer and Hock, 1999), and pol III thought to be limited to separate, discrete sites within the nucleus (Pombo et al., 1999). Although neither has been reported from somatic Cajal bodies, we have seen strong reactions in the oocyte with five antibodies: two against the two largest subunits of pol I (RPA194 and RPA127), two against specific subunits of pol III (RPC62 and RPC53), and one against a subunit shared by pol I and pol III (RPC19). It is possible that the high rate of transcription and the large store of polymerases in the oocyte nucleus facilitate detection of pol I and pol III in the Cajal bodies, and that a closer study of somatic nuclei will reveal a comparable localization.
Although the existence of some transcription in somatic Cajal bodies is difficult to rule out because of their proximity to transcriptionally active regions, oocyte Cajal bodies clearly do not contain DNA and do not incorporate RNA precursors. Why then should they contain inactive forms of all three polymerases? A possible clue may lie in homologies among the enzymes. The two largest subunits of pol I, II, and III share sequence similarities with each other and with the two subunits of bacterial RNA polymerase. Furthermore, five subunits of pol I, II, and III are identical in yeast and are highly conserved in other eukaryotes (reviewed in Carles and Riva, 1998). An attractive hypothesis is that the three multicomponent polymerases share parts of the same assembly pathway in the Cajal body, perhaps based ultimately on a common evolutionary history.
Transcription and Processing Factors in Cajal Bodies
Many factors involved in RNA transcription and processing have been described from oocyte and somatic Cajal bodies. Figure 10 lists those so far detected in amphibian oocytes, and a similar list for somatic cells was recently published by Matera (1999). A particularly important analysis of somatic Cajal bodies has been carried out by Schul and colleagues (Schul et al., 1996, 1998, 1999; Schul, 1998). They have shown that various transcription and processing factors are localized in and around Cajal bodies but are not always completely coincident with them. They found that several proteins required for snRNA transcription (PTFγ, TBP, and a subset of pol II) associate with coiled bodies adjacent to the genes for U1 and U2 snRNAs. They postulate that factors are distributed from the Cajal body to the associated genes to facilitate and regulate expression of these genes. In a similar manner, the RNA 3′-processing factors CstF64 and CPSF100 occur in and around Cajal bodies in a region they refer to as cleavage bodies. Cleavage bodies show striking cell cycle changes, being colocalized with Cajal bodies in G1, adjacent to them in the S phase, and less defined or absent in G2. Furthermore, when transcription is inhibited, the processing factors are found tightly associated with Cajal bodies. Cleavage bodies and their associated Cajal bodies occur next to histone genes but do not overlap U1 and U2 snRNA genes. These studies all point to the importance of the Cajal bodies in supplying or modifying factors required for transcription.
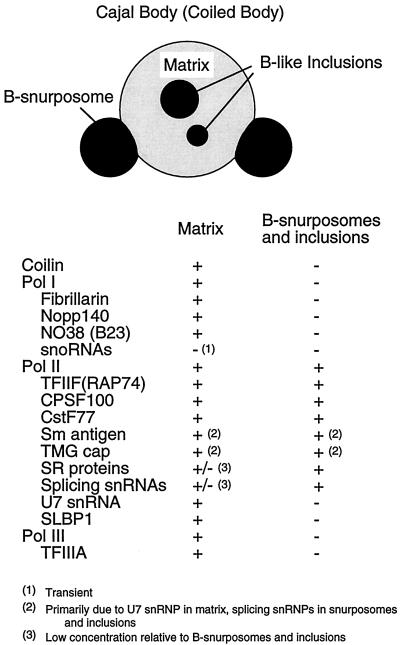
Figure 10.
Diagram of an oocyte Cajal body and a list of its known molecular components. The Cajal body consists of a spherical matrix, a variable number of B-snurposomes attached to its surface, and B-like inclusions.
Oocyte Cajal bodies also contain two pol II processing factors devoted specifically to 3′ cleavage of histone pre-mRNA; namely, the U7 snRNP (Wu et al., 1996) and the stem–loop binding protein SLBP1 (Abbott et al., 1999). U7 snRNA has also been demonstrated in the Cajal bodies of somatic nuclei (Frey and Matera, 1995).
Cajal bodies contain fibrillarin (Raška et al., 1991), Nopp140 (Meier and Blobel, 1992), NAP57 (Meier and Blobel, 1994), and NO38 (B23), proteins that are found primarily in nucleoli and are probably involved in processing and modifying pre-rRNA. Fibrillarin is associated with U3, U8, and U13 snoRNAs (Tyc and Steitz, 1989), and with many additional snoRNAs that serve as guides for 2′-O-methylation of pre-rRNA (reviewed by Smith and Steitz, 1997; Weinstein and Steitz, 1999). Although we do not detect snoRNAs in oocyte Cajal bodies by in situ hybridization, U3, U8, and U14 snoRNAs have been reported from somatic Cajal bodies (Bauer et al., 1994; Jiménez-García et al., 1994; Samarsky et al., 1998; Shaw et al., 1998), and injection experiments suggest that U3 and U8 transit the oocyte Cajal bodies on their way to the nucleoli (Narayanan et al., 1999).
The only specific pol III transcription factor so far identified in Cajal bodies is TFIIIA.
Targeting of Transcription and Processing Components to Cajal Bodies
If components of the transcriptional machinery are assembled into large complexes in the Cajal bodies and then distributed to other parts of the nucleus, one should be able to follow their movement by appropriate kinetic experiments. In injection experiments, some molecules should be targeted rapidly to the Cajal bodies, whereas other might appear there more slowly, depending on pool sizes and details of the assembly pathway.
We have shown that U1, U2, and U5 splicing snRNAs are initially targeted to the matrix of oocyte Cajal bodies, despite their low steady-state concentration in this compartment. They slowly appear in the B-snurposomes and chromosomes, which are sites of higher concentration (Figure 7, A–C). Furthermore, a truncated form of U1 accumulates in the matrix of Cajal bodies, suggesting that it is unable to exit properly (Figure 3B). The cleavage factor CstF77 (Figure 8) and the U1-specific snRNP C protein (Jantsch and Gall, 1992) are also targeted initially to oocyte Cajal bodies, as are U7 snRNA (Wu et al., 1996) and the stem–loop binding protein SLBP1 (Abbott et al., 1999), both of which are involved in histone pre-mRNA 3′ processing. The elongation factor TFIIS is likewise targeted rapidly and specifically to oocyte Cajal bodies of the newt Triturus vulgaris (Morgan, personal communication).
After injection into the Xenopus GV, U3 and U8 snoRNAs appear almost simultaneously in Cajal bodies and the fibrillar region of the nucleoli (Figure 7D), and mutant forms of the RNAs that fail to accumulate in the nucleoli are detained in the Cajal bodies (Narayanan et al., 1999). Evidence from cultured monkey cells (Samarsky et al., 1998) and maize (Shaw et al., 1998) suggests that U14 passes through Cajal bodies on the way to the nucleolus. The nucleolar protein NO38 (B23) is targeted to both Cajal bodies and nucleoli in Xenopus oocytes. A truncated form of the molecule is targeted to the Cajal bodies but not the nucleoli, suggesting that the protein normally transits through the Cajal bodies on the way to the nucleoli (Peculis and Gall, 1992).
In summary, a number of factors involved in processing pol I and pol II transcripts and one subunit of the core pol II itself are targeted initially to Cajal bodies. The model presented here suggests that the same should be true for a large number of additional factors.
Notes on Terminology
Cajal Body
In 1903 the Spanish neurobiologist Santiago Ramón y Cajal described a small spherical body in the nuclei of mammalian neurons (Cajal, 1903). Like the larger nucleolus with which it was sometimes associated, Cajal’s accessory body became blackened when tissues were “stained” with silver nitrate to outline the delicate neurofilaments. In the ensuing years accessory bodies were rediscovered several times and given different names. In amphibian oocytes they were described as spheres or sphere organelles (Gall, 1954; Callan and Lloyd, 1960; Callan, 1986). In insects they were called Binnenkörper (Bier et al., 1967). In thin sections of mammalian cells their heterogeneous morphology suggested a coiled-up rope, and the name coiled body was applied to them (Monneron and Bernhard, 1969). Coiled body has been used in most recent literature dealing with cultured mammalian cells. In hopes of standardizing the terminology, we suggest that these structures be renamed Cajal bodies in honor of the first investigator to describe them. This name change is particularly appropriate for another reason. In 1906 Ramón y Cajal and Camillo Golgi shared the Nobel Prize for their observations on the architecture of the nervous system. Incidental to his work on neurons, Golgi described the cytoplasmic organelle that now bears his name. An appropriate symmetry would be achieved by honoring Cajal’s discovery of an equally important nuclear organelle.
Transcriptosome
As defined here, pol II transcriptosome is a morphological term to describe the 20- to 30-nm particles that make up the bulk of the B-snurposomes in the amphibian GV. Its composition is inferred from immunostaining and in situ hybridization on the assumption that there is only one type of particle in the B-snurposomes, and that each transcriptosome contains at least one molecule of every component in the B-snurposomes. Pol II transcriptosomes are probably identical to interchromatin granules. The name is intended to imply that the particle participates in many or all aspects of forming a pol II transcript (transcription, splicing, cleavage, and polyadenylation). The relationship of the pol II transcriptosome to the pol II holoenzyme remains to be determined. The term transcriptosome was first used by Halle and Meisterernst (1996) to describe an “omnipotent transcription complex” that might be part of an immobile transcription factory.
ACKNOWLEDGMENTS
We thank the following for antibodies: D. Bentley (anti-TFIIF, CstF77, and CPSF100), C. Dreyer (mAb P7 1A4), X.-D. Fu and T. Maniatis (mAb SC35), W. Keller (serum 2685), S. Munro (mAb 9E10), K.M. Pollard (mAb 17C12), R.Roeder (anti-RPC19, RPC53, RPC62, and TFIIIA), L. Rothblum (anti-RPA194 and RPA127), M. Schmidt-Zachmann (mAbs No114 and No185), J. Steitz (mAb Y12), R. Tuma and M. Roth (mAb H1), S. Warren (mAbs H5 and H14), and Z. Wu (serum C236). A clone of CstF77 was kindly provided by D. Bentley. We thank Michael Sepanski for performing the electron microscopy. We are especially indebted to Robert Roeder for helpful discussion about polymerases and transcription factors. This work was supported by research grant GM-33397 from the National Institute of General Medical Sciences. J.G.G. is American Cancer Society Professor of Developmental Genetics.
REFERENCES
- Abbott J, Marzluff WF, Gall JG. The stem loop binding protein (SLBP1) is present in coiled bodies of the Xenopus germinal vesicle. Mol Biol Cell. 1999;10:487–499. doi: 10.1091/mbc.10.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade LEC, Chan EKL, Raška I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie J-P, Cavaille J. Guiding ribose methylation of rRNA. Trends Biochem Sci. 1997;22:257–261. doi: 10.1016/s0968-0004(97)01057-8. [DOI] [PubMed] [Google Scholar]
- Bauer DW, Murphy C, Wu Z, Wu C-H H, Gall JG. In vitro assembly of coiled bodies in Xenopus egg extract. Mol Biol Cell. 1994;5:633–644. doi: 10.1091/mbc.5.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini M, Gall JG. Coilin can form a complex with the U7 small nuclear ribonucleoprotein. Mol Biol Cell. 1998;9:2987–3001. doi: 10.1091/mbc.9.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr Opin Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- Beyer AL, McKnight SL, Miller OL. Transcriptional units in eukaryotic chromosomes. Mol Genet. 1979;3:117–175. [Google Scholar]
- Bier K, Kunz W, Ribbert D. Struktur und Funktion der Oocytenchromosomen und Nukleolen sowie der Extra-DNS während der Oogenese panoistischer und meroistischer Insekten. Chromosoma. 1967;23:214–254. doi: 10.1007/BF00331114. [DOI] [PubMed] [Google Scholar]
- Birnstiel ML, Schaufele FJ. Structure and function of minor snRNPs. In: Birnstiel ML, editor. Structure and Function of Major and Minor Small Nuclear Ribonucleoproteins. Berlin: Springer-Verlag; 1988. pp. 155–182. [Google Scholar]
- Bohmann K, Ferreira J, Santama N, Weis K, Lamond AI. Molecular analysis of the coiled body. J Cell Sci Suppl. 1995;19:107–113. doi: 10.1242/jcs.1995.supplement_19.16. [DOI] [PubMed] [Google Scholar]
- Bregman DB, Du L, van der Zee S, Warren SL. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Dawid IB. Specific gene amplification in oocytes. Science. 1968;160:272–280. doi: 10.1126/science.160.3825.272. [DOI] [PubMed] [Google Scholar]
- Bucci S, Nardi I, Mancino G, Fiume L. Incorporation of tritiated uridine in nuclei of Triturus oocytes treated with α-amanitin. Exp Cell Res. 1971;69:462–465. doi: 10.1016/0014-4827(71)90255-2. [DOI] [PubMed] [Google Scholar]
- Busch H, Smetana K. The Nucleolus. New York: Academic Press; 1970. [Google Scholar]
- Cajal S, Ramón y. Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab Lab Invest Biol. 1903;2:129–221. [Google Scholar]
- Callan HG. Molecular Biology, Biochemistry, and Biophysics. Vol. 36. Berlin: Springer-Verlag; 1986. Lampbrush chromosomes; pp. 1–254. [PubMed] [Google Scholar]
- Callan HG, Gall JG. Association of RNA with the B and C snurposomes of Xenopus oocyte nuclei. Chromosoma. 1991;101:69–82. doi: 10.1007/BF00357056. [DOI] [PubMed] [Google Scholar]
- Callan HG, Gall JG, Berg CA. The lampbrush chromosomes of Xenopus laevis: preparation, identification, and distribution of 5S DNA sequences. Chromosoma. 1987;95:236–250. doi: 10.1007/BF00294780. [DOI] [PubMed] [Google Scholar]
- Callan HG, Lloyd L. Lampbrush chromosomes of crested newts Triturus cristatus (Laurenti) Philos Trans R Soc Lond B Biol Sci. 1960;243:135–219. [Google Scholar]
- Carles C, Riva M. Yeast RNA polymerase I subunits and genes. In: Paule MR, editor. Transcription of rRNA Genes by Eukaryotic RNA Polymerase I. Berlin: Springer-Verlag; 1998. pp. 9–38. [Google Scholar]
- Davidson EH. Gene Activity in Early Development. 3rd ed. Orlando, FL: Academic Press; 1986. [Google Scholar]
- de Jong L, Grande MA, Mattern KA, Schul W, van Driel R. Nuclear domains involved in RNA synthesis, RNA processing and replication. Crit Rev Eukaryot Gene Expr. 1996;6:215–246. doi: 10.1615/critreveukargeneexpr.v6.i2-3.60. [DOI] [PubMed] [Google Scholar]
- Dumont JN. Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory maintained animals. J Morphol. 1972;136:153–180. doi: 10.1002/jmor.1051360203. [DOI] [PubMed] [Google Scholar]
- Engelke DR, Ng S-Y, Shastry BS, Roeder RG. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980;19:717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S, Puvion E. The ultrastructural visualization of nucleolar and extranucleolar RNA synthesis and distribution. Int Rev Cytol. 1980;65:255–299. doi: 10.1016/s0074-7696(08)61962-2. [DOI] [PubMed] [Google Scholar]
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Gall JG. Lampbrush chromosomes from oocyte nuclei of the newt. J Morphol. 1954;94:283–352. [Google Scholar]
- Gall JG. Chromosomal differentiation. In: McElroy WD, Glass B, editors. A Symposium on the Chemical Basis of Development. Baltimore: Johns Hopkins Press; 1958. pp. 103–135. [Google Scholar]
- Gall JG. Differential synthesis of the genes for rRNA during amphibian oogenesis. Proc Natl Acad Sci USA. 1968;60:553–560. doi: 10.1073/pnas.60.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. Spread preparation of Xenopus germinal vesicle contents. In: Spector D, Goldman R, Leinwand L, editors. Cells: A Laboratory Manual. Vol. 1. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. pp. 52.1–52.4. [Google Scholar]
- Gall JG, Callan HG. H3 Uridine incorporation in lampbrush chromosomes. Proc Natl Acad Sci USA. 1962;48:562–570. doi: 10.1073/pnas.48.4.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Callan HG, Wu Z, Murphy C. Lampbrush chromosomes. In: Kay BK, Peng HB, editors. Xenopus laevis: Practical Uses in Cell and Molecular Biology. Vol. 36. San Diego: Academic Press; 1991. pp. 149–166. [PubMed] [Google Scholar]
- Gall JG, Tsvetkov A, Wu Z, Murphy C. Is the sphere organelle/coiled body a universal nuclear component? Dev Genet. 1995;16:25–35. doi: 10.1002/dvg.1020160107. [DOI] [PubMed] [Google Scholar]
- Grande MA, van der Kraan I, de Jong L, van Driel R. Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J Cell Sci. 1997;110:1781–1791. doi: 10.1242/jcs.110.15.1781. [DOI] [PubMed] [Google Scholar]
- Greenblatt J. RNA polymerase II holoenzyme and transcriptional regulation. Curr Opin Cell Biol. 1997;9:310–319. doi: 10.1016/s0955-0674(97)80002-6. [DOI] [PubMed] [Google Scholar]
- Greenleaf A. Positive patches and negative noodles: linking RNA processing to transcription? Trends Biochem Sci. 1993;18:117–119. doi: 10.1016/0968-0004(93)90016-g. [DOI] [PubMed] [Google Scholar]
- Halle J-P, Meisterernst M. Gene expression: increasing evidence for a transcriptosome. Trends Genet. 1996;12:161–163. doi: 10.1016/0168-9525(96)30035-8. [DOI] [PubMed] [Google Scholar]
- Hannan RD, Hempel WM, Cavanaugh A, Arino T, Dimitrov SI, Moss T, Rothblum L. Affinity purification of mammalian RNA polymerase I. J Biol Chem. 1998;273:1257–1267. doi: 10.1074/jbc.273.2.1257. [DOI] [PubMed] [Google Scholar]
- Izawa M, Allfrey VG, Mirsky AL. The relationship between RNA synthesis and loop structure in lampbrush chromosomes. Proc Natl Acad Sci USA. 1963;49:544–551. doi: 10.1073/pnas.49.4.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch MF, Gall JG. Assembly and localization of the U1-specific snRNP C protein in the amphibian oocyte. J Cell Biol. 1992;119:1037–1046. doi: 10.1083/jcb.119.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Hauri H-P, Keller W. Characterization of cleavage and polyadenylation specificity factor and cloning of its 100-kilodalton subunit. Mol Cell Biol. 1994;14:8183–8190. doi: 10.1128/mcb.14.12.8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-García LF, Segura-Valdez MdL, Ochs RL, Rothblum LI, Hannan R, Spector DL. Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-mRNA transcription after mitosis. Mol Biol Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P, Mannervik M, Tora L, Carmo-Fonseca M. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J Cell Biol. 1996;133:225–234. doi: 10.1083/jcb.133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay BK. Appendix B. Injection of ocytes and embryos. In: Kay BK, Peng HB, editors. Xenopus laevis: Practical Uses in Cell and Molecular Biology. Vol. 36. San Diego: Academic Press; 1991. pp. 663–669. [PubMed] [Google Scholar]
- Kazmaier M, Tebb G, Mattaj IW. Functional characterization of X. laevis U5 snRNA genes. EMBO J. 1987;6:3071–3078. doi: 10.1002/j.1460-2075.1987.tb02614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Du L, Bregman DB, Warren SL. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix J-C, Azzouz R, Boucher D, Abbadie C, Pyne CK, Charlemagne J. Monoclonal antibodies to lampbrush chromosome antigens of Pleurodeles waltlii. Chromosoma. 1985;92:69–80. doi: 10.1007/BF00327246. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lange TS, Borovjagin A, Gerbi SA. Nucleolar localization elements (NoLEs) in U8 snoRNA differ from sequences required for RNA processing. RNA. 1998a;4:789–800. doi: 10.1017/s1355838298980438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange TS, Borovjagin A, Maxwell ES, Gerbi SA. Conserved boxes C and D are essential nucleolar localization elements of U8 and U14 snoRNAs. EMBO J. 1998b;17:3176–3187. doi: 10.1093/emboj/17.11.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange TS, Ezrokhi M, Borovjagin AV, Rivera-León R, North MT, Gerbi SA. Nucleolar localization elements of Xenopus laevis U3 small nucleolar RNA. Mol Biol Cell. 1998c;9:2973–2985. doi: 10.1091/mbc.9.10.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: Probes for molecular biology and autoimmune disease. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor HC. Pattern of incorporation of [3H] uridine into RNA of amphibian oocyte nucleoli. J Cell Sci. 1967;2:145–150. doi: 10.1242/jcs.2.2.145. [DOI] [PubMed] [Google Scholar]
- Macgregor HC. The nucleolus and its genes in amphibian oogenesis. Biol Rev Camb Philos Soc. 1972;47:177–210. doi: 10.1111/j.1469-185x.1972.tb00972.x. [DOI] [PubMed] [Google Scholar]
- Matera AG. Of coiled bodies, gems, and salmon. J Cell Biochem. 1998;70:181–192. [PubMed] [Google Scholar]
- Matera AG. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- Matera AG, Tycowski KT, Steitz JA, Ward DC. Organization of small nucleolar ribonucleoproteins (snoRNPs) by fluorescence in situ hybridization and immunocytochemistry. Mol Biol Cell. 1994;5:1289–1299. doi: 10.1091/mbc.5.12.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW. UsnRNP assembly and transport. In: Birnstiel ML, editor. Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Berlin: Springer-Verlag; 1988. pp. 100–114. [Google Scholar]
- Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- Meier UT, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- Meier UT, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messmer B, Dreyer C. Requirements for nuclear translocation and nucleolar accumulation of nucleolin of Xenopus laevis. Eur J Cell Biol. 1993;61:369–382. [PubMed] [Google Scholar]
- Miller OL. Structure and composition of peripheral nucleoli of salamander oocytes. In: Vincent WS, Miller OL, Drets ME, Saez FA, editors. The Nucleolus—Its Structure and Function. Vol. 23. Bethesda, MD: National Cancer Institute; 1966. pp. 53–66. [PubMed] [Google Scholar]
- Miller OL, Beatty BR, Hamkalo BA, Thomas CA. Electron microscopic visualization of transcription. Cold Spring Harbor Symp Quant Biol. 1970;35:505–512. [Google Scholar]
- Miller OL, Hamkalo BA. Visualization of RNA synthesis on chromosomes. Int Rev Cytol. 1972;33:1–25. doi: 10.1016/s0074-7696(08)61446-1. [DOI] [PubMed] [Google Scholar]
- Mintz PJ, Patterson SD, Neuwald AF, Spahr CS, Spector DL. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 1999;18:4308–4320. doi: 10.1093/emboj/18.15.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Keller W. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr Opin Cell Biol. 1999;11:352–357. doi: 10.1016/S0955-0674(99)80049-0. [DOI] [PubMed] [Google Scholar]
- Misteli T, Spector DL. The cellular organization of gene expression. Curr Opin Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- Monneron A, Bernhard W. Fine structural organization of the interphase nucleus in some mammalian cells. J Ultrastruct Res. 1969;27:266–288. doi: 10.1016/s0022-5320(69)80017-1. [DOI] [PubMed] [Google Scholar]
- Myer VE, Young RA. RNA polymerase II holoenzymes and subcomplexes. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- Narayanan A, Speckmann W, Terns R, Terns MP. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol Biol Cell. 1999;10:2131–2147. doi: 10.1091/mbc.10.7.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z-Q, Prives C. Assembly of functional U1 and U2 human-amphibian hybrid snRNPs in Xenopus laevis oocytes. Science. 1988;241:1328–1331. doi: 10.1126/science.2970672. [DOI] [PubMed] [Google Scholar]
- Parvin JD, Young RA. Regulatory targets in the RNA polymerase II holoenzyme. Curr Opin Genet Dev. 1998;8:565–570. doi: 10.1016/s0959-437x(98)80012-9. [DOI] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- Paule MR. Transcription of rRNA Genes by Eukaryotic RNA Polymerase I. Berlin: Springer; 1998. [Google Scholar]
- Peculis BA, Gall JG. Localization of the nucleolar protein NO38 in amphibian oocytes. J Cell Biol. 1992;116:1–14. doi: 10.1083/jcb.116.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HRB, Brown DD. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci USA. 1980;77:4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K, Lee D, Casiano C, Bluthner M, Johnston M, Tan E. The autoimmunity-inducing xenobiotic mercury interacts with the autoantigen fibrillarin and modifies its molecular and antigenic properties. J Immunol. 1997;158:3521–3528. [PubMed] [Google Scholar]
- Pombo A, Jackson DA, Hollinshead M, Wang Z, Roeder RG, Cook PR. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 1999;18:2241–2253. doi: 10.1093/emboj/18.8.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raška I, Andrade LEC, Ochs RL, Chan EKL, Chang C-M, Roos G, Tan EM. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res. 1991;195:27–37. doi: 10.1016/0014-4827(91)90496-h. [DOI] [PubMed] [Google Scholar]
- Roeder RG. Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harbor Symp Quant Biol. 1998;63:201–218. doi: 10.1101/sqb.1998.63.201. [DOI] [PubMed] [Google Scholar]
- Roth MB, Gall JG. Monoclonal antibodies that recognize transcription unit proteins on newt lampbrush chromosomes. J Cell Biol. 1987;105:1047–1054. doi: 10.1083/jcb.105.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MB, Gall JG. Targeting of a chromosomal protein to the nucleus and to lampbrush chromosome loops. Proc Natl Acad Sci USA. 1989;86:1269–1272. doi: 10.1073/pnas.86.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MB, Murphy C, Gall JG. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J Cell Biol. 1990;111:2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MB, Zahler AM, Stolk JA. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothblum L. Isolation of nucleoli. In: Spector D, Goldman R, Leinwand L, editors. Cells: A Laboratory Manual. Vol. 1. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. pp. 48.1–48.9. [Google Scholar]
- Samarsky DA, Fournier MJ, Singer RH, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Hock R. Structure and function of the nucleolus. Curr Opin Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- Schmidt-Zachmann M, Hügle B, Scheer U, Franke WW. Identification and localization of a novel nucleolar protein of high molecular weight by a monoclonal antibody. Exp Cell Res. 1984;153:327–346. doi: 10.1016/0014-4827(84)90604-9. [DOI] [PubMed] [Google Scholar]
- Schmidt-Zachmann MS, Hügle-Dörr B, Franke WW. A constitutive nucleolar protein identified as a member of the nucleoplasmin family. EMBO J. 1987;6:1881–1890. doi: 10.1002/j.1460-2075.1987.tb02447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schul W. The spatial and functional organization of transcription and RNA processing in relation to nuclear domains. Ph.D. Thesis. Amsterdam: University of Amsterdam; 1998. [Google Scholar]
- Schul W, Groenhaut B, Koberna K, Takagaki Y, Jenny A, Manders EMM, Raška I, van Driel R, de Jong L. The RNA 3′ cleavage factors CstF 64 kDa and CPSF 100 kDa are concentrated in nuclear domains closely associated with coiled bodies and newly synthesized RNA. EMBO J. 1996;15:2883–2892. [PMC free article] [PubMed] [Google Scholar]
- Schul W, van der Kraan I, Matera AG, van Driel R, de Jong L. Nuclear domains enriched in RNA 3′ processing factors associate with coiled bodies and histone genes in a cell cycle-dependent manner. Mol Biol Cell. 1999;10:3815–3824. doi: 10.1091/mbc.10.11.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schul W, van Driel R, de Jong L. Coiled bodies and U2 snRNA genes adjacent to coiled bodies are enriched in factors required for snRNA transcription. Mol Biol Cell. 1998;9:1025–1036. doi: 10.1091/mbc.9.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz LD, Kay BK, Gall JG. In vitro RNA synthesis in oocyte nuclei of the newt Notophthalmus. Chromosoma. 1981;82:171–187. doi: 10.1007/BF00286102. [DOI] [PubMed] [Google Scholar]
- Seither P, Iben S, Grummt I. Mammalian RNA polymerase I exists as a holoenzyme with associated basal transcription factors. J Mol Biol. 1998;275:43–53. doi: 10.1006/jmbi.1997.1434. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Beven AF, Leader DJ, Brown JWS. Localization and processing from a polycistronic precursor of novel snoRNAs in maize. J Cell Sci. 1998;111:2121–2128. doi: 10.1242/jcs.111.15.2121. [DOI] [PubMed] [Google Scholar]
- Smith CM, Steitz JA. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Smith HO, Tabiti K, Schaffner G, Soldati D, Albrecht U, Birnstiel ML. Two-step affinity purification of U7 small nuclear ribonucleoprotein particles using complementary biotinylated 2′-O-methyl oligoribonucleotides. Proc Natl Acad Sci USA. 1991;88:9784–9788. doi: 10.1073/pnas.88.21.9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ. Pre-mRNA processing and the CTD of RNA polymerase II: the tail that wags the dog? Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. A polyadenylation factor subunit is the human homologue of the Drosophila suppressor of forked protein. Nature. 1994;372:471–474. doi: 10.1038/372471a0. [DOI] [PubMed] [Google Scholar]
- Thompson NE, Steinberg TH, Aronson DB, Burgess RR. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J Biol Chem. 1989;264:11511–11520. [PubMed] [Google Scholar]
- Tsvetkov A, Jantsch M, Wu Z, Murphy C, Gall JG. Transcription on lampbrush chromosome loops in the absence of U2 snRNA. Mol Biol Cell. 1992;3:249–261. doi: 10.1091/mbc.3.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma R, Stolk JA, Roth MB. Identification and characterization of a sphere organelle protein. J Cell Biol. 1993;122:767–773. doi: 10.1083/jcb.122.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc K, Steitz JA. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent WS, Miller OL. In: International Symposium on the Nucleolus: its Structure and Function. Vincent WS, Miller OL, editors. Montevideo, Uruguay: National Cancer Institute; 1965. [Google Scholar]
- Wallace RA, Jared DW, Dumont JN, Sega MW. Protein incorporation by isolated amphibian oocytes: III. Optimum incubation conditions. J Exp Zool. 1973;184:321–333. doi: 10.1002/jez.1401840305. [DOI] [PubMed] [Google Scholar]
- Wang Z, Luo T, Roeder RG. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 1997;11:2371–2382. doi: 10.1101/gad.11.18.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein LB, Steitz JA. Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- Wu C-HH, Gall JG. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc Natl Acad Sci USA. 1993;90:6257–6259. doi: 10.1073/pnas.90.13.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-HH, Murphy C, Gall JG. The Sm binding site targets U7 snRNA to coiled bodies (spheres) of amphibian oocytes. RNA. 1996;2:811–823. [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Murphy C, Callan HG, Gall JG. Small nuclear ribonucleoproteins and heterogeneous nuclear ribonucleoproteins in the amphibian germinal vesicle: loops, spheres, and snurposomes. J Cell Biol. 1991;113:465–483. doi: 10.1083/jcb.113.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Murphy C, Wu C-HH, Tsvetkov A, Gall JG. Snurposomes and coiled bodies. Cold Spring Harbor Symp Quant Biol. 1994;58:747–754. doi: 10.1101/sqb.1993.058.01.082. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]