Abstract
During human immunodeficiency virus type 1 (HIV-1) infection, there is a strong positive correlation between CCL2 levels and HIV viral load. To determine whether CCL2 alters HIV-1 infection of resting CD4+ T cells, we infected purified resting CD4+ T cells after incubation with CCL2. We show that CCL2 up-regulates CXCR4 on resting CD4+ T cells in a CCR2-dependent mechanism, and that this augmentation of CXCR4 expression by CCL2 increases the ability of these cells to be chemoattracted to CXCR4 using gp120 and renders them more permissive to X4-tropic HIV-1 infection. Thus, CCL2 has the capacity to render a large population of lymphocytes more susceptible to HIV-1 late in the course of infection.
Human immunodeficiency virus type 1 (HIV-1)2 establishes a persistent long lived and latent infection of resting CD4+ T cells that eludes immune surveillance and current drug therapy, and it is the major barrier to the treatment of HIV. Chemokine (C-C motif) ligand 2 (CCL2; also known as MCP-1 (monocyte chemoattractant protein 1)) is a pro-inflammatory chemokine induced during many inflammatory and autoimmune diseases and during bacterial and viral infections. During HIV-1 infection, there is a strong positive correlation between CCL2 levels and HIV viral load with higher expression of CCL2 mRNA and elevated serum CCL2 concentrations being found in viremic (viral load >100,000 RNA copies/ml plasma) compared with aviremic (viral load <50 copies/ml) HIV-1-infected patients (1). CCL2 is produced from infected macrophages (2), and extracellular secreted HIV-1 proteins such as Tat and gp120 can induce the production of CCL2 from uninfected bystander cells either directly or indirectly by up-regulating tumor necrosis factor production (3–5). The cognate receptor for CCL2 is chemokine (C-C motif) receptor 2 (CCR2). CCL2 chemoattracts CCR2+ cells such as monocytes (6), activated and memory T cells (7), and natural killer cells (8) to the site of inflammation and infection, rendering them more susceptible to HIV-1 infection. This provides an ideal environment for HIV-1 to replicate in freshly recruited target cells that leads to the appearance of high viral load. As with other C-C chemokines, CCL2 plays a role in maintaining immune homeostasis. However, unlike the other C-C chemokines, CCL2 regulates T helper (Th) cell differentiation by polarizing Th0 cells toward a Th2 phenotype as opposed to a Th1 phenotype (9). Conversely, CCL2, along with CCL3, CCL4, and CCL5, is able to increase the replication of X4-tropic strains in activated CD4+ T cells (10).
HIV-1 isolates are distinguishable by their main coreceptor, chemokine (CXC motif) receptor-4 (CXCR4) or CCR5, they use for cell entry (11). Thus, the corresponding phenotypes of the viruses are X4, R5, or R5X4 if they use CXCR4, CCR5, or both coreceptors, respectively. R5 viruses predominate primary HIV-1 infection for all subtypes A, B, D, and CRF_AE, whereas X4 or R5X4 viruses emerge in about 50% of late HIV disease (reviewed in Ref. 12), accompanied by a marked increase in CD4+ T cell depletion (13, 14). Subtype C is the exception with R5 variants dominating all stages of disease, including late stage AIDS, with only limited X4 variants being described or characterized (15–19).
CXCR4 is a seven-transmembrane receptor coupled to a pertussis toxin (PTX)-sensitive heterotrimeric Gi protein, which modulates the levels of intracellular cAMP by inhibiting the activity of adenylate cyclase. It also links to phosphoinositide 3-kinase, which is intrinsically linked to cell motility by promoting reorganization of the actin cytoskeleton (20). CXCR4 also activates Src kinase, which, in conjunction with other protein-tyrosine kinases of the Syk, Tec, and focal adhesion kinase families, promotes the activation of mitogen-activated protein kinases (MAPK) and Ras-related GTPases that change the transcriptional profile of the cell and promote actin remodeling (20). IL-4 specifically enhances cell surface expression of CXCR4 on resting CD4+ T cells (21), whereas stimulation of resting CD4+ T cells with phytohemagglutinin (PHA), anti-CD3, or anti-CD28 antibodies leads to down-regulation. Th2 cells express variable levels of CXCR4; however, CXCR4 is undetectable on polarized Th1 cells (21).
To test our hypothesis that the chemotactic and polarizing nature of CCL2 drives the recruitment of target cells to the site of HIV infection favoring viral infection and eventually a higher viral load in infected individuals, we investigated the modulatory effects of CCL2 on CXCR4 expression on resting CD4+ T cells and tested the effect of CCL2-induced CXCR4 expression on gp120-induced chemotaxis and HIV-1 infection.
EXPERIMENTAL PROCEDURES
Reagents
Cytokines and Chemokines—The human recombinant cytokine interleukin-2 (IL-2) was purchased from Roche Applied Science. The human recombinant chemokines (CXC motif) ligand-12 (CXCL12) and CCL2 were purchased from R & D Systems (Minneapolis, MN) and Invitrogen, respectively.
Chemicals and Proteins—The CXCR4 antagonist AMD3100, the extracellular signal-regulated kinase (Erk) 1/2 inhibitor U0126, the CCR2b antagonist RS102895, pertussis toxin (PTX) from Bordetella pertussis, and phytohemagglutinin (PHA) from Phaseolus vulgaris were purchased from Sigma. The possible cytotoxic effect of the different inhibitors and antagonists was tested at the concentrations used in the experiments by the trypan blue dye exclusion assay, and none was found to be cytotoxic (viability was >99%). HIV-1MN gp120 was purchased from Immunodiagnostics (Woburn, MA). HIV-1SF162 gp120 was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health. Anti-CD3 antibody (clone HIT3a) was purchased from eBioscience (San Diego, CA). Anti-CD4-peridin-chlorophyll protein, anti-CD3-fluorescein isothiocyanate (FITC), anti-CD3, anti-CXCR4-phycoerythrin (PE), anti-CCR5-allophycocyanin (APC), anti-CD14-APC, anti-CD14-FITC, anti-CD25-APC, anti-CD69-APC, anti-human leukocyte antigen-DR (HLA-DR)-PE, and anti-phospho-Erk 1/2 (T202/Y204)-Alexa Fluor 647 were purchased from Pharmingen. Anti-plectin (C-20) and anti-β-actin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and Sigma, respectively.
CD4+ T Cells—Human peripheral blood mononuclear cells (PBMC) were isolated from the heparinized blood of healthy HIV-1 seronegative donors by density centrifugation over Ficoll-Hypaque (Amersham Biosciences). CD4+ T cells were isolated from PBMC by negative selection using the CD4+ T cell isolation kit from Miltenyi Biotec (Auburn, CA) in accordance with the manufacturer's instructions. The final cultures of CD4+ T cells were always >95% pure, as determined by two-color flow cytometry analysis using anti-CD4-PerCp and anti-CD3-FITC. After purification, cells were resuspended in AIM-V medium with human serum albumin (AIM-V; Invitrogen) at 1 × 106 CD4+ T cells/ml for 24 h to allow for maximal unstimulated expression of CXCR4 before use.
Viruses—The following viruses were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health: HIV-1Lai was from Dr. Jean-Marie Bechet and Dr. Luc Montagnier (22, 23), and HIV-193In905 was from Dr. Robert Bollinger and the UNAIDS Network for HIV Isolation and Characterization, and the DAIDS, NIAID (24). Both were propagated on PHA/IL-2-activated PBMC, treated with RNase-free DNase I (Invitrogen), and purified using Vivaspin 20 columns with a 300,000 molecular weight cutoff (Sartorius Stedim Biotech, Edgewood, NY). The 50% tissue culture infective dose (TCID50) was determined by serial dilution and calculated using the Spearman-Kärber method as described elsewhere (25). The HIV-1 p24 antigen concentration in culture supernatants was determined using the Alliance HIV-1 p24 antigen enzyme-linked immunosorbent assay kit (PerkinElmer Life Sciences).
Flow Cytometry
All flow cytometry was performed on a FACSCalibur flow cytometer (BD Biosciences). All cytogram analysis was performed using CellQuest Pro®software (BD Biosciences).
CXCR4, CCR5, CD25, CD69, HLA-DR, and CD45RO surface expression was investigated by flow cytometry. CD4+ T cells (106/ml) were incubated with increasing concentrations of CCL2 at 37 °C, 5% CO2 in AIM-V for 4 h. Cells were then harvested and stained with appropriate antibodies, fixed in Dulbecco's phosphate-buffered saline (DPBS) supplemented with 4% (w/v) paraformaldehyde, and analyzed. Total CXCR4 content was evaluated by surface staining for CXCR4, permeabilizing with Cytofix-Cytoperm (BD Biosciences), and then re-staining for CXCR4. Nonspecific fluorescence was assessed by using isotype-matched controls. Results are expressed as follows: ((mean fluorescence intensity in treated cells)/(mean fluorescence intensity in untreated cells)) × 100.
For intracellular Erk 1/2 MAPK phosphorylation, CCL2-conditioned cells or unconditioned controls were harvested and treated with gp120 for 2 min. The reaction was stopped by adding an equal volume of glacial DPBS supplemented with 2% (w/v) paraformaldehyde. Cells were permeabilized in 90% glacial methanol overnight at 4 °C, washed twice in DPBS supplemented with 1% (w/v) bovine serum albumin and 0.1% (w/v) NaN3, and stained with anti-phospho-Erk 1/2 (T202/Y204)-Alexa Fluor 647.
Filamentous actin (F-actin) polymerization following the addition of gp120 was assessed mainly as described previously (26). Briefly, CD4+ T cells (107/ml) were incubated in RPMI 1640 medium containing 20 mm Hepes (both Invitrogen) and gp120MN. Every 15 s, 100 μl of cell suspension was added to 400 μl of DPBS supplemented with 4.5% (w/v) paraformaldehyde, 100 nm FITC-labeled phalloidin, and 125 μg/ml l-α-lysophosphatidylcholine (both Sigma). Results are expressed as ((median fluorescence intensity after addition of ligand)/(median fluorescence intensity before addition of ligand)) × 100.
Receptor internalization was measured by retention of anti-CXCR4 antibody binding following elimination of cell surface-bound antibody via acid wash as described previously (27). Receptor internalization was quantified as the fraction of total anti-CXCR4 fluorescence intensity (pH 7 wash) retained following acid wash (pH 4).
Western Blot Analysis
CD4+ T cells were treated with CCL2 for 4 h, washed three times with ice-cold DPBS, and lysed in CelLytic M (Sigma). Cell debris was removed by centrifugation (15,000 × g, 15 min). 40 μg per sample was then heated at 70 °C for 10 min in NuPAGE lithium dodecyl sulfate sample buffer containing reducing agent and separated by 3–8% Tris acetate-polyacrylamide gel (Invitrogen) and transferred to nitrocellulose membranes (Fisher). Membranes were blocked by 1 h of incubation at room temperature with DPBS supplemented with 0.1% (v/v) polysorbate 20 (Sigma) and 5% (w/v) dried nonfat milk (Genesee Scientific, San Diego) and then probed with polyclonal antibodies against plectin followed by detection using the WesternBreeze chemiluminescence kit (Invitrogen). Membranes were subsequently stripped and reprobed with anti-β-actin antibody to confirm equal loading. The relative density of the protein bands was analyzed using the freely available ImageJ (National Institutes of Health).
Real Time-PCR
CXCR4 mRNA expression was measured by real time PCR. Target cells were harvested and total cellular RNA prepared with the RNeasy mini kit using the optional DNase step in accordance with the manufacturer's directions (Qiagen, Valencia, CA). CXCR4 mRNA expression in relation to RNA polymerase II (RPII) expression (internal standard) was determined using the LightCycler System and the FastStart RNA Master SYBR Green I kit (Roche Applied Science) according to the manufacturer's instructions. Primers used were as follows: CXCR4 sense 5′-CAGCGGTTACCATGGAGGG-3′ and antisense 5′-TTCCTCGGTGTAGTTATCTGAAGTGT-3′; RPII sense 5′-GCACCACGTCCAATGACAT-3′ and antisense 5′-GTGCGGCTGCTTCCATAA-3′. Amplification was performed for 45 cycles, with the following cycle parameters: 5 s of denaturation at 95 °C, 12 s of primer annealing at 52 °C, and 15 s of fragment elongation at 72 °C.
The quantity of HIV-1 long terminal repeat (LTR) DNA in relation to RPII (internal standard) was determined using the LightCycler System and the FastStart DNA Master SYBR Green I kit (Roche Applied Science). PCRs were carried out in a 20-μl mixture composed of 3 mm MgCl2, 0.5 μm of each primer, 5 μl of sample, and 1-fold LightCycler FastStart DNA Master SYBR Green I. The primers for HIV-1 amplify a 128-bp fragment in the LTR R/U5 region, which was designed to detect early steps in reverse transcription: LTR sense 5′-CTCTCTGGCTAACTAGG-3′ and antisense 5′-ACTGACTAAAAGGGTCT-3′. The reaction mixture was initially incubated at 95 °C for 10 min to denature the DNA. Amplification was performed for 45 cycles, with the following cycle parameters: 10 s of denaturation at 95 °C, 10 s of primer annealing at 62 °C, and 15 s of fragment elongation at 72 °C.
Quantification and melting curve were analyzed with Light-Cycler analysis software, RelQuant (Roche Applied Science). All results are expressed as the ratio between the copy number of the target gene and the copy number of RPII and normalized so that CXCR4 mRNA expression and HIV-1 LTR in unconditioned cells equals 1.00.
Chemotaxis
The cell migration of CD4+ T cells toward a gp120 gradient after 4 h of conditioning with CCL2 in AIM-V was evaluated using 24-well Transwell migration chambers with 5-μm pore size polyvinylpyrrolidone-free polycarbonate filters (Corning Life Sciences, Lowell, MA) as described previously (5, 28) using gp120 in the basal chamber and 5 × 106 cells/ml in the apical chamber. Data are expressed as the percentage of migrated cells and are representative of three independent experiments carried out in triplicate.
HIV Infection Assay
After 4 h of conditioning with CCL2, CD4+ T cells were infected with X4-tropic HIV-1Lai or the R5-tropic HIV-193In905 at a multiplicity of infection of 0.01 for 3 h and then washed three times with DPBS. Cells were then cultured in RPMI 1640 medium supplemented with 15% (v/v) fetal bovine serum, 2 mm glutamine, and antibiotics (all Invitrogen). After 14 h, cells were either harvested and total DNA prepared with the QIAamp DNA mini kit in accordance with the manufacturer's directions (Qiagen, Valencia, CA) or, alternatively, media were supplemented with 20 units/ml IL-2, and cells were cultured for a further 96 h. Supernatants were then collected and assayed using the Alliance HIV-1 p24 antigen enzyme-linked immunosorbent assay kit (PerkinElmer Life Sciences).
Statistics
The results are expressed as the means ± S.E. of three or more experiments performed in triplicate. All p values correspond to two-tailed t tests.
RESULTS
CXCR4 Antibody-binding Sites Are Increased on CCL2-treated Resting CD4 CD4+ T Cells—We first examined whether CCL2 could activate or regulate surface expression of receptors on CD4+ T cells that had been freshly isolated from healthy donors and cultured for 24 h as described under “Experimental Procedures.” CD4+ T cells were exposed to CCL2 for 4 h, harvested, stained, and analyzed by flow cytometry for the surface expression of CD4, CD3, CD25, CD45RO, CD25, CD69, HLA-DR, CXCR4, and CCR5. We observed a CCL2-specific dose-dependent increase in the surface expression of CXCR4 (Fig. 1A). Flow cytometry demonstrated that stimulation with CCL2 did not alter the expression of any activation markers (HLA-DR, CD25, or CD69) and had no effect on the surface expression of CD4, CD3, CCR5, or CD45RO (data not shown). Furthermore, although natural variations in CXCR4 surface expression exist between individuals, we observed a uniform up-regulation across all donors tested.
FIGURE 1.
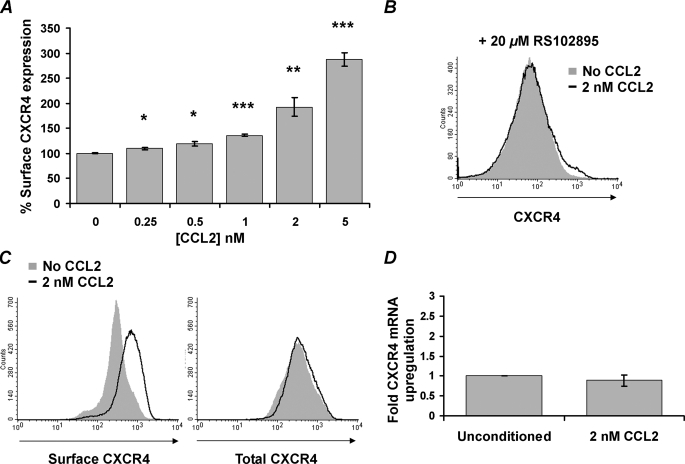
CCL2 induces up-regulation of surface CXCR4 expression on resting CD4+ T cells through a CCR2-dependent mechanism that does not require de novo protein synthesis. A, purified resting CD4+ T cells from HIV-negative subjects were incubated with increasing concentrations of CCL2. After 4 h, cells were harvested and stained for surface CXCR4. CCL2 induced a dose-dependent increase in surface expression of CXCR4. Data are expressed as means ± S.E. of the mean fluorescence intensity calculated from three independent experiments. B, purified resting CD4+ T cells were pretreated with 20 μm RS102895 and cultured for 4 h in the presence of 2 nm CCL2 (black line histogram) or with vehicle control (solid gray histogram). Cells were then harvested and stained for surface CXCR4. Histograms are shown for a representative donor. C, purified resting CD4+ T cells were incubated with 2 nm CCL2 (black line histogram) or with vehicle control (solid gray histogram) for 4 h. Cells were then stained for surface CXCR4 or permeabilized and stained for total CXCR4. Histograms are shown from a representative donor. D, CXCR4 mRNA content was evaluated using real time PCR after 4 h of incubation with 2 nm CCL2. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
We tested the hypothesis that CCL2 triggers the up-regulation of surface CXCR4 expression through a CCR2-dependent mechanism by incubating the cells with RS102895, a potent and specific spiropiperidine class inhibitor for CCR2b. This resulted in the complete inhibition of surface CXCR4 up-regulation (Fig. 1B). To determine whether the CCL2-induced up-regulation of surface CXCR4 was because of de novo protein synthesis or merely the relocation of pre-existing intracellular receptor to the cell surface, CD4+ T cells were incubated with 2 nm CCL2, and the expression of both surface and total (intracellular and surface) CXCR4 was quantified by flow cytometry (Fig. 1C), and CXCR4 mRNA was quantified by real time PCR (Fig. 1D). Although cell surface expression of CXCR4 increases significantly (1.92-fold increase, p < 0.0007), there was no significant increase of either total CXCR4 (mean 100% versus 97.96%, p = 0.5735; Fig. 1C) or CXCR4 mRNA (fold increase 1 versus 0.89, p = 0.4662; Fig. 1D).
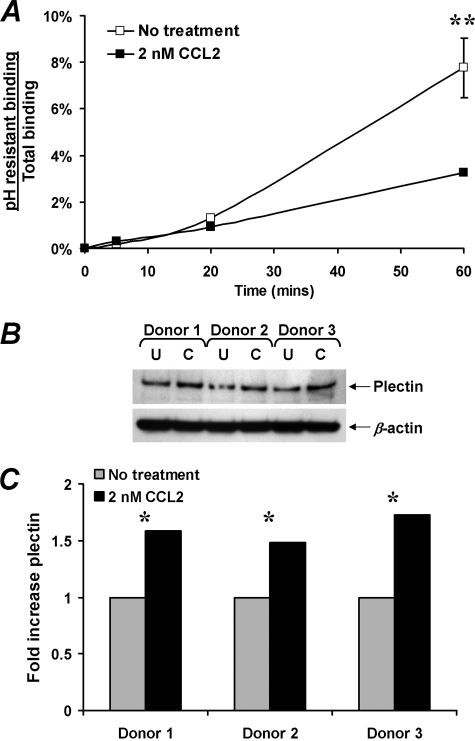
CCL2 Reduces CXCR4 Internalization—To determine whether altered receptor trafficking might play a role in CCL2 up-regulation of CXCR4 cell surface expression, receptor internalization rates were quantified by flow cytometric analysis of internalized anti-CXCR4 antibody following removal of surface-bound antibody by acid washing. Residual antibody binding reflects internalized receptors not exposed to extracellular low pH (29). Low pH washing efficiently removed cell surface-bound antibody as demonstrated by the abrogation of the fluorescent signal of both anti-CD4-PerCp and anti-CXCR4-PE (data not shown). After 1 h of incubation at 37 °C, acid-resistant anti-CXCR4 antibody binding averaged 7% of total CXCR4 fluorescence intensity (Fig. 2A). Parallel incubation in the presence of 2 nm CCL2 decreased acid-resistant anti-CXCR4 Ab binding by 42%, to an average of 3.2% of total fluorescence intensity at 60 min (p = 0.03). Thus, CCL2 appears to up-regulate cell surface expression of CXCR4 in part by reducing receptor internalization rates rather than by increasing the size of the total CXCR4 receptor pool.
FIGURE 2.
Effect of CCL2 on CXCR4 internalization and on plectin content. A, CXCR4 internalization was quantified by flow cytometric assessment of anti-CXCR4 antibody binding following acid stripping of cell surface-bound antibody. Acid-resistant binding (internalized CXCR4) is expressed as a fraction of total antibody binding over 1 h of incubation at 37 °C. CCL2 reduced CXCR4 internalization rates by 42% across four experiments (**, p = 0.03). B, effect of CCL2 on plectin expression. Western blot was used to quantify plectin levels in resting CD4+ T cells cultured for 4 h in the presence of 0 (U) or 2 nm CCL2 (C). Parallel determination of β-actin verified equivalent protein loading. C, densitometric analysis of the protein bands revealed that CCL2 conditioning of resting CD4+ T cells significantly increased plectin content (*, p = 0.01).
In the absence of ligand binding, the precise mechanisms that regulate CXCR4 signaling and intracellular trafficking are not well understood. However, recent studies have suggested a role for plectin, a cytolinker protein that plays an important role as a scaffolding platform for proteins involved in cellular signaling, and in CXCR4 signaling and trafficking (30). Therefore, to determine whether CCL2 has an effect on plectin expression, plectin levels were quantified by Western blot (Fig. 2B). We found that CCL2 significantly increased the expression of plectin in CD4+ T cells (mean fold increase 1 versus 1.6, p = 0.01; Fig. 2C). These data suggest that plectin may play a role in CCL2-dependent up-regulation of surface CXCR4 expression.
CCL2 Increases CXCR4 Signaling, Leading to an Increase in X4 gp120-induced Actin Polymerization and Chemotaxis—Using a Transwell assay, we tested the hypothesis that CCL2-induced surface CXCR4 up-regulation would enhance CD4+ T cell chemotaxis toward gp120. Initially, we compared the chemotactic effect of 200 nm X4 gp120MN, the reported Kd value of X4-tropic gp120 for CXCR4 (31), on unconditioned resting CD4+ T cells with known CXCR4 expression levels. We observed that CD4+ T cells with greater CXCR4 surface expression had a higher chemotactic index than those with lower surface expression, but the difference was not significant (data not shown). We then compared the percentage of unconditioned resting CD4+ T cells that migrated toward gp120 to those with prior conditioning with 1 nm CCL2 for 4 h. CD4+ T cell migration toward 200 nm X4 gp120MN was enhanced 43% by prior exposure to 1 nm CCL2 (p < 0.0001), whereas migration toward R5 gp120SF162 remained unaffected (Fig. 3). Preincubation of CD4+ T cells with 20 μm RS102895 before CCL2 exposure reduced migration toward X4 gp120MN to 34%, the same as for non-CCL2 treated X4 gp120MN-specific migration. All chemotactic activity toward gp120MN was abrogated by pretreating the cells with 1 μm AMD3100 or 1 μg/ml PTX, indicating a requirement for CXCR4 and Gαi proteins in X4 gp120MN-induced migration. All chemotactic responses to the R5 gp120SF162 were at significantly inferior levels to that of gp120MN (p ≤ 0.0001) and were unaffected by treatment with either AMD3100 or PTX. To establish whether anti-CD3 stimulation would inhibit the migration of CD4+ T cells toward X4 gp120MN, the response of CD4+ T cells pretreated with 10 μg/ml anti-CD3 antibody was evaluated. The response was reduced to 2% of the migration of cells in response to X4 gp120MN without prior anti-CD3 treatment (p < 0.0001).
FIGURE 3.
CD4+ T cell migration induced by 200 nm gp120 protein alone or with prior conditioning with 1 nm CCL2 and/or inhibitors. Bars represent the mean ± S.E. number of migrated cells of three independent experiments performed in triplicate. Cells not pretreated with an inhibitor or antibody are represented by white bars. In some instances the cells were also treated with 20 μm RS102895 (black bars), 1 μm AMD3100 (gray bars), 1 μg/ml PTX (white striped bars), or 10 μg/ml anti-CD3 antibody (gray striped bars). Experiments were performed three times in triplicate. Chemokinesis was minimal when equal concentrations of gp120 were added to both the apical and basal chambers and has been subtracted from these histograms. X4 gp120MN induced chemotaxis of CD4+ T cells and was augmented by preconditioning with 1 nm CCL2. The augmentation was abrogated in the presence of RS102895 (p < 0.0001). Treatment with AMD3100, PTX, or anti-CD3 antibody inhibited X4 gp120MN induced chemotaxis. R5 gp120SF162 induced minimal chemotaxis and was unaffected by all inhibitor and stimulus treatments. *, p < 0.0001.
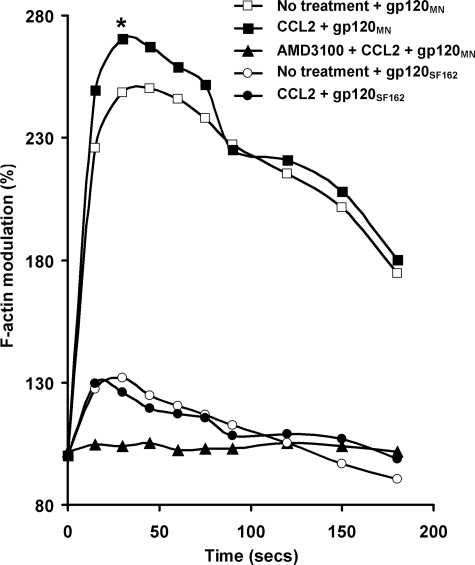
Cell polarization and motility are governed by actin cytoskeleton rearrangements. Actin turnover is also involved in HIV-1 entry, which requires disintegration and reconstitution of the membrane structure. Thus, by inducing F-actin polymerization, CXCR4 can regulate both X4 HIV-1 entry and chemotaxis. CD4+ T cells conditioned with 1 nm CCL2 for 4 h were treated with 200 nm gp120 and probed with FITC-phalloidin for F-actin polymerization at various time points. F-actin polymerization was rapidly induced following the addition of 200 nm X4 gp120MN. Kinetics in all instances was rapid with the peak response achieved within 30 s post-stimulation and was completely abrogated in the presence of AMD3100, indicating a dependence on signaling through CXCR4. CCL2 conditioning increased the gp120MN-induced F-actin polymerization (p = 0.022; Fig. 4). The R5-tropic gp120SF162 also induced rapid polymerization of F-actin, although we observed no difference between the CCL2-conditioned and nonconditioned cells (1.32-fold increase versus 1.33-fold increase, p = 0.89). The peak response achieved using gp120SF162 in nonconditioned cells was significantly less compared with gp120MN alone (2.48-fold increase versus 1.32-fold increase, p = 0.033; Fig. 4).
FIGURE 4.
CCL2 augments X4 gp120MN-induced F-actin polymerization in resting CD4+ T cells. Using FITC-phalloidin as a probe for intracellular F-actin, the effects of CCL2 pre-conditioning on F-actin polymerization in resting CD4+ T cells was assessed by flow cytometry. Results show the kinetics of F-actin polymerization following stimulation of unconditioned cells with R5 gp120SF162 (open circles) or X4 gp120MN (open squares). The change in F-actin content is expressed as a relative fold change in the median fluorescence intensity, when the base-line fluorescence intensity before addition of ligand (time 0) is expressed as 100%. CCL2 preconditioning of resting CD4+ T cells significantly increased the content of X4 gp120-induced F-actin (solid squares; *, p = 0.022) but had no effect on R5 gp120-induced F-actin (solid circles; p = 0.89). AMD3100 inhibited the increase in X4 gp120-induced F-actin content (solid triangles). All data presented are representative of three independent experiments.
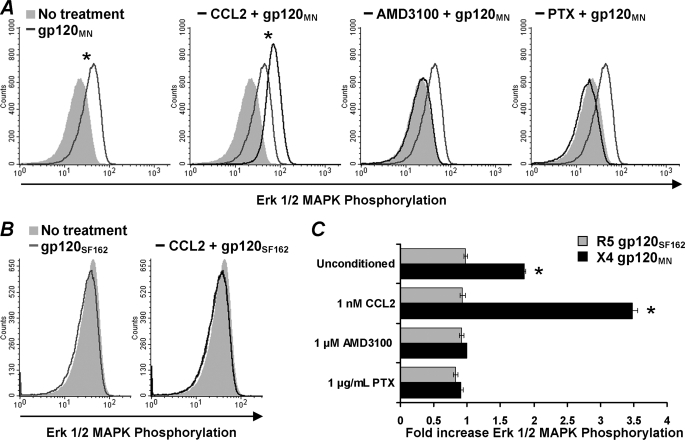
CCL2 Enhances X4 gp120-induced Erk 1/2 Phosphorylation in Primary CD4+ T Cells—Erk 1/2 has been implicated in regulating chemotaxis in some systems (32, 33). Therefore, we analyzed whether gp120 could induce the phosphorylation of Erk 1/2 in resting unstimulated CD4+ T cells. At both 40 and 80 nm, we observed no effect of either gp120MN or gp120SF162 (data not shown). However, at 200 nm, the reported Kd value of X4-tropic gp120 for CXCR4 (31), X4 gp120MN induced significant phosphorylation of Erk 1/2 MAPK (1.86-fold increase, p < 0.0001; Fig. 5). Furthermore, Erk 1/2 MAPK phosphorylation was greater in cells conditioned with 1 nm CCL2 than in unconditioned cells (1.86-fold versus 3.47-fold increase, p < 0.0001; Fig. 5). X4 gp120MN-induced phosphorylation of Erk 1/2 MAPK was abrogated if cells were pretreated with either AMD3100, a specific CXCR4 antagonist, or PTX, which irreversibly inhibits the Gi subunits of heterotrimeric G proteins by ADP-ribosylation, indicating the inhibition of the Ras/MAPK pathway and a requirement for CXCR4 in X4 gp120MN-induced Erk 1/2 phosphorylation; conditioning with CCL2 had no effect on this result (Fig. 5). Conversely, the R5-tropic gp120SF162 had no effect on Erk 1/2 MAPK phosphorylation at either its Kd for CCR5, 6 nm (34; data not shown) or at 200 nm (Fig. 5).
FIGURE 5.
CCL2 augments X4 gp120-induced Erk 1/2 phosphorylation in primary resting CD4+ T cells. Primary resting CD4+ T cells were stimulated with gp120 for 2 min, fixed, permeabilized/ and stained with a fluorescently tagged antibody specific for phosphorylated Erk 1/2 and then analyzed by flow cytometry. A, Erk 1/2 phosphorylation induced by X4 tropic gp120MN. Histograms shown are from a representative donor. Solid gray histogram in each image corresponds to cells not stimulated with 200 nm gp120MN. Light gray line corresponds to unconditioned cells stimulated with 200 nm gp120MN. Black line corresponds to pretreatment with either 1 nm CCL2, 1 μm AMD3100, or 1 μg/ml PTX. B, Erk 1/2 phosphorylation induced by R5-tropic gp120SF162. Histograms shown are from a representative donor. Solid gray histogram in each image corresponds to cells not stimulated with 200 nm gp120SF162. Light gray line corresponds to unconditioned cells stimulated with 200 nm gp120SF162. Black line in -CCL2 corresponds to pretreatment with 1 nm CCL2. C, bars represents the mean ± S.E. of the relative change in fluorescence intensity indicative of Erk 1/2 phosphorylation after stimulation with X4 gp120MN (black bars) or R5 gp120SF162 (gray bars) from three independent donors carried out in triplicate. The change in Erk 1/2 phosphorylation is expressed as the relative fold change in the mean fluorescence intensity, with the base-line fluorescence intensity before the addition of gp120 expressed as 1.00. CCL2 preconditioning of resting CD4+ T cells significantly increased X4 gp120-induced phosphorylation of Erk 1/2 (p < 0.0001) but had no effect on R5 gp120-induced events. AMD3100 and PTX inhibited Erk 1/2 phosphorylation. *, p < 0.0001.
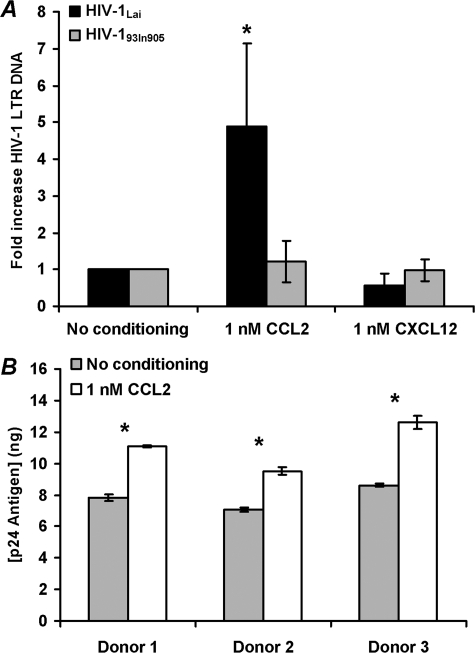
CCL2 Increases Entry of X4-tropic HIV-1Lai but Not R5-tropic HIV-193In905 into Resting CD4+ T Cells—The previous results suggest that CCL2 conditioning of resting CD4+ T cells selectively affects X4-tropic HIV-1 gp120-CXCR4 interactions. Therefore, we tested the hypothesis that increased surface expression of CXCR4 will enhance X4 HIV-1 entry into resting CD4+ T cells while having no effect on R5 HIV-1 entry. To test this hypothesis directly, we measured viral entry by using PCR. Highly purified resting CD4+ T cells were conditioned with 1 nm CCL2 or vehicle control for 4 h before infection with HIV-1 (multiplicity of infection of 0.01) for 3 h. After an additional 14 h culture in fresh medium, samples were analyzed by real time PCR for the presence and quantity of strong-stop HIV-1 DNA (with LTR R/U5 primers), an early product of reverse transcription. CCL2 conditioning significantly enhanced X4-tropic HIV-1IIIB infection 5-fold (p = 0.039; Fig. 6A). Conversely, there was no enhancement of R5-tropic HIV-193In905 infection (p = 0.36).
FIGURE 6.
CCL2 augments X4 HIV-1 infection of primary resting CD4+ T cells. A, purified resting CD4+ T cells were conditioned with 1 nm CCL2, 1 nm CXCL12, or left unconditioned for 4 h before infection with X4-tropic HIV-1Lai (black bars) or the R5-tropic HIV-193In905 (gray bars) at a multiplicity of infection of 0.01 for 3 h. Cells were then cultured for 14 h without further stimulation. Entry of HIV-1 was evaluated by real time PCR using primers specific for the LTR R/U5 region as described under “Experimental Procedures” (means ± S.E.; n = 3). CCL2 conditioning significantly augmented X4-tropic HIV-1Lai infection of resting CD4+ T cells (*, p < 0.05) and had no effect on R5-tropic HIV-193In905 infection. B, cells infected with X4-tropic HIV-1Lai as described above were further cultured in the presence of 20 units/ml IL-2. After 96 h, supernatants were collected from unconditioned (gray bars) and CCL2-conditioned (white bars) infected cells and p24-quantified. CCL2 conditioning significantly augmented X4-tropic HIV-1Lai infection of resting CD4+ T cells (*, p < 0.05).
Virus replication was then induced using 20 units/ml IL-2 for a further 96 h, and p24 antigen was quantified at the end of this period. The increase in viral DNA found in CCL2-conditioned cells with the X4-tropic HIV-1IIIB correlated with a significant 35–46% increase in p24 antigen concentration at 4 days post-infection (p < 0.05; Fig. 6B).
We then tested the hypothesis that Erk 1/2 phosphorylation is important to the infection of primary resting CD4+ T cells with X4 HIV-1 by incubating the cells with U0126, a selective inhibitor of Erk 1/2, prior to infection. This resulted in the inhibition of viral replication at a point before the reverse transcription step (data not shown). Treatment of the cells with CCL2 had no effect on this inhibition.
DISCUSSION
In this study, we show that CCL2 up-regulates the expression of CXCR4 on the surface of resting CD4+ T cells through a dose- and CCR2-dependent mechanism that does not require de novo protein synthesis. Furthermore, we show that surface CXCR4 levels on resting CD4+ T cells are positively associated with both the efficiency of X4-tropic but not R5-tropic gp120 to chemoattract resting CD4+ T cells and of X4 but not R5 HIV-1 entry into resting CD4+ T cells. CXCR4 functional receptor protein is detectable in a diverse range of cells and, with its natural ligand CXCL12, is involved in the regulation of numerous biological processes, including fundamental roles in the development of numerous organs during embryonic development, proliferation, survival, and the retention of primitive hematopoietic progenitor cells in the bone marrow. The CXCR4/CXCL12 pair have been shown to be critically involved in inflammatory conditions, such as rheumatoid arthritis, and they are often highly overexpressed in cancers contributing to tumor metastasis, as well as CXCR4 acting as a coreceptor for X4-tropic HIV infection.
The CCL2-mediated CXCR4 up-regulation was not accompanied by the up-regulation of other cell markers such as HLA-DR, CD69, CD25, CD45RO, CCR5, CD4, and CD3. Moreover, the inability of CCL2 to up-regulate the activation markers CD69, CD25, and HLA-DR suggests that the CCL2-induced up-regulation of CXCR4 is not dependent upon activation of these cells. The efficient inhibitory effect of RS102895 on the CCL2-induced increase in the CXCR4 surface expression suggests that the CCL2-mediated up-regulation of CXCR4 is through a CCR2b-dependent mechanism. Furthermore, the inability of CCL2 to induce an increase in total CXCR4 receptor or an increase in CXCR4 mRNA, combined with the reduction in CXCR4 internalization rates and increase in plectin content, suggests that CCL2-mediated CXCR4 up-regulation is because of the relocation of preexisting receptor to the cell surface combined with a decrease in internalization rates that does not require de novo CXCR4 synthesis. CXCR4, like many G protein-coupled receptors, including the α1a-adrenoreceptor and the μ-opioid receptor, actively and continuously undergoes internalization in the absence of ligand, which fine-tunes the threshold at which cells sense ligand. The C-terminal cytoplasmic domain of CXCR4 is known to regulate its function and spatiotemporal expression (35) and is involved in the plectin complex formation (30). However, little is known about the mechanism underlying constitutive internalization and surface expression of CXCR4 compared with internalization mediated by its ligand CXCL12. The present data indicate that CCL2 up-regulates CXCR4 surface expression primarily by altering receptor trafficking through up-regulation of plectin. However, the precise mechanisms by which CXCR4 signaling and intracellular trafficking are regulated remain not fully understood.
We next determined whether CCL2 modulates the functional responses of CXCR4. X4 tropic gp120MN, when used as a ligand, induced CXCR4 signaling leading to F-actin polymerization, Erk 1/2 phosphorylation, and a chemotactic response more efficiently in CCL2-conditioned resting CD4+ T cells than in unconditioned cells. The response was CXCR4- and Gαi protein-specific as pretreating the cells with either AMD3100 or PTX abrogated the response; thus X4 tropic gp120MN functions as a full CXCR4 agonist. Treating CD4+ T cells with anti-CD3 antibody induced a marginal down-regulation of CXCR4 surface expression (data not shown), mediated via protein kinase C activation that phosphorylates CXCR4, followed by receptor internalization (36). However, it is unlikely that this down-regulation accounted for the almost complete inhibition of gp120MN-induced chemotaxis that we observed. Previous studies have shown that CXCR4 physically interacts with the TCR in response to signaling via CXCL12 and uses TCR signaling machinery from signal initiation to signal termination. CD3-mediated activation of protein kinase C leads to phospholipase C-β-3 phosphorylation, which in turn inhibits the activation of Gqα (37). However, it is also possible that the signaling pathways of both the TCR and CXCR4 use the same Gqα subunit. Therefore, by stimulating the cells with anti-CD3 antibody prior to the gp120 chemotaxis assay, CXCR4 is transiently desensitized, rendering the cell insensitive to the agonistic effects of the gp120MN-CXCR4 interaction.
In addition to the surface up-regulation of CXCR4 by CCL2, the enhanced X4 tropic gp120MN-induced chemotaxis signaling events that accompanied it suggested that CCL2 might increase the susceptibility of resting CD4+ T cells to infection by X4 HIV-1. The best characterized latently infected cells are resting CD4+ T cells that are formed while in an activated state prior to returning to a resting state (38). However, HIV-1 can be isolated from CD45RA+ naive CD4+ T cells in HIV-1-infected organ cultures and from HIV-1-infected individuals (39, 40). Moreover, resting CD4+ T cells can be latently infected in vivo without the requirement for a secondary stimulus (41). Together, this suggests that an activation step is not required for either HIV-1 entry or for HIV-1 integration. In this study we show that pretreatment of resting CD4+ T cells from peripheral blood with CCL2 increased the efficiency of X4 but not R5-tropic viral entry.
Genetic variations of CCL2 and CCR2 play a role in HIV-1 pathogenesis. In European-Americans, homozygosity for the CCL2-2518G allele, i.e. the GA/GA haplotype pair, is associated with accelerated progression to AIDS and death in infected adults (42). Moreover, in vitro studies show that PBMCs from individuals homozygous or heterozygous for CCL2-2518G produce larger amounts of CCL2 than PBMCs from individuals homozygous for CCL2-2518A (43) and that this correlated with in vivo sera studies (42). Interestingly, in an in vitro model of HIV-1 infection, blocking endogenous CCL2 production decreased viral replication (2). Furthermore, higher CCL2 levels correlate with higher HIV load (1) and enhanced leukocyte trafficking to tissues (42), possibly playing a role in the etiology of HIV-1-associated dementia. The CCR2-V64I allele also has a significant influence on the rate of HIV-1 disease progression in adults, with the presence of CCR2-V64I associated with a 2–3-year delay in progression to AIDS and death, although it has no impact upon HIV-1 transmission where CCR5 using variants are dominant (44). Interestingly, although PBMCs isolated from CCR2-V64I heterologous donors have normal levels of both CCR5 and CCR2, and the calcium flux mediated by CCL2 through CCR2-V64I was unaffected, they have reduced surface expression of CXCR4 (45). It is possible that in late stage HIV disease, the increase in CCL2 expression and secretion up-regulates the surface expression of CXCR4 on resting CD4+ T cells, increasing their permissiveness to X4 HIV-1 infection and contributing to the switch from R5 to X4 using virus and more rapid disease progression.
In summary, CCL2 up-regulates CXCR4 expression on resting CD4+ T cells through a CCR2-dependent mechanism that augments the ability of these cells to be chemoattracted to X4 gp120 and renders them more permissive to X4-tropic HIV-1 entry. Thus, CCL2 has the capacity to render a large population of lymphocytes more susceptible to X4 HIV-1 late in the course of infection.
Acknowledgments
We thank Carol Mundy and Dennis Young for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant AI068632 (NIAID). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HIV-1, human immunodeficiency virus type 1; HIV, human immunodeficiency virus; APC, allophycocyanin; CCL, chemokine (C-C motif) ligand; CCR2, chemokine (C-C motif) receptor 2; CXCL12, chemokine (CXC motif) ligand 12; CXCR4, chemokine (CXC motif) receptor 4; Erk, extracellular signal-regulated kinase; F-actin, filamentous actin; FITC, fluorescein isothiocyanate; HLA-DR, human leukocyte antigen-DR; IL-2 intereukin-2; LTR, long terminal repeat; MAPK, mitogen-activated protein kinase; PBMC, peripheral blood mononuclear cells; DPBS, Dulbecco's phosphate-buffered saline; PE, phycoerythrin; PHA, phytohemagglutinin; PTX, pertussis toxin; RPII, RNA polymerase II; TCR, T cell receptor.
References
- 1.Ansari, A. W., Bhatnagar, N., Dittrich-Breiholz, O., Kracht, M., Schmidt, R. E., and Heiken, H. (2006) Int. Immunol. 18 1443-1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mengozzi, M., De Filippi, C., Transidico, P., Biswas, P., Cota, M., Ghezzi, S., Vicenzi, E., Mantovani, A., Sozzani, S., and Poli, G. (1999) Blood 93 1851-1857 [PubMed] [Google Scholar]
- 3.Park, I. W., Wang, J. F., and Groopman, J. E. (2001) Blood 97 352-358 [DOI] [PubMed] [Google Scholar]
- 4.Fantuzzi, L., Canini, I., Belardelli, F., and Gessani, S. (2001) J. Immunol. 166 5381-5387 [DOI] [PubMed] [Google Scholar]
- 5.Campbell, G. R., Watkins, J. D., Singh, K. K., Loret, E. P., and Spector, S. A. (2007) J. Virol. 81 5919-5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsushima, K., Larsen, C. G., DuBois, G. C., and Oppenheim, J. J. (1989) J. Exp. Med. 169 1485-1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr, M. W., Roth, S. J., Luther, E., Rose, S. S., and Springer, T. A. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 3652-3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maghazachi, A. A., al-Aoukaty, A., and Schall, T. J. (1994) J. Immunol. 153 4969-4977 [PubMed] [Google Scholar]
- 9.Gu, L., Tseng, S., Horner, R. M., Tam, C., Loda, M., and Rollins, B. J. (2000) Nature 404 407-411 [DOI] [PubMed] [Google Scholar]
- 10.Kinter, A., Catanzaro, A., Monaco, J., Ruiz, M., Justement, J., Moir, S., Arthos, J., Oliva, A., Ehler, L., Mizell, S., Jackson, R., Ostrowski, M., Hoxie, J., Offord, R., and Fauci, A. S. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 11880-11885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berger, E. A. (1997) AIDS 11 Suppl. A, S3-S16 [PubMed] [Google Scholar]
- 12.Philpott, S. M. (2003) Curr. HIV. Res. 1 217-227 [DOI] [PubMed] [Google Scholar]
- 13.Koot, M., van't Wout, A. B., Kootstra, N. A., de Goede, R. E., Tersmette, M., and Schuitemaker, H. (1996) J. Infect. Dis. 173 349-354 [DOI] [PubMed] [Google Scholar]
- 14.Glushakova, S., Baibakov, B., Zimmerberg, J., and Margolis, L. B. (1997) AIDS Res. Hum. Retroviruses 13 461-471 [DOI] [PubMed] [Google Scholar]
- 15.Björndal, A., Deng, H., Jansson, M., Fiore, J. R., Colognesi, C., Karlsson, A., Albert, J., Scarlatti, G., Littman, D. R., and Fenyö, E. M. (1997) J. Virol. 71 7478-7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ping, L. H., Nelson, J. A., Hoffman, I. F., Schock, J., Lamers, S. L., Goodman, M., Vernazza, P., Kazembe, P., Maida, M., Zimba, D., Goodenow, M. M., Eron, J. J., Jr., Fiscus, S. A., Cohen, M. S., and Swanstrom, R. (1999) J. Virol. 73 6271-6281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abebe, A., Demissie, D., Goudsmit, J., Brouwer, M., Kuiken, C. L., Pollakis, G., Schuitemaker, H., Fontanet, A. L., and Rinke de Wit, T. F. (1999) AIDS 13 1305-1311 [DOI] [PubMed] [Google Scholar]
- 18.Batra, M., Tien, P. C., Shafer, R. W., Contag, C. H., and Katzenstein, D. A. (2000) AIDS Res. Hum. Retroviruses 16 973-979 [DOI] [PubMed] [Google Scholar]
- 19.Johnston, E. R., Zijenah, L. S., Mutetwa, S., Kantor, R., Kittinunvorakoon, C., and Katzenstein, D. A., (2003) J. Virol. 77 7682-7688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sotsios, Y., and Ward, S. G. (2000) Immunol. Rev. 177 217-235 [DOI] [PubMed] [Google Scholar]
- 21.Jourdan, P., Abbal, C., Noraz, N., Hori, T., Uchiyama, T., Vendrell, J. P., Bousquet, J., Taylor, N., Pène, J., and Yssel, H. (1998) J. Immunol. 160 4153-4157 [PubMed] [Google Scholar]
- 22.Barre-Sinoussi, F., Chermann, J. C., Rey, F., Nugeyre, M. T., Chamaret, S., Gruest, J., Dauguet, C., Axler-Blin, C., Vezinet-Brun, F., Rouzioux, C., Rozenbaum, W., and Montagnier, L. (1983) Science 220 868-871 [DOI] [PubMed] [Google Scholar]
- 23.Wain-Hobson, S., Vartanian, J. P., Henry, M., Chenciner, N., Cheynier, R., Delassus, S., Martins, L. P., Sala, M., Nugeyre, M. T., Guetard, D., Klatzmann, D., Gluckman, J. C., Rozenbaum, W., Barre-Sinoussi, F., and Montagnier, L. (1991) Science 252 961-965 [DOI] [PubMed] [Google Scholar]
- 24.Lole, K. S., Bollinger, R. C., Paranjape, R. S., Gadkari, D., Kulkarni, S. S., Novak, N. G., Ingersoll, R., Sheppard, H. W., and Ray, S. C. (1999)) J. Virol. 73 152-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Japour, A. J., Mayers, D. L., Johnson, V. A., Kuritzkes, D. R., Beckett, L. A., Arduino, J. M., Lane, J., Black, R. J., Reichelderfer, P. S., D'Aquila, R. T., Crumpacker, C. S., and the RV-43 Study Group and The AIDS Clinical Trials Group Virology Committee Resistance Working Group (1993) Antimicrob. Agents Chemother. 37 1095-1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balabanian, K., Harriague, J., Decrion, C., Lagane, B., Shorte, S., Baleux, F., Virelizier, J. L., Arenzana-Seisdedos, F., and Chakrabarti, L. A. (2004) J. Immunol. 173 7150-7160 [DOI] [PubMed] [Google Scholar]
- 27.Cole, S. W., Jamieson, B. D., and Zack, J. A. (1999) J. Immunol. 162 1392-1400 [PubMed] [Google Scholar]
- 28.Patrussi, L., Ulivieri, C., Lucherini, O. M., Paccani, S. R., Gamberucci, A., Lanfrancone, L., Pelicci, P. G., and Baldari, C. T. (2007) Blood 110 1730-1738 [DOI] [PubMed] [Google Scholar]
- 29.Pelchen-Matthews, A., Armes, J. E., and Marsh, M. (1989) EMBO J. 8 3641-3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding, Y., Zhang, L., Goodwin, J. S., Wang, Z., Liu, B., Zhang, J., and Fan, G. H. (2008) Exp. Cell Res. 314 590-602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babcock, G. J., Mirzabekov, T., Wojtowicz, W., and Sodroski, J. (2001) J. Biol. Chem. 276 38433-38440 [DOI] [PubMed] [Google Scholar]
- 32.Riol-Blanco, L., Sánchez-Sánchez, N., Torres, A., Tejedor, A., Narumiya, S., Corbí, A. L., Sánchez-Mateos, P., and Rodríguez-Fernández, J. L. (2005) J. Immunol. 174 4070-4080 [DOI] [PubMed] [Google Scholar]
- 33.Kumar, A., Humphreys, T. D., Kremer, K. N., Bramati, P. S., Bradfield, L., Edgar, C. E., and Hedin, K. E. (2006) Immunity 25 213-224 [DOI] [PubMed] [Google Scholar]
- 34.Wu, L., Gerard, N. P., Wyatt, R., Choe, H., Parolin, C., Ruffing, N., Borsetti, A., Cardoso, A. A., Desjardin, E., Newman, W., Gerard, C., and Sodroski, J. (1996) Nature 384 179-183 [DOI] [PubMed] [Google Scholar]
- 35.Futahashi, Y., Komano, J., Urano, E., Aoki, T., Hamatake, M., Miyauchi, K., Yoshida, T., Koyanagi, Y., Matsuda, Z., and Yamamoto, N. (2007) Cancer Sci. 98 373-379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peacock, J. W., and Jirik, F. R. (1999) J. Immunol. 162 215-223 [PubMed] [Google Scholar]
- 37.Stanners, J., Kabouridis, P. S., McGuire, K. L., and Tsoukas, C. D. (1995) J. Biol. Chem. 270 30635-30642 [DOI] [PubMed] [Google Scholar]
- 38.Han, Y., Wind-Rotolo, M., Yang, H. C., Siliciano, J. D., and Siliciano, R. F. (2007) Nat. Rev. Microbiol. 5 95-106 [DOI] [PubMed] [Google Scholar]
- 39.Eckstein, D. A., Penn, M. L., Korin, Y. D., Scripture-Adams, D. D., Zack, J. A., Kreisberg, J. F., Roederer, M., Sherman, M. P., Chin, P. S., and Goldsmith, M. A. (2001) Immunity 15 671-682 [DOI] [PubMed] [Google Scholar]
- 40.Kinter, A. L., Umscheid, C. A., Arthos, J., Cicala, C., Lin, Y., Jackson, R., Donoghue, E., Ehler, L., Adelsberger, J., Rabin, R. L., and Fauci, A. S. (2003) J. Immunol. 170 2449-2455 [DOI] [PubMed] [Google Scholar]
- 41.Agosto, L. M., Yu, J. J., Dai, J., Kaletsky, R., Monie, D., and O'Doherty, U. (2007) Virology 368 60-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzalez, E., Rovin, B. H., Sen, L., Cooke, G., Dhanda, R., Mummidi, S., Kulkarni, H., Bamshad, M. J., Telles, V., Anderson, S. A., Walter, E. A., Stephan, K. T., Deucher, M., Mangano, A., Bologna, R., Ahuja, S. S., Dolan, M. J., and Ahuja, S. K. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13795-13800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rovin, B. H., Lu, L., and Saxena, R. (1999) Biochem. Biophys. Res. Commun. 259 344-348 [DOI] [PubMed] [Google Scholar]
- 44.Smith, M. W., Dean, M., Carrington, M., Winkler, C., Huttley, G. A., Lomb, D. A., Goedert, J. J., O'Brien, T. R., Jacobson, L. P., Kaslow, R., Buchbinder, S., Vittinghoff, E., Vlahov, D., Hoots, K., Hilgartner, M. W., and O'Brien, S. J. (1997) Science 277 959-965 [DOI] [PubMed] [Google Scholar]
- 45.Lee, B., Doranz, B. J., Rana, S., Yi, Y., Mellado, M., Frade, J. M., Martinez-A., C., O'Brien, S. J., Dean, M., Collman, R. G., and Doms, R. W. (1998) J. Virol. 72 7450-7458 [DOI] [PMC free article] [PubMed] [Google Scholar]