Abstract
Prokaryotic toxin-antitoxin modules are involved in major physiological events set in motion under stress conditions. The toxin Doc (death on curing) from the phd/doc module on phage P1 hosts the C-terminal domain of its antitoxin partner Phd (prevents host death) through fold complementation. This Phd domain is intrinsically disordered in solution and folds into an α-helix upon binding to Doc. The details of the interactions reveal the molecular basis for the inhibitory action of the antitoxin. The complex resembles the Fic (filamentation induced by cAMP) proteins and suggests a possible evolutionary origin for the phd/doc operon. Doc induces growth arrest of Escherichia coli cells in a reversible manner, by targeting the protein synthesis machinery. Moreover, Doc activates the endogenous E. coli RelE mRNA interferase but does not require this or any other known chromosomal toxin-antitoxin locus for its action in vivo.
Small operons encoding a toxin and its antitoxin are common in the genomes of bacteria and archaea and are also found on certain plasmids and bacteriophages. These so-called toxin-antitoxin (TA)2 modules have been proposed to regulate the pace of metabolism and may induce a state of dormancy in case of nutritional stress (1–3). TA modules are highly abundant in opportunistic pathogens such as Mycobacterium tuberculosis (4), and their presence has been linked to persistence (5).
On plasmids, TA modules act as addiction systems, aiding plasmid maintenance in the bacterial population by post-segregational killing (6), filling in a function related to apoptosis and programmed cell death in eukaryotes (7). Related effects have been observed for chromosome-located TA systems as some of them have been shown to diminish large scale genome reductions in the absence of selection (8). In the presence of the plasmid, both toxin and antitoxin are expressed, leading to a steady state equilibrium where the antitoxin counteracts the effect of the toxin. In its free state, the antitoxin, usually a modular protein that contains a functional intrinsically disordered region (9–11), is under constant proteolytic attack. The toxin-antitoxin complex acts as an autorepressor for the TA operon, ensuring that only small amounts of the proteins are present in the cell. Upon plasmid loss, the antitoxin is degraded by a specific intracellular protease, releasing the toxin. Without the possibility of replenishing the antitoxin population, the toxin action becomes irreversible, resulting in cell death.
The phd/doc operon encodes a TA module aiding the maintenance of the plasmid-prophage P1 in Escherichia coli (12). Doc is an inhibitor of translation elongation through its association with the 30 S ribosomal subunit in a way similar to the antibiotic hygromycin (13). The action of Doc is suppressed by the antitoxin Phd, which consists of two domains. Its C-terminal domain (residues 52–73) harbors the interaction site with Doc and on its own prevents Doc-mediated growth arrest (14, 15). The N-terminal region (residues 1–51) of Phd is a dimerization/DNA-binding domain that binds to the operator site of the phd/doc operon. Phd forms a heterotrimeric complex with Doc (16). Operator binding and repression of the phd/doc operon by Phd are enhanced by the presence of Doc in a cooperative manner (17–19).
Here we present the crystal structure of a non-toxic version of Doc (DocH66Y, an H66Y mutant of Doc) in complex with the C-terminal domain of Phd (Phd52–73Se, a peptide corresponding to residues 52–73 of Phd with Se-Met substituted for Leu-52 and Leu-70) and provide further information on the interplay between Doc and endogenous chromosomal TA modules. The structure reveals a new all α-helical fold, leads to new insights into the molecular action of Doc and its interaction with Phd, and suggests a possible origin for the phd/doc module.
EXPERIMENTAL PROCEDURES
Crystal Structure Determination—Purification of DocH66Y and crystallization of DocH66Y and of its complex with Phd52–73Se (C-terminal 22 residues of Phd with Leu-52 and Leu-70 substituted to Se-Met) are reported elsewhere.3 After a failed attempt to phase the structure of DocH66Y-Phd52–73Se using the two Se-methionines present in Phd52–73Se, a crystal was soaked for about 3 min in a cryo-protecting solution (0.2 m NaCl, 0.1 m sodium acetate, pH 4.6, and 35% 2-methyl-2,4-pentanediol) enriched with 1.5 m of NaBr. Data were collected at the K-edge of bromine and at a high energy remote wavelength (Table 1). The data sets were indexed, integrated, and merged using the HKL suite of programs (20). Substructure calculation with ShelxD (21) detected 17 potential bromine sites. Phases were calculated by two-wavelength multiwavelength anomalous dispersion using the software pipeline AutoRickshaw (22). The experimentally phased electron density map allowed automated building of the model with ARP/wARP (23) after 2-fold symmetry averaging of the electron density based upon non-crystallographic symmetry parameters derived from the known heavy atom positions. The resulting model was highly complete and refined using REFMAC5 (24) to an Rfree factor of 19.8% and a conventional R-factor of 18.3%. Details of data collection and refinement statistics are given in Table 1.
TABLE 1.
Data collection and refinement statistics r.m.s., root mean square.
| Data set | DocH66Y-Phd52–73Se-Peak | DocH66Y-Phd52–73Se-Remote |
|---|---|---|
| Beamline | X12 (EMBL Hamburg) | X12 (EMBL Hamburg) |
| Wavelength (Å) | 0.9189 | 0.9116 |
| Space group | C2 | C2 |
| Unit cell | ||
| a (Å) | 110.9 | 110.9 |
| b (Å) | 38.2 | 38.2 |
| c (Å) | 63.7 | 63.7 |
| β (°) | 99.3 | 99.3 |
| Resolution limits (Å) | 15.0–1.7 | 15.0–1.7 |
| Number of measured reflections | 642571 | 649052 |
| Number of unique reflections | 29371 | 29397 |
| Completeness | 96.7 | 95.7 |
| Rmergea | 0.09 | 0.09 |
| <I/σ(I)> | 9.0 | 8.3 |
| R-factorb | 18.2 | |
| Rfree factor | 19.8 | |
| Ramachandran profile | ||
| Core | 100% | |
| Other allowed | 0.0% | |
| Outliers | 0.0% | |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.01 | |
| Bond angles (°) | 1.24 | |
| Number of atoms | ||
| Protein | 2207 | |
| Water | 228 | |
| Other | 16 | |
| B-factors (Å2) | ||
| From Wilson plot | 19.4 | |
| All atoms | 22.3 | |
| Protein atoms | 20.7 | |
| Water atoms | 30.5 | |
| Other atoms | 24.5 | |
| PDB entry | 3dd7 |
Rmerge = Σhkl Σi|Ihkli,i – 〈Ihkl〉|Σhkl ΣiIhkl,i
R-factor = Σhkl||Fobs (hkl)|–k|Fcalc(hkl)||/Σhkl|Fobs(hkl)|
CD Spectropolarimetry—Far UV-CD spectra were recorded on a J-715 spectropolarimeter (Jasco). Scans were taken using a 0.1-cm cuvette. The temperature of the cuvette was monitored using a probe, and a water bath was used for maintaining the temperature of the sample constant. The measurements were performed at 298 K, in 50 mm Tris (pH 7.5), 150 mm sodium chloride. Spectra of DocH66Y and Phd52–73Se were taken using a protein concentration of 60 μm. For the spectrum of the complex, DocH66Y and Phd52–73Se were mixed in equimolar ratio to a final concentration of 60 μm of the complex and preincubated for 5 min before taking the spectrum. The mean residue ellipticities ([θ], degrees cm2 mol-1) were obtained from the raw data (θ, ellipticity) after correcting for the buffer solution, according to [θ] = θ·Mw/(n·c·l), where Mw is the molecular weight, c is the mass concentration, l is the optical path length, and n is the number of amino acid residues.
Northern Blotting and Primer Extension Analyses—Strains and plasmids used in this work are described in the supplemental Materials and Methods and are summarized in supplemental Table S1. Cells were grown in LB at 37 °C. At an A450 of 0.5, the cultures were diluted 10 times and grown to an OD of 0.5. Transcription of the toxins was induced by the addition of arabinose to 0.2%. To inhibit translation, chloramphenicol (50 μg/ml) was added. For Northern analysis, total RNA was fractionated by PAGE (6% bis-acrylamide), blotted to a Zeta-Probe nylon membrane, and hybridized with a single-stranded 32P-labeled riboprobe, complementary to the RNA. The radioactive probe was generated using linearized plasmid DNA of pSC333 constructing probes for lpp mRNA. Semiquantitative primer extension analysis was performed essentially according to the method previously described (25). The stop codons of mRNAs originating from pKW254T derivatives were mapped with the primer pKW71D-3#PE, which is complementary to the linker RNA of pKW254T. The primers lpp 21 and lpp 26 were used to map the 5′ end of lpp mRNA. The 5′ end of dksA mRNA was mapped using the primer dksA PE1. The complete list of oligonucleotides used is given in supplemental Table S2.
RESULTS
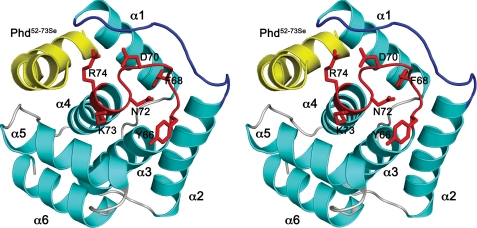
Structure of DocH66Y—Because of difficulties in producing sufficient amounts of wild-type Doc, we used the less toxic mutant H66Y (19) for structure determination. DocH66Y was crystallized in complex with a peptide encompassing the C-terminal 22 amino acids of Phd with Se-Met substituted for Leu-52 and Leu-70 (Phd52–73Se). This fragment was chosen based upon previous work that delineates the toxin-binding domain of Phd (14, 15). The Doc protein shows an all-α-helical fold consisting of six α-helices (Fig. 1). It can be described as the stacking of three consecutive helix-loop-helix elements. Helices α3 and α4 are central in the structure and carry a high proportion of aliphatic side chains to pack α1 and α2 on one side and α5 and α6 on the other.
FIGURE 1.
Structure of Doc. A, stereo view of the DocH66Y-Phd52–73Se complex. Helices of DocH66Y are shown in cyan, and loop structures are shown in gray. The α-helices are labeled. The loop α3-α4 containing the conserved sequence motif HXFX(D/E)(A/G)N(K/G)R is highlighted in red, and its side chains are shown as sticks. Loop α1-α2 is highlighted in blue. The bound Phd52–73Se fragment is shown in yellow.
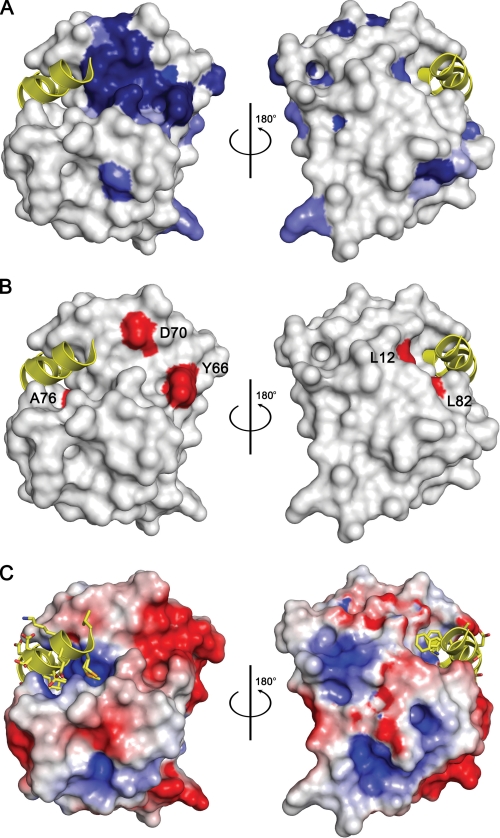
Multiple sequence alignment of Doc family members reveals a single highly conserved motif, HXFX(D/E)(A/G)N(K/G)R. It is located in the loop α3-α4 (residues 66–74) (Fig. 1), and its conformation can be described as two consecutive NEST motifs. These are short structural elements that are often used as anion-binding sites (26) (supplemental Fig. S2a). This sequence motif forms the core of a single patch of conserved surface residues (Fig. 2A) that further includes residues from the loop α1-α2 as well as His-13. The latter plays a structural role for establishing the correct conformation of the conserved sequence motif. In addition, a number of mutations known to eliminate Doc toxicity but that retain co-repression activity (H66Y, H66R, and D70N) map within this motif (Fig. 2B) (19). This is a clear indication for a functional role of this region, likely an interaction site.
FIGURE 2.
Residue conservation and Phd-binding site of Doc. A, surface representation of DocH66Y. Residues that are conserved in Doc sequences are colored blue with the intensity of the color reflecting the degree of conservation: darkest blue represents full conservation in all sequences, and lightest blue represents 50% conservation (residue conservation calculated according to Livingstone and Barton (39) and based upon the sequence alignment shown in supplemental Fig. S1). Two views are shown 180° apart. The bound fragment of Phd is shown as a yellow helix ribbon. B, mapping of toxicity-eliminating mutations on the surface of Doc. Residues that lead to a non-toxic form of P1 Doc (19) are colored red and labeled. Asp-60 and Tyr-66 are part of the conserved sequence motif. Leu-12 and Leu-82 are within the Phd52–73Se-binding site. However, the corresponding mutations L12P and L82P are likely to disrupt helices α1 and α4, respectively, in agreement with the observation that both lead to a protein that loses its toxicity as well as its regulatory activity. The orientations are the same as in panel A. C, electrostatic surface potential mapped on the surface of Doc. Negatively charged regions are colored red, and positively charged regions are colored blue. The bound fragment of Phd is shown as a yellow helix ribbon. The orientations are the same as in panel A.
Interactions with Phd—The C-terminal domain of Phd on its own is sufficient to protect against Doc (14, 15). Phd52–73Se binds into a groove of DocH66Y of which helix α4 forms the base and that is flanked by helix α1 on one side and the loop α4-α5 on the other side (Fig. 1A). This binding site is adjacent to the hotspot of conserved residues on the surface of Doc but by itself is not highly conserved (Fig. 2A). The binding groove has an approximate volume of 2050 Å3 and contains a large number of positively charged side chains (Arg-2, Arg-19, Lys-73, Arg-74, Arg-85, and Arg-86) around a hydrophobic center. As such, it forms a positively charged patch on the otherwise overall negatively charged surface of Doc (Fig. 2C).
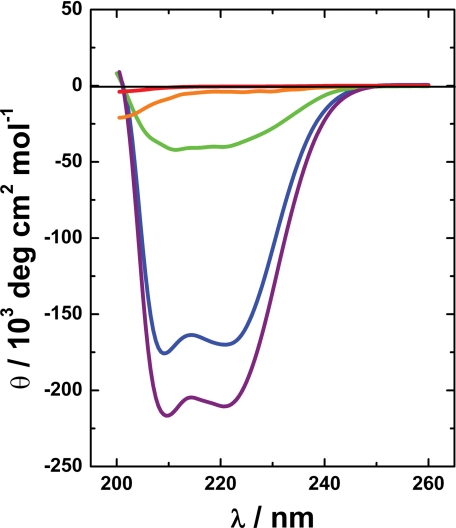
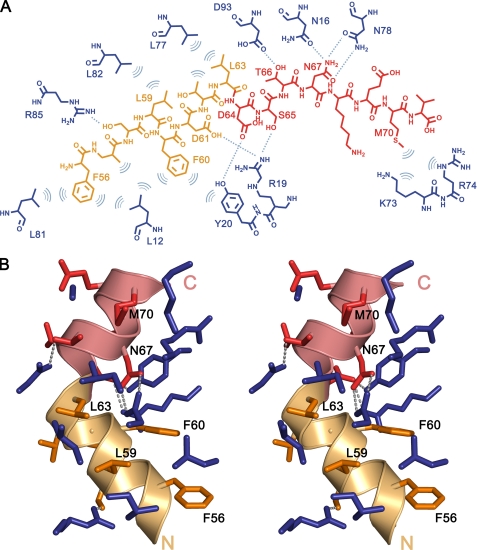
Far UV CD experiments show that Phd52–73Se is intrinsically unstructured in its isolated state but gains an appreciable amount of α-helix upon binding to Doc (Fig. 3). In the crystal structure, Phd52–73Se adopts an α-helical conformation when bound to Doc and shields about 820 Å2 of the Doc surface from the solvent. The interactions between Phd52–73Se and DocH66Y are mediated entirely by side chain atoms (Fig. 4A). A kink in the α-helix divides Phd52–73Se into a hydrophobic N-terminal segment (residues 54–62) and a predominantly negatively charged C-terminal segment (residues 64–73) (Fig. 4B). In its folded state, bound to Doc, the N-terminal segment of Phd52–73Se is distinctly amphipathic with its hydrophobic side interacting with Doc. The side chains Leu-59, Phe-60, and Leu-63 of Phd52–73Se become completely buried on this hydrophobic surface, creating an extension of the hydrophobic core of Doc. Phe-56, which is more exposed, extends this set of interactions by packing against Leu-12 and Leu-81 of Doc (Fig. 4).
FIGURE 3.
Solution structures of Phd52–73Se and DocH66Y. The far UV CD spectrum of isolated Phd52–73Se 60 μm (red) and 300 μm (orange) is characteristic of an intrinsically unstructured protein with a weak minimum around 200 nm. Consistent with its crystal structure, the spectrum of DocH66Y in blue (60 μm) is dominated by a high α-helical content, which is further increased in the DocH66Y-Phd52–73Se complex (purple). The difference spectrum between the complex and free DocH66Y (in green) shows that Phd52–73Se in its bound state is mostly α-helical with the typical minima at 210 and 221 nm.
FIGURE 4.
Interactions between Phd52–73Se and DocH66Y. A, schematic drawing of interacting residues in DocH66Y (blue) and Phd52–73Se. The N-terminal α-helical region of the peptide (residues 52–63), predominantly hydrophobic, is colored in orange. The C-terminal region of the peptide, predominantly hydrophilic, is colored in red. Hydrogen bonds (cutoff level 3.5 Å) are shown as dashed lines, and hydrophobic interactions are represented as arcs. B, stereo view of the DocH66Y-Phd52–73Se complex interface. Residues are colored as in panel A. Phd52–73Se residues that are buried in the interface are indicated. Hydrogen bonds are shown as dashed lines.
The C-terminal segment of Phd52–73Se is highly hydrophilic and provides only a single hydrophobic residue (Leu-70 in Phd, Se-Met-70 in Phd52–73Se) to the binding interface. This residue makes extensive contacts with a small hydrophobic cavity on the Doc surface (Fig. 4B). The Phd-Doc contact surface in this region shows a high degree of charge complementarity with several negatively charged side chains of Phd52–73Se (Glu-55, Asp-61, and Asp-64) interacting favorably with positive residues in the Phd-binding groove of DocH66Y (Arg-19 and Arg-85). Most striking in this region of Phd52–73Se are the interactions involving Asn-67. This residue is completely buried in the interface, its side chain protruding inside a small hydrophilic pocket where it makes complementary hydrogen bonds with the side chain of Asn-16 and Asn-78 of Doc (Fig. 4).
It should be noted here that although the C terminus of Phd52–73Se is adjacent to the surface cluster of conserved residues, the conserved sequence motif of Doc is not part of the Phd-binding site. This indicates that Phd52–73Se counteracts the toxic activity of Doc either by inducing a conformational change in Doc or by sterically preventing Doc to interact with the ribosome. Both mechanisms have been proposed earlier on for other TA modules (27, 28).
Doc Has an Incomplete Fic Fold—Structural similarity searches against the Structural Classification of Proteins-(SCOP) data base using the DALI server failed to reveal any protein with significant similarity to Doc. However, all Doc homologues possesses a conserved central motif (see above) of 9 residues that is shared with two other protein families: the bacterial cAMP-induced filamentation protein (Fic) and a domain of the eukaryotic Huntingtin Yeast Protein E (HYPE) protein (29). Sequence similarity between Doc and these other two protein families outside this 9-residue region is very weak with overall sequence identities below 15%.
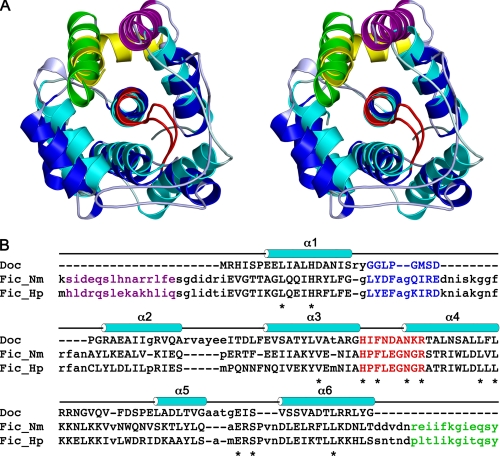
A query of the Protein Data Bank with the conserved central motif of Doc resulted in the identification of two proteins that show a high structural similarity to Doc. These otherwise undescribed recent depositions from the Midwest Center for Structural Genomics are crystal structures of two Fic proteins: Fic_Hp from Helicobacter pylori (PDB entry 2F6S) and Fic_Nm from Neisseria meningitidis (PDB entry 2G03). Fic_Nm is the most closely related to Doc with an root mean square deviation of 2.3 Å for 105 matching Cα atoms (Z score 6.7) (Fig. 5A). A structure-based sequence alignment is given in Fig. 5B.
FIGURE 5.
Doc has an incomplete Fic-like fold. A, stereo view of the superposition between DocH66Y and Fic from N. meningitidis (Fic_Nm). DocH66Y is shown in cyan, and the bound Phd52–73Se fragment is shown in yellow. Fic_Nm is shown in blue, and its C-terminal helix is shown in green. The loops α3-α4 containing the conserved sequence motif are highlighted in red for both proteins. The N-terminal α-helix of Fic, which has no counterpart in Doc, is shown in purple. B, structure-based sequence alignment of Doc with Fic_Nm (PDB entry 2G03) and Fic_Hp (PDB entry 2F6S). The proteins were aligned to Doc using SALIGN (40). Residues with a structural match to Doc are shown in uppercase, and those that are structurally divergent from Doc and do not allow for a 1:1 match are shown in lowercase. The α-helices in Doc are indicated above the sequence. Residues corresponding to the conserved sequence motif in loop α3-α4 are highlighted in red, and conserved residues are indicated with asterisks. The loop α1-α2 is highlighted in blue. Residues from the C-terminal α-helix from the Fic proteins are shown in green, and those of the N-terminal α-helix are shown in purple.
Fic in most respects resembles the architecture of Doc, including very similar conformations for the conserved central motif in loop α3-α4, suggesting a common evolutionary origin (supplemental Fig. S2). Also, His-13, the residue that in Doc anchors the conserved central motif, is conserved and makes equivalent interactions in all structures (supplemental Fig. S2). Nevertheless, Fic differs from Doc by the presence of an extra N-terminal α-helix, by an insertion in the loop α1-α2, and most importantly, by an additional C-terminal α-helix (Fig. 5A). The latter adopts a position and conformation in the Fic structure that is strikingly similar to the position and conformation of the Phd peptide in the Doc-peptide complex (Fig. 5A). Removing Phd52–73Se from the complex unveils a large hydrophobic patch that extends toward the hydrophobic core. This is suggestive of an incomplete protein, and indeed, free Doc is poorly soluble and prone to aggregation and misfolding.
Based upon these observations, we propose that Doc evolved from a Fic-like ancestor of which the C-terminal α-helix was transferred to a DNA-binding domain, thereby generating the antitoxin Phd. Upon binding, the antitoxin donates an α-helix to form the complex thereby complementing the fold.
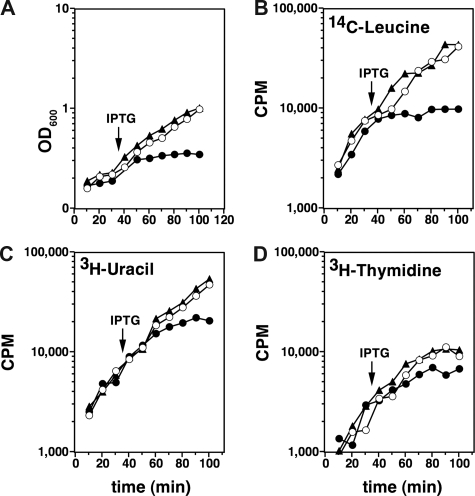
Doc Induces Growth Arrest but Not Cell Death—Induction of Doc leads to growth arrest within the doubling time of E. coli. However, the cells do not lyse and remain motile for several hours after induction, when examined under a light microscope. This indicates an intact cell membrane and a working proton motive force. We do not observe filamentation in cells in which Doc has been activated, although filamentation but not induction of the SOS pathway was reported earlier (12). The observed growth arrest is reversible as cells replated in the absence of IPTG are capable of colony formation for several hours after the start of IPTG induction. We further confirmed that Doc arrests bulk protein synthesis but not RNA or DNA synthesis (Fig. 6). Additionally, we observed that Doc inhibits protein synthesis in a cell-free expression system and that Phd prevents this inhibition (data not shown).
FIGURE 6.
Effect of Doc on growth and macromolecular synthesis. Open circles correspond to BR7046 (carrying wild-type Doc) without induction by IPTG. Filled circles correspond to BR7046 with induction of Doc by IPTG (time of induction indicated by an arrow). Filled triangles correspond to BR7044 carrying Ptac-docH66Y and induced at the same moment as BR7046. A, induction of Doc leads to a quick cessation of growth. B, the effect of Doc on protein synthesis as measured by incorporation of 14C leucine is pronounced and parallels the cessation of growth. C and D, the effects of Doc on RNA and DNA synthesis measured by the incorporation of [3H]uracil and [3H]thymidine, respectively, are less pronounced and somewhat lag behind the effects on growth and protein synthesis, indicative of a secondary rather than a primary effect. See supplemental Material and Methods for further details.
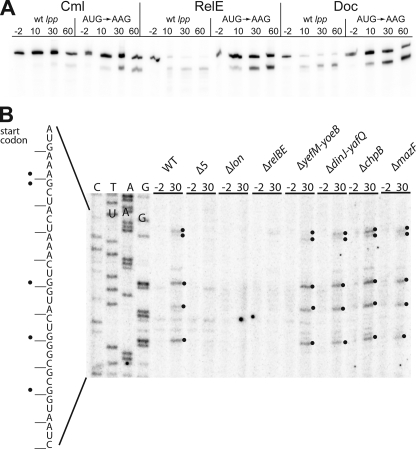
Doc Induces RelE-mediated Cleavage of Model mRNAs—A recent study showed that Doc expression in E. coli strain BL21(DE3) led to mRNA stabilization (13). By contrast, we observe destabilization of two different model mRNAs (lpp and dksA) after induction of doc in E. coli strain MG1655 (Fig. 7A and supplemental Fig. S3). Primer extension analysis after doc induction of lpp and dskA mRNAs reveals cleavage patterns very similar to that induced by RelE, especially just downstream of the start codons (Fig. 7B; supplemental Figs. S3 and S4). However, in contrast to RelE, Doc does not induce mRNA cleavages near the stop codons. Strikingly, non-translated versions of the lpp and dskA mRNAs (start codons changed to AAG) are not affected by induction of doc (Fig. 7A and supplemental Figs. S3 and S4). Thus, like RelE-induced cleavage, Doc-induced mRNA cleavage depends on translation. These results suggest that the cleavage is due to the activation of endogenous TA loci rather than a direct effect of Doc itself. Indeed, Doc-induced mRNA cleavage is not observed in a strain deficient in the five major E. coli TA modules (mazEF, chpBIK, relBE, yoeB/yefM, and dinJ/yafQ-MG1655Δ5) (Fig. 7B, lanes 7–8). Moreover, expression of Doc in a strain that lacks relBE (MG1655ΔrelBE) also fails to induce Doc-mediated mRNA cleavage (Fig. 7B, lanes 11–12), indicating that ectopic production of Doc activates endogenous RelE. A previous report (30) indicated that Doc induces MazF activity. However, we did not observe mRNA cleavage at ACA sites, the signature sequence of MazF-mediated mRNA cleavage (31). Neither do we observe any influence of deleting mazEF on the cleavage pattern. Therefore our results do not confirm that Doc activates MazF. RelE has previously been described to be activated during nutritional stress due to Lon-mediated degradation of RelB antitoxin. The Doc-induced cleavage sites depend on Lon (Fig. 6B, lanes 9 and 10). These results are consistent with the proposal that the Doc-mediated inhibition of translation leads to RelE activation via Lon-dependent decay of RelB. Thus, the mRNA decay seen after induction of doc appears to be an indirect consequence of Lon-dependent activation of RelE. Consistent with this hypothesis is the observation by Liu et al. (13) that mRNA is not destabilized by induction of doc in E. coli strain BL21(DE3). BL21(DE3) is optimized for protein production and lacks the Lon and OmpT proteases (32).
FIGURE 7.
Induction of RelE activity by Doc. A, Doc induces cleavage of a translated model lpp mRNA. Cells of MG1655Δlpp/MG3323 (pBAD-relE) and MG1655Δlpp/pMCD3303 (pBAD-doc) containing either of the plasmids pSC710 (wild-type (wt) lpp) or pSC711 (ATG start codon of lpp changed to AAG) were grown exponentially in LB medium at 37 °C. To induce transcription of the toxin genes, arabinose (0.2%) was added at time 0. As a control, the strains MG1655Δlpp/pSC710 and MG1655Δlpp/pSC711 were treated with chloramphenicol (Cml) (50 μg/ml) at time 0 to inhibit translation. Total RNA samples were fractionated by PAGE, and lpp mRNA was visualized by Northern blotting analysis. Numbers are time points of cell sampling relative to inhibition of translation by either chloramphenicol or arabinose. B, overproduction of Doc induces RelE-dependent mRNA cleavage. Primer extension analysis of lpp mRNA after transcriptional induction of doc from pMCD3303 (pBAD-doc) in the following strains: MG1655 (wild type), Δ5 (SC301467), Δlon, ΔrelBE (SC31), ΔyefM-yoeB (SC36), ΔdinJ-yafQ (SC37), ΔmazF (SC30), and ΔchpB (SC31). The strains were grown in LB medium to an A450 nm of 0.5, and total RNA was prepared from bacterial samples taken at the indicated time points (-2 and 30 min) after the addition of 0.2% arabinose. Primer extension was performed as described under “Experimental Procedures” using 32P-labeled primer lpp21. Significant cleavage sites in the RNA are marked with black dots on the gel and in the corresponding sequence to the left.
DISCUSSION
The phd/doc locus of prophage P1 is an archetype member of a family of toxin-antitoxin modules found also on bacterial chromosomes. Although first reported in 1993 (12), phd/doc has remained less understood than the well known ccdAB, relBE and mazEF modules. Here we show that the toxin Doc adopts an all-α-helical fold different from the folds of other TA toxins with known structure. Doc contains a single highly conserved surface patch distinct from its interaction site with the antitoxin Phd. The significance of this conserved surface region is corroborated by the observation that Doc resembles members of another family of bacterial proteins called Fic, that a signature sequence present in the conserved surface region of Doc is also conserved in Fic, and that mutations within this motif affect Doc toxicity. This is indicative of a functionally relevant site and suggests that the mechanisms of action of Doc and Fic are related.
Doc has been shown to inhibit protein synthesis and to associate with the 70 S ribosome and with the 30 S ribosomal subunit (13). The mechanism of action of Fic is currently unknown. The strong structural similarities between both proteins and the presence of a highly conserved and functionally important region raise the possibility that Fic is also capable of halting translation on the ribosome or to modify ribosomal activity. Available data indicate that Fic has a role in cell division (33), most likely under the tight control of the cell division machinery. Mutations of Fic, such as G55R (34), may distort this control, leading to its accidental activation (12, 33). We suggest that Doc evolved from a mutant of a Fic-like ancestor defective in regulation of its activity. Likely, the C terminus of such an ancestor of Doc was transferred to a DNA-binding domain. This resulted in a novel regulatory system that by its ability to be used for conditional killing or growth arrest can function as a plasmid addiction module and as a TA stress operon. The existence of several open reading frames of doc-like genes fused to genes encoding DNA-binding domains among different bacterial genomes favors this hypothesis. Further studies are nevertheless required to substantiate this hypothesis.
During complex formation, Phd donates its C-terminal domain to complement the truncated Fic-like fold of Doc. Fold complementation has been described as one of the major mechanisms by which proteins can bind peptides in a β-strand conformation (35). As a result of the complex formation, the added β-strand complements for a missing secondary structure element in an otherwise incomplete fold. The completion of an Ig-like β-sandwich in the subunit-subunit and chaperone-subunit interactions in bacterial pili assembled by the chaperoneusher pathway and the addition of the hepatitis C virus NS4A cofactor peptide to the N-terminal β-sheet in the NS3 protease that complements a chymotrypsin-like fold are classical examples of this mechanism (36, 37). To our knowledge, this mechanism has not been observed before for α-helical peptide ligands binding to all-α-helical proteins. The amphipathic nature of the α-helix of Phd and the hydrophobic surface patch at the center of the Phd-binding site on Doc are likely remnants of the Fic-like ancestor of Doc, which had its hydrophobic core partially disrupted when it lost its C terminus.
Our in vivo experiments indicate that the ectopic overexpression of Doc can induce the mRNA interferase activity of RelE, a chromosomal TA toxin of E. coli. Activation of RelE, together with MazF, is triggered by stress conditions and is probably part of the general response of the cell to internal alarm signals. RelE activation may be an attempt of the cells to relieve the stress induced by Doc-mediated translation arrest. Indeed, RelE activity correlates with tmRNA activity (38). Thus, mRNA degradation on Doc-arrested ribosomes may mimic translation quality control systems that are invoked when termination of translation fails.
Supplementary Material
Acknowledgments
We acknowledge the use of beam time at the European Molecular Biology Laboratory (EMBL) beamlines at the Deutsches Elektronen-Synchrotron (DESY) synchrotron (Hamburg, Germany).
The atomic coordinates and structure factors (code 3dd7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant 2 R15 GM67668-03 from the NIGMS (to R. D. M.) and a National Institutes of Health Intramural Research Training Award grant. This work was also supported by grants from the VIB, the Fonds voor Wetenschappelijk Onderzoek Vlaanderen, the Centre for mRNP Biogenesis and Metabolism of the Danish National Research Foundation, and the Onderzoeksraad of the Vrije Universiteit Brussel. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Materials and Methods, four supplemental figures, two supplemental tables, and supplemental references.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: TA, toxin-antitoxin; PDB, Protein Data Bank; IPTG, isopropyl-1-thio-β-d-galactopyranoside.
A. Garcia-Pino and R. Loris, manuscript in preparation.
References
- 1.Buts, L., Lah, J., Dao-Thi, M. H., Wyns, L., and Loris, R. (2005) Trends Biochem. Sci 30 672-679 [DOI] [PubMed] [Google Scholar]
- 2.Gerdes, K., Christensen, S. K., and Lobner-Olesen, A. (2005) Nat. Rev. Microbiol. 3 371-382 [DOI] [PubMed] [Google Scholar]
- 3.Pedersen, K., Christensen, S. K., and Gerdes, K. (2002) Mol. Microbiol. 45 501-510 [DOI] [PubMed] [Google Scholar]
- 4.Pandey, D. P., and Gerdes, K. (2005) Nucleic Acids Res. 33 966-976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shah, D., Zhang, Z., Khodursky, A., Kaldalu, N., Kurg, K., and Lewis, K. (2006) BMC Microbiol. 6 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerdes, K., Rasmussen, P. B., and Molin, S. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 3116-3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yarmolinsky, M. B. (1995) Science 267 836-837 [DOI] [PubMed] [Google Scholar]
- 8.Szekeres, S., Dauti, M., Wilde, C., Mazel, D., and Rowe-Magnus, D. A. (2007) Mol. Microbiol. 63 1588-1605 [DOI] [PubMed] [Google Scholar]
- 9.Loris, R., Marianovsky, I., Lah, J., Laeremans, T., Engelberg-Kulka, H., Glaser, G., Muyldermans, S., and Wyns, L. (2003) J. Biol. Chem. 278 28252-28257 [DOI] [PubMed] [Google Scholar]
- 10.Madl, T., Van Melderen, L., Mine, N., Respondek, M., Oberer, M., Keller, W., Khatai, L., and Zangger, K. (2006) J. Mol. Biol. 364 170-185 [DOI] [PubMed] [Google Scholar]
- 11.Oberer, M., Zangger, K., Gruber, K., and Keller, W. (2007) Protein Sci. 16 1676-1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehnherr, H., Maguin, E., Jafri, S., and Yarmolinsky, M. B. (1993) J. Mol. Biol. 233 414-428 [DOI] [PubMed] [Google Scholar]
- 13.Liu, M., Zhang, Y., Inouye, M., and Woychik, N. A. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 5885-5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKinley, J. E., and Magnuson, R. D. (2005) J. Bacteriol. 187 765-770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith, J. A., and Magnuson, R. D. (2004) J. Bacteriol. 186 2692-2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gazit, E., and Sauer, R. T. (1999) J. Biol. Chem. 274 16813-16818 [DOI] [PubMed] [Google Scholar]
- 17.Gazit, E., and Sauer, R. T. (1999) J. Biol. Chem. 274 2652-2657 [DOI] [PubMed] [Google Scholar]
- 18.Magnuson, R., Lehnherr, H., Mukhopadhyay, G., and Yarmolinsky, M. B. (1996) J. Biol. Chem. 271 18705-18710 [DOI] [PubMed] [Google Scholar]
- 19.Magnuson, R., and Yarmolinsky, M. B. (1998) J. Bacteriol. 180 6342-6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 21.Schneider, T. R., and Sheldrick, G. M. (2002) Acta Crystallogr. Sect. D Biol. Crystallogr. 58 1772-1779 [DOI] [PubMed] [Google Scholar]
- 22.Panjikar, S., Parthasarathy, V., Lamzin, V. S., Weiss, M. S., and Tucker, P. A. (2005) Acta Crystallogr. Sect. D Biol. Crystallogr. 61 449-457 [DOI] [PubMed] [Google Scholar]
- 23.Morris, R. J., Perrakis, A., and Lamzin, V. S. (2003) Methods Enzymol. 374 229-244 [DOI] [PubMed] [Google Scholar]
- 24.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 25.Dam, M., and Gerdes, K. (1994) J. Mol. Biol. 236 1289-1298 [DOI] [PubMed] [Google Scholar]
- 26.Watson, J. D., and Milner-White, E. J. (2002) J. Mol. Biol. 315 171-182 [DOI] [PubMed] [Google Scholar]
- 27.Kamada, K., and Hanaoka, F. (2005) Mol. Cell 19 497-509 [DOI] [PubMed] [Google Scholar]
- 28.Takagi, H., Kakuta, Y., Okada, T., Yao, M., Tanaka, I., and Kimura, M. (2005) Nat. Struct. Mol. Biol. 12 327-331 [DOI] [PubMed] [Google Scholar]
- 29.Anantharaman, V., and Aravind, L. (2003) Genome Biol. 4 R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hazan, R., Sat, B., Reches, M., and Engelberg-Kulka, H. (2001) J. Bacteriol. 183 2046-2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, Y., Zhang, J., Hoeflich, K. P., Ikura, M., Qing, G., and Inouye, M. (2003) Mol. Cell 12 913-923 [DOI] [PubMed] [Google Scholar]
- 32.Studier, F. W., and Moffatt, B. A. (1986) J. Mol. Biol. 189 113-130 [DOI] [PubMed] [Google Scholar]
- 33.Utsumi, R., Nakamoto, Y., Kawamukai, M., Himeno, M., and Komano, T. (1982) J. Bacteriol. 151 807-812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawamukai, M., Matsuda, H., Fujii, W., Utsumi, R., and Komano, T. (1989) J. Bacteriol. 171 4525-4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Remaut, H., and Waksman, G. (2006) Trends Biochem. Sci. 31 436-444 [DOI] [PubMed] [Google Scholar]
- 36.Sauer, F. G., Futterer, K., Pinkner, J. S., Dodson, K. W., Hultgren, S. J., and Waksman, G. (1999) Science 285 1058-1061 [DOI] [PubMed] [Google Scholar]
- 37.Yan, Y., Li, Y., Munshi, S., Sardana, V., Cole, J. L., Sardana, M., Steinkuehler, C., Tomei, L., De Francesco, R., Kuo, L. C., and Chen, Z. (1998) Protein Sci. 7 837-847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christensen, S. K., and Gerdes, K. (2003) Mol. Microbiol. 48 1389-1400 [DOI] [PubMed] [Google Scholar]
- 39.Livingstone, C. D., and Barton, G. J. (1993) Comput. Appl. Biosci. 9 745-756 [DOI] [PubMed] [Google Scholar]
- 40.Marti-Renom, M. A., Madhusudhan, M. S., and Sali, A. (2004) Protein Sci. 13 1071-1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.