Abstract
Claudin family transmembrane proteins play an important role in tight junction structure and function in epithelial cells. Among the 24 isoforms identified in mice and humans, claudin-4 and -3 serve as the receptor for Clostridium perfringens enterotoxin (Cpe). The second extracellular loop (Ecl2) of claudin-4 is responsible for the binding to the C-terminal 30 amino acids of Cpe (Cpe30). To define the structural constraints for the claudin-4/Cpe30 interaction, a surface plasmon resonance (SPR) method was developed. GST fusions with claudin-4 revealed that Ecl2 with the downstream transmembrane domain of claudin-4 reconstituted the basic structural requirement for optimal binding activity to Cpe30, with affinity in the nanomolar range. Two 12-mer peptides selected by phage display against claudin-4-transfected CHO cells and a 12-mer Cpe mutant peptide also showed significant affinity for claudin-4 with this SPR assay, suggesting that a short peptide can establish stable contact with Ecl2 with nanomolar affinity. Alignment of these short peptides unveiled a common Ecl2 binding motif: <XX(Y/W)(X)3 or 4Y(Y/X)(L/I)XX>. Whereas the short peptides bound native claudin-4 on transfected CHO cells in pull-down assays, only the larger Cpe30 peptide affected trans-epithelial electrical resistance (TER) in peptide-treated Caco-2BBe monolayers. Importantly, Cpe30 retained its binding to claudin-4 when fused to the C terminus of influenza hemagglutinin, demonstrating that its binding activity can be maintained in a different biochemical context. These studies may help in the design of assays for membrane receptor interactions with soluble ligands, and in applying new targeting ligands to delivering attached “cargo” proteins.
Claudin-4 (Cldn-4)2 is a tight junction transmembrane protein important in establishing trans-epithelial electrical resistance (TER) in the mucosal epithelial barrier (1, 2). It belongs to the claudin protein family with ∼24 members so far identified in the human and features two extracellular loops (Ecl1 and Ecl2) and four transmembrane domains (3). In addition to its function in tight junction formation, Cldn-4 (along with claudin-3) also functions as a receptor for Clostridium perfringens enterotoxin (Cpe). Treatment of epithelial cells at the basolateral side with C-Cpe (the non-toxic C terminus of Cpe) causes a decrease in TER and increase in paracellular permeability, concomitant with Cldn-4 endocytosis and degradation (2, 4). Immunofluorescence staining, Western blotting, and gene expression analysis demonstrate that Cldn-4 is highly expressed in colon, nasopharynx surface epithelia cells as well as on M-cells in Peyer's patch (5, 6, 7). Reports also show that Cldn-4 can be overexpressed in a variety of cancers including breast, prostate, and colorectal, suggesting a potential for therapies through Cldn-4 targeting (8).
Studies to date on the molecular mechanisms of Cldn-4/Cpe interaction have shown that the second extracellular domain (Ecl2) of Cldn-4 interacts with the last C-terminal 17 amino acids of Cpe (9). The structure of C-Cpe (amino acids 194–319) (10) reveals that two β-strands and an intervening large surface loop provide the structure that binds Cldn-4, with the three tyrosine residues (Tyr-306, Tyr-310, and Tyr-316) in this loop providing important contacts (11). An alanine scan also identified Tyr-306 and Leu-315 as indispensable for Cldn-4 binding and modulation of TJ function (12). The affinity of C-Cpe binding to Cldn-4 has been measured at micro- to subnanomolar levels by various approaches (2, 4).
Though the equilibrium affinity for Cpe binding to Cldn-4 has been studied, the kinetics for binding have not been reported. Because targeting in vivo using Cldn-4-binding peptides will be affected by the kinetics of the interaction, we sought more detailed information on the behavior of various Cpe-related peptides in their interactions with the Ecl2 of Cldn-4. In the present report, we identified several short Cldn-4-binding peptides and studied their binding by surface plasmon resonance (SPR). This technology enables quantitation of on- and off-rates as well as the equilibrium affinity (KD). In this method, a ligand or receptor is bound to a sensor chip, and its binding partner (analyte) in solution is allowed to flow across the sensor surface. Binding of the analyte is measured in real time as a change in resonance units (RU) detected by the sensor chip. Off-rates are similarly measured by the decrease in RU as analyte is washed off by buffer. The kinetics of binding is calculated by the rates of change in RU over time, so steeper slopes indicate faster on- or off-rates. While binding kinetics can be easily measured for soluble proteins, transmembrane protein conformation is more difficult to reproduce on a sensor surface, so different approaches are necessary. Thus, because Cldn-4 Ecl2 is formed by an extracellular loop with two transmembrane domains, we developed an approach using GST fusions with portions of claudin-4 to generate analytes that could reasonably mimic the behavior of the native proteins. From these studies, we were able to identify a Cldn-4 Ecl2 binding motif, and we were able to establish that a peptide as short as 12 amino acids was still able to bind Cldn-4 Ecl2 with nanomolar affinity. These findings will facilitate the application of Cldn-4-binding peptide for targeting in a variety of settings, including mucosal immunization and cancer therapy.
EXPERIMENTAL PROCEDURES
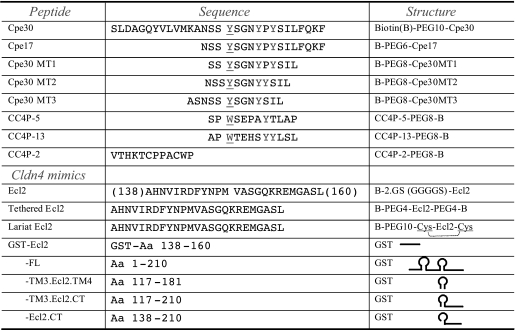
Materials—Peptides used in this study were synthesized by Abgent and AnaSpec. Peptides were synthesized by solid-phase synthesis procedure, purified by reverse phase HPLC to >98% purity. The detailed sequences of peptides were summarized in Table 1. For immobilization to streptavidin (SA) sensor chip (Biacore, GE), peptides were biotinylated at N or C terminus with GS (GGGGSGGGGS) or PEG (8- or 10-mer) linker to increase the accessibility of peptide. Peptides were dissolved in a small amount of solvent (e.g. DMSO at 5–10% final concentration) and brought up to specific concentrations by H2O; the pH was adjusted to neutral by phosphate buffer. Primers for subcloning were synthesized by IDT.
TABLE 1.
Peptides and Claudin-4 mimics used in this study Peptides were synthesized as a biotinylated form for conjugation to the streptavidin sensor chip; PEG or GS linker was used as a spacer to increase the accessibility of peptide. Claudin-4 deletion mutants were expressed as a GST recombinant protein to improve the solubility of this transmembrane protein. The dashed bracket in the “Lariat Ecl2” structure indicates the intramolecular disulfide bond.
Screening Cldn4-binding Peptides by Phage Display Library—A random 12-mer peptide phage display library (cat. E8110S, NEB) was used for screening. The library size is about 2.7 × 109 transformants. To select phage on the basis of binding to Cldn4 extracellular domains, CHO cells transfected with full-length Cldn-4 (CHO-Cldn-4) or a GFP-Cldn-4 (GFP-Cldn-4) fusion protein gene were employed for phage library selection. GFP-Cldn-4-transfected cells were sorted by flow cytometry for high green fluorescence resulting in a line with high GFP-Cldn-4 expression. The screening was performed according to the manufacturer's instruction with some modifications. Four rounds of panning were done, where the 1st round was selected by CHO-Cldn4 cells (“positive selection”). The 2nd round used CHO control cells (“negative selection”) to remove phage specific for CHO determinants. This second, negatively selected library was followed by a 3rd positive selection round with CHO-Cldn-4 cells, and an additional 4th negative selection round with CHO control cells. Phage clones were then isolated for sequencing of display peptide sequences, and abundant or repeated sequences were selected as candidate clones. Two positive binding peptides (CC4P-13, CC4P-5) (Table 1) were selected after 4 rounds of screening. The peptides were then synthesized for SPR assays.
Protein Expression and Purification—(a) Claudin-4: mouse claudin-4 (cldn4, NM_00903) was subcloned into pGEX4T-2 by PCR with high fidelity DNA polymerase Pfu (Stratagene). The primers for each deletion mutant were: full-length clnd4 by F1 (forward primer 1): 5′-GGATCCGCGATGGCGTCTATGGGAC-3′; R1 (reverse primer 1): 5′-CTCGAGTTACACATAGTTGCTGGCGGGG-3′; Ecl2 by F2: 5′-GGATCCATCATGATCACCGCCGGAG-3′; R2: 5′-CTCGAGTCAGAGGAGGCCTCCTCC-3′; TM3.Ecl2.CT by primer F2 and R1; and Ecl2.CT by F3: 5′-GGATCCTGGACCGCTCACAACG-3′ and primer R1. The underlined sequences are BamHI and XhoI restriction sites. The constructs were confirmed by DNA sequencing. GST-Cldn4/pGEX4T-2 construct was transformed into Escherichia coli (BL21, pLysS) for protein expression. The soluble protein was purified by glutathione-agarose affinity chromatography (Pierce), and the co-purified GST protein was separated by gel filtration chromatography on FPLC with Superdex 200 column. For use as the analyte in Biacore assay, GST-Cldn4 was balanced to HBS-EP buffer by Microcon (Millipore) centrifugation.
(b) Hemagglutinin (HA): HA from influenza A virus (A/Puerto Rico/8/34/Mount Sinai, AF389118) was used to produce recombinant HA protein. It was subcloned into pENTR3C vector via BamHI (5′) and EcoRV (3′) by PCR, and recombined into BaculoDirect Linear DNA (BaculoDirect™ Baculovirus expression system, Invitrogen). The resultant expression virus was used to express protein in insect cell (Sf9). The C-terminal 37 amino acids of HA containing the transmembrane domain were removed to enable secretion of soluble protein, and a trimerization sequence (ts, from Fibritin-C) was inserted to facilitate efficient trimerization of HA. Cpe30 (the C-terminal 30 amino acids of the C. perfringens enterotoxin) was introduced to the C terminus with upstream GS linkers (Fig. 6A). This recombinant HA (HA-Cpe30) was C-terminally His-tagged, and purified by HisPur Cobalt (Pierce) affinity chromatography.
FIGURE 6.
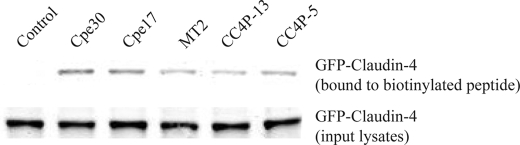
Pull-down assay for peptide binding to native claudin-4. Biotinylated peptides were added to CHO cells transfected with GFP-Claudin-4 fusion protein. Biotin-peptide complexes were pulled-down from cell lysates by Neutravidin beads, and the precipitated protein was tested for GFP-Claudin-4 by Western blot. Western blots of input cell lysates are also shown for reference. Data are representative of three separate experiments.
Measurement of Binding Kinetics by SPR—The Biacore X100 system (Biacore, GE) was used in this study. Streptavidin (SA) sensor chips were used to immobilize the biotinylated peptides. HBS-EP buffer (10 mm HEPES, 0.15 m NaCl, 3 mm EDTA, 0.005% Surfactant P20, pH7.4) was used as the binding buffer. General procedures were according to the manufacturer's instructions. Biotinylated peptide (e.g. Cpe30) was immobilized to the SA chip after first conditioning the chip surface with 50 mm NaOH, 1 m NaCl. The immobilized amount of the ligand was ∼500 RU to ensure efficient binding. The control channel was treated in the same way as assay channel but without peptide immobilized. The analyte was a GST-Cldn-4 protein comprised of GST fused at the C-terminal end to a fragment of Cldn4 from the second extracellular domain (Ecl2) through the C terminus. The titration was measured with purified GST-Cldn-4 from 10 to 100 nm in HBS-EP. GST protein only was employed as the control analyte. The binding was carried out at 25 °C with a flow rate at 30 μl/min, and data were collected for 2 min of association and 3 min of dissociation. In the other assay, an HA conformation sensitive antibody (H36) was immobilized to CM5 sensor chip by amine-coupling reaction; then recombinant HA protein was indirectly captured as the ligand to interact with GST-Cldn4.R4 analyte in HBS-EP buffer as above. All peptides used as ligands were synthesized with a PEG or GGGGSGGGGS peptide linker between the ligand peptide and a terminal biotin, to provide flexibility and accessibility. To analyze the data, the assay channel was subtracted by the control channel to eliminate nonspecific interaction. Multiple sensorgrams from different concentrations of analyte were overlaid and aligned, and kinetic constants were calculated by BIAevaluation 3.1 software with nonlinear fitting, the 1:1 (Langmuir) binding model was used, where KD = Kd/Ka.
Pull-down Assay for Peptide Binding to Native Claudin-4— GFP-Cldn-4-transfected CHO cells were incubated with 50 μm biotinylated peptides in culture media (F-12) with 1% bovine serum albumin for 1 h at 4°C. Cells were washed once with phosphate-buffered saline, then lysed (lysing buffer: 50 mm sodium phosphate, 150 mm NaCl, 0.1% Nonidet P-40, 0.05% deoxycholate, pH 7.4, plus protease inhibitors), and biotinylated peptides were extracted using Neutravidin beads. The pelleted protein complexes were assayed for bound GFP-Cldn-4 by Western blot specific for Cldn-4. The control peptide used was a 12-mer from a phage display clone (sequence: WHKHRVSPEIEW) established from SPR analysis to have no measurable affinity for Cldn4.
TER Assays—Caco-2BBe intestinal epithelial cells were cultured on transwell filters (0.4-micron pore) for 7 days when tight junction formation established high trans-epithelial electrical resistance. Peptides (100 or 200 μm) were added to either the basal or apical sides of the monolayers. TER readings (Endohm 6/EVOM ohmmeter, World Precision Instruments) were taken before, 4 h, and 24 h after peptide treatment (in some cases 14 h readings were also taken). A final reading was taken after washing the peptides out to confirm recovery of the epithelial cells. Triplicate cultures were used for each condition.
RESULTS
Claudin-4 Ecl2 in the Context of Transmembrane Domains Provides the Conformation for Cpe30 Binding—Claudin-4 was initially cloned as the Cpe receptor (CPE-R); consequently, Cpe, as a naturally occurring ligand, has proven to be a useful molecular probe to study the function of Cldn-4. To measure the binding affinity, many methods have been utilized. When 125I-C-Cpe (Aa 184–319) was used to interact with claudins overexpressed in mouse L cells, it was found that C-Cpe does not bind to Cldn-1 and Cldn-2, whereas Cldn-3 and Cldn-4 show specific binding with calculated affinity (KA from Scatchard plot) of 8.4 × 107 m-1 and 1.1 × 108 m-1, respectively (2). In another study, a far-Western approach was employed to measure the binding of purified C-Cpe to GST-Cldn3 Ecl2 protein separated by SDS-PAGE and blotted onto nitrocellulose membrane; here, the KA was calculated to be 1.0 × 108 m-1. Claudin-6, 7, 8, and 14 were also tested for C-Cpe binding by this method, with affinities estimated from submicro- to micromolar levels (4).
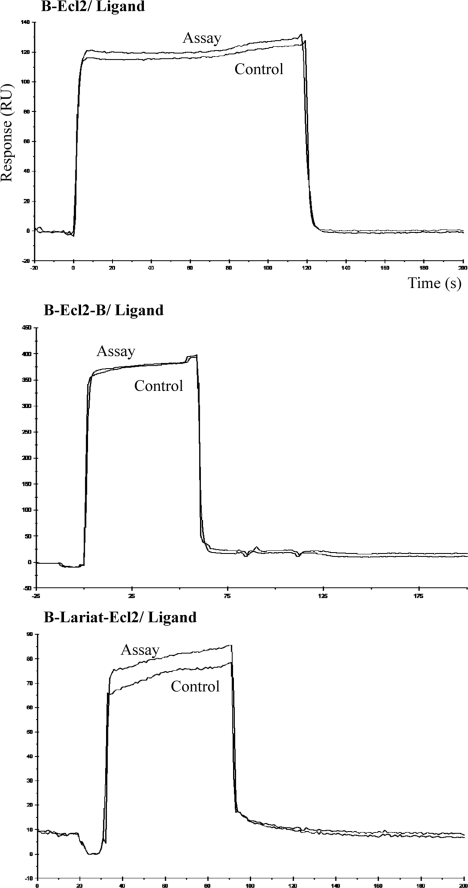
These above methods only measured the binding affinity at the end point, and binding kinetics were not determined. Because binding on- and off-rates may be important parameters in the behavior of the peptides in vivo, a new SPR method was developed for this study. Our first strategy was to directly use Ecl2 peptide bound to an SA chip to interact with binding peptides. The Ecl2 sequence alone was synthesized as a biotinylated peptide (Table 1), immobilized to SA chip as the ligand, then tested against the free Cpe30 peptide as the analyte. No specific binding was observed from this assay (Fig. 1, upper panel). In an attempt to better model the conformation of the Cldn-4 extracellular domain, the Ecl2 peptide was synthesized with biotin moieties at both the N and C termini. Our rationale was that such a loop would potentially be tethered to an SA chip by binding to a single tetravalent streptavidin molecule; this could present Ecl2 in a more physiological “loop” conformation. However, the result still failed to bind to Cpe30 (Fig. 1, middle panel). Another attempt at mimicking the natural Ecl2 loop was done by synthesis of Ecl2 as a cyclic peptide with a disulfide bond between two cysteines flanking the ends; this loop also failed to bind (Fig. 1, lower panel). In these attempts, the constraints on the Ecl2 loop may have failed to permit proper folding of the peptide.
FIGURE 1.
Claudin-4 Ecl2-only does not bind to Cpe30. The Claudin-4 Ecl2 domain was synthesized as a biotinylated peptide as described in Table 1. Biotin-Ecl2 peptides were bound to streptavidin (SA) sensor chips at three different configurations; B-Ecl2 was a linear structure, B-Ecl2-B was tethered to streptavidin by biotins at both the N and C termini, and B-lariat-Ecl2 contained a loop via disulfide bond formed between two cysteines flanking the Ecl2 loop. The ligands were tested against Cpe30 peptide as the analyte. The control channel was treated the same as the assay channel, but without B-peptide. The sensorgrams with 5 μm Cpe30 are shown.
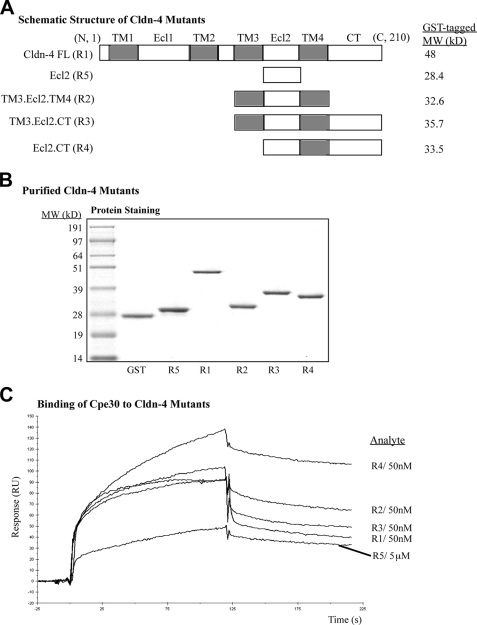
Our results above indicated that the isolated Cldn-4 Ecl2 alone is not sufficient for binding to Cpe30, consistent with other in vitro studies (10). It is likely that when Cldn-4 is in the cell membrane in vivo, the upstream and downstream transmembrane domains (TM) provide additional structural contexts to maintain Ecl2 conformation; these TMs may therefore be helpful in establishing the correct Ecl2 conformation in vitro. Thus, we began studies using Ecl2 in the context of other Cldn-4 domains as shown in Fig. 2A. The small molecular weight of Cldn-4 with 4 TMs and a strong tendency of oligomerization pose a difficult solubility problem for this protein (4). To enhance the solubility of these constructs, GST was fused to the N terminus; this was helpful, though there was still significant protein in the inclusion body when expressed in BL21 (pLysS) E. coli strain. Soluble protein was purified by glutathione-agarose affinity chromatography with further separation by gel filtration. These recombinant proteins were then found to be at a purity sufficient for SPR assays (Fig. 2B).
FIGURE 2.
Ecl2 with the downstream transmembrane domain is sufficient enough to express optimal binding activity to Cpe30. Biotin-Cpe30 was immobilized on an SA chip as the ligand, and Cldn-4 deletion mutant proteins fused to GST were tested as analytes. A, schematic diagram of full-length claudin-4 and its mutants, with the structural domains labeled above. B, SDS-PAGE analysis of purified GST-Cldn-4 mutant fusions. C, overlay of the sensorgrams from different analytes at medium concentration. GST-only protein did not bind Cpe30 (not shown). Specific binding was identified by subtraction of the control channel from the assay channel; this processing was applied to the rest of experiments in this study.
Assays were then performed using B-Cpe30 as the ligand bound to an SA chip, and the GST-Cldn4 recombinant proteins were used as the analytes. The higher molecular weight of these analytes was also beneficial in increasing the sensitivity of the SPR assay. When these GST cldn-4 fusion proteins were tested against Cpe30, high affinity specific binding was observed. The assay channel showed a typical association and dissociation response by comparison to the control channel, which only reflected fluctuations in analyte concentration. The subtracted sensorgrams at a medium concentration of each analyte were overlaid and shown in Fig. 2C. We found that all of the Cldn-4 variants shown exhibited binding activity, suggesting that Ecl2 in the context of neighboring cldn-4 domains displayed better conformation than as a separated peptide. Sensorgrams from all fusion proteins behaved normally except for the fusion containing the full-length Cldn-4. This protein showed a slight decrease in the late stages of association, raising the possibility of heterogeneous interaction from interactions with Ecl1 or other parts of Cldn-4 sequence.
The TM3.Ecl2.TM4 construct showed a consistent binding to Cpe30, indicating that the C-terminal tail of Cldn-4 was not required for interaction with ligand, even though this domain is thought to be involved in the signaling functions of Cldn-4 in vivo (15, 16). In practice however, it was difficult to use this protein due to poor solubility. By removing the TM3 and adding the C-terminal tail (the R4 construct), the fusion protein now displayed optimal binding activity to Cpe30. Presumably, the GST moiety at the N terminus could serve as an anchor similar to TM3. Therefore, the R4 construct was chosen as the best mimic of Ecl2 for the rest of this study. It was interesting to note that GST-Ecl2 without any TM domain also exhibited a detectable binding activity, albeit at much lower affinity. This may in part be due to GST dimerization, helping Ecl2 form a partial “loop” conformation.
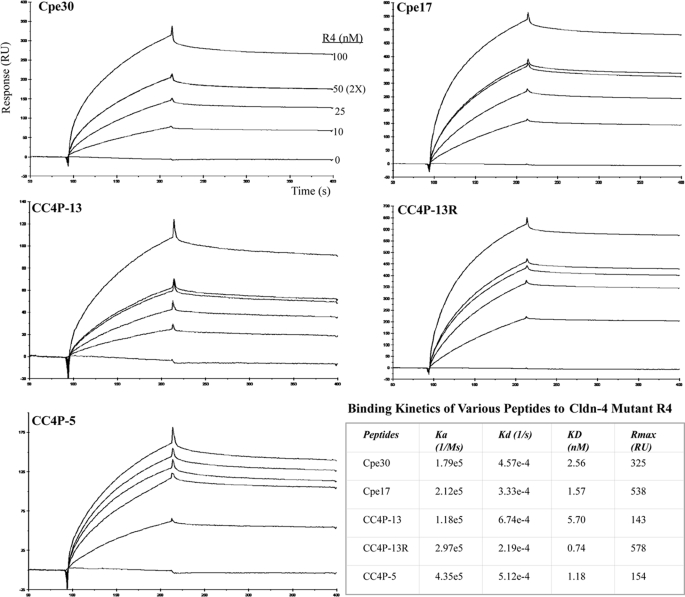
Claudin-4-binding Peptides Exhibit Different Kinetics but Share a Common Binding Motif—With the establishment of this SPR method, we first tested it to measure the kinetics of binding of Cpe30 and Cpe17 to Cldn-4 Ecl2. Previously reported deletion experiments already demonstrated that the last 30 amino acids of Cpe were responsible for the binding to Cldn-4 (15). Loss of the 17 terminal amino acids also caused the loss of binding, raising the possibility that this region contained the minimal binding domain. Therefore, we tested B-Cpe30 and Cpe17 immobilized to an SA chip as the ligand, and a series of concentrations of GST-Cldn-4.Ecl2 (R4) as the analyte was applied to measure binding kinetics. Our results demonstrated a strong specific binding; the subtracted sensorgrams were overlaid as shown in Fig. 3. GST protein as a control did not bind Cpe30 (not shown), confirming that the specific binding was from the Cldn-4 Ecl2. The 50 nm analyte concentration was repeated twice to ensure the reproducibility of the assay. The calculated equilibrium affinities (KD) for Cpe30 and Cpe17 were 2.56 nm and 1.57 nm, respectively, which was in the typical range of receptor-ligand binding. Kinetics analysis showed the on-rates were similar for Cpe30 and Cpe17, but Cpe17 had a slightly slower off-rate, giving it a higher calculated equilibrium affinity.
FIGURE 3.
Binding kinetics of peptides to GST-Cldn-4 Ecl2 mutant R4. Biotinylated peptides shown in the figure were immobilized to an SA chip with a PEG linker as the ligands; the GST-Cldn-4.R4 was used as the analyte from 10 to 100 nm to measure the kinetics of binding. The processed specific binding sensorgrams are presented for each ligand. Curves at several analyte concentrations were generated to ensure accurate calculated kinetics, and the 50 nm analyte concentration was repeated twice to confirm reproducibility. The binding kinetics was analyzed by BIAevalution 3.1 software with 1:1 (Langmuir) binding mode, and the binding constants are summarized in the table. CC4P-2 peptide had no specific binding to GST.Cldn-4-Ecl2, as the assay and control channels responded similarly to the different concentrations of analyte (not shown).
This SPR method also provided a useful tool to evaluate candidate peptides selected by phage display. To generate new short peptides with Cldn-4 binding activity, a phage display approach was used. A large library of random 12-mer display peptides allowed us to select for peptides able to bind Cldn-4 under physiological conditions in vitro. Claudin-4 overexpressed in CHO cells was used as the bait for selection using alternating rounds of positive selection on transfected CHO cells, and negative selection against non-transfected cells. Starting after the third round, phage clones were sequenced, and sequences showing enrichment with successive rounds of selection were considered to be candidate binding clones. Peptides encoded by these sequences were then synthesized (with a PEG spacer, ending with a biotin for binding to the SA chip) and tested for specific binding to cldn-4. Three abundant peptides (CC4P-13, -5, and -2) were identified after 4 rounds of screening (Table 1). CC4P-13 was biotinylated at the C terminus to reproduce the orientation of the peptide when displayed on the surface of the phage particle. By contrast, CC4P-13R was biotinylated at the N terminus; this orientation is similar to that in the recombinant HA fusion protein described below. The assays were performed as for Cpe30, and the sensorgrams shown in Fig. 3 demonstrated a clear specific binding of CC4P-13 and CC4P-5 peptides to Ecl2. The KD was measured at the nanomolar level, with CC4P-13R showing the slowest off-rate. CC4P-2 showed no binding to Ecl2 by this SPR (not shown). This could be due to several factors; this clone might be specific for CHO determinants and simply survived negative selection, it could be specific for the Cldn-4 Ecl1 domain, or other unknown factors enabled its enrichment.
In any case, our SPR assay proved to be helpful in determining the specific binding and the detailed kinetics of several Cldn-4-binding peptides. Interestingly, the orientation of biotinylation did not affect the binding of CC4P-13 to Ecl2, reinforcing the notion that this peptide was a true Cldn-4 ligand. Such adaptable peptides could have utility in a variety of other contexts.
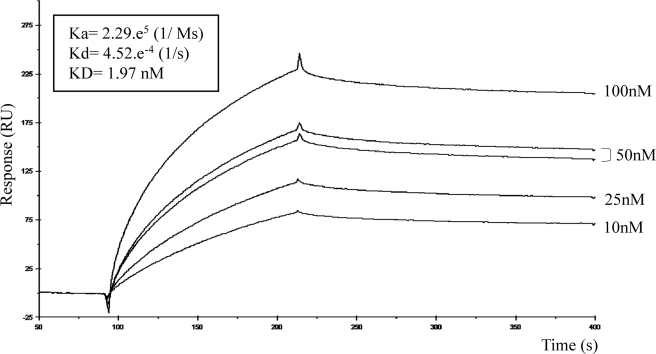
One research report showed that the binding activity of Cpe30 was abolished upon removal of the last five amino acids (17), so it was likely that Cpe17 binding activity was also contained in this terminal sequence. Because we were able to identify functional 12-mer peptides, we looked for the possibility of a common binding motif for Cldn-4 Ecl2. Based on the CC4P-13 and CC4P-5 similarities to Cpe30 and Cpe17, the basic Cpe30 sequence was reduced to a 12-mer. This was done by removing the last 4 amino acids and the N-terminal 13 amino acids not included in Cpe17, then the spacing between the Tyr-306, Tyr-310, and Tyr-312 was modified with minimal changes in amino acid composition (Table 1). Three candidate mutant 12-mers (MT1 through MT3) were tested using the same SPR assay as in Fig. 3. MT2 was found to possess binding activity to Ecl2 with high affinity (Fig. 4), although MT1 and MT3 showed no specific binding (not shown).
FIGURE 4.
Cpe30 mutant MT2 exhibits specific binding to GST-Cldn-4.R4. Cpe30 was reduced to 12 amino acid peptides as indicated in Table 1 (Cpe30MT1 through MT3), and tested against GST-Clnd-4.R4 as described in the legend to Fig. 3. Cpe30 MT2 was found to interact with Cldn-4 Ecl2; the sensorgrams are shown here. Cpe30 MT1 and MT3 did not bind Cldn-4 Ecl2 (not shown).
Comparing the sequences among these positive binding peptides revealed an interesting pattern. As shown in Fig. 5, all of the peptides contained tyrosine or tryptophan at the position corresponding to the Tyr-306 in Cpe, and after a 3–4 amino acids spacer there were 1 or 2 tyrosines followed by a leucine or isoleucine. This pattern constituted an apparent Ecl2 binding motif; we propose that the first Tyr or Trp is the priming or docking site and the remaining Tyr and Leu/Ile residues participate in the chemical bonding of the contact. Reports demonstrated the importance of the three Tyr in C-Cpe (11), and a recent study indicated that the leucine is also critical for the binding (12), but no study has been done directly with a peptide as short as 12 amino acids, even with the last 4 amino acids of Cpe30 eliminated.
FIGURE 5.
Claudin-4 Ecl2 binding motif of peptide ligands. The Cpe17 sequence was aligned with several Ecl2-binding peptides found to have significant affinity. The structure-based alignment was performed according to the crystal structure of the C terminus of Cpe (10). A common binding motif was deduced, with the underlined tyrosine or tryptophan constituting a structural requirement for docking of peptide into the Ecl2 cleft.
Biological Activity of Claudin-4-binding Peptides—Although the 12-mer peptides were selected on native claudin-4 on transfected CHO cells, we confirmed the binding of these peptides by synthesizing biotinylated peptides, and binding them directly to GFP-Claudin-4-transfected CHO cells. The bound biotinylated peptides pulled down with Neutravidin were confirmed to be bound to GFP-Claudin-4 by Western blot, while control peptide (established to have no Cldn4 binding by SPR) showed minimal pull-down of GFP-Claudin-4 (Fig. 6). The enterotoxin peptides Cpe30 and Cpe17 showed the strongest binding. The MT2, CC4P-13, and CC4P-5 peptides also pulled-down GFP-Cldn-4, but it appeared to be consistently lower than with Cpe30 and Cpe17, despite the similar SPR affinity measurements of the peptide.
Binding to the native protein in vivo could also affect the biological function of claudin-4 in tight junctions, so we tested the effect of the soluble peptides on TER of Caco-2BBe epithelial monolayers. As might be predicted from previous reports (2, 4), basal but not apical addition of Cpe30 significantly reduced the TER by up to 25–30% detectable as early as 4 h after addition, persisting more than 24 h (Fig. 7A); however, the shorter peptide Cpe17 had no effect on TER (Fig. 7B). Similarly, MT2, CC4P-13, and CC4P-5 had no detectable effect on TER after 24 h whether applied to the apical or basolateral side of the monolayers (Fig. 7B and data not shown). Thus, while the shorter peptides were confirmed to have high affinity binding to native claudin-4 by SPR and in vivo pull-down assays, such assays did not predict their effect on epithelial tight junctions, as these effects may be related instead to the size of the peptide or ability to form larger secondary protein complexes.
FIGURE 7.
Claudin-4-binding peptide effects on TER in Caco-2BBe monolayers. Peptides were added to apical or basal sides of transwell cultures with established Caco-2BBe monolayers, and TER measurements were taken as described under “Experimental Procedures.” A, Cpe30 showed up to 25–30% reduction in TER but only when added to the basal side of the monolayers; shown here are results after 14 h of treatment. B, Cpe17, a shorter version of Cpe30, and CC4P-13 had no effect on TER, here shown after 24 h of treatment. Similarly, no effects on TER were seen with MT2, and CC4P-5 after 24 h (not shown).
Claudin-4-binding Peptide Retains Binding Activity in Recombinant Protein—Peptides with the ability to bind to Cldn-4 in vivo could have many useful applications, including targeting delivery vehicles to tumors overexpressing Cldn-4, or targeting vaccine antigens to mucosal M cells. For this purpose, it would be important to know whether the peptide can still function in other contexts, such as part of a recombinant fusion protein. Here, we tested whether the attachment of a targeting peptide to the C-terminal end of a recombinant influenza hemagglutinin (HA) would still retain targeting specificity.
To test this question, Cpe30 was integrated into the C terminus of HA as shown in Fig. 8A. The recombinant fusion protein (HA-ts-Cpe30) was produced by an insect expression system and purified for our SPR assay. The recombinant protein was captured on a CM5 chip using an anti-HA antibody (H36), which is specific for a conformational determinant on the globular head of the HA trimer. In this orientation, the HA trimer is presented with the C-terminal tail and Cpe30 domain displayed outwards for interaction with analyte. The binding to GST-Cldn-4.R4 was tested as in Fig. 3. We found that the HA fusion protein with Cpe30 exhibited strong specific binding to Ecl2, though the association seemed to be a heterogeneous reaction and the on-rate was slower than free Cpe30 peptide (Fig. 8B). This is possibly due to the decreased surface accessibility caused by the C-terminal His tag. Cpe30 in HA protein could also be affected by flanking sequences to some extent, even though it was predicted to be exposed on the protein surface (Protean software (Lasergene), not shown). When HA protein without Cpe30 was used as the ligand, no specific interaction was detectable. In sum, the addition of a Cldn-4-binding peptide to a recombinant globular protein still showed binding activity even when in a heterologous context.
FIGURE 8.
Cpe30 in recombinant influenza HA retains binding activity to Cldn-4 Ecl2. C-terminally His-tagged HA-ts-Cpe30 and HA shown in A were expressed in baculovirus and purified to homogeneity. HA-specific antibody (H36) was immobilized to a CM5 chip by amine coupling, which was used to capture HA-ts-Cpe30 or HA protein as the ligand. Cldn-4 Ecl2.R4 was used as the analyte. B, overlay of sensorgrams of HA-ts-Cpe30 with different concentrations of Cldn-4.R4 show the specific interaction. C, HA protein without Cpe30 showed no binding to Cldn-4 Ecl2; the figure shows that the assay and control channels responded similarly at different concentrations of analyte.
DISCUSSION
In this study we found that we can use a soluble fusion protein between GST and the C-terminal domains of Cldn-4 to measure high affinity specific binding with peptides from both the C. perfringens enterotoxin and a phage display library selected against native Cldn-4. Thus, it appears that the interaction of a transmembrane protein and its ligand can be mimicked under certain membrane-free conditions. In addition, we found that the binding of short peptides, including the 12 amino acid phage display peptides, could be measured with affinities in the nanomolar range, close to the estimated affinity for the intact Cpe binding to Cldn-4. Finally, as a matter for potential practical application, peptides such as Cpe30 could still bind the target claudin-4 domain even when the Cpe30 peptide was produced as a recombinant fusion protein in the context of a large globular protein such as influenza hemagglutinin.
In other studies on binding of the C. perfringens enterotoxin to Cldn-4, a variety of methods were used, but all examined equilibrium binding, and found a range of affinity values, often lower than our measurements using SPR. One challenge with estimating the binding of a ligand to a transmembrane protein receptor is developing a simple system in which few other interactions can affect binding. Thus, binding to receptors on transfected cells may be subject to the presence of a broad array of additional cellular proteins, and the dynamics of cell surface proteins in the plasma membrane can interfere with estimates of equilibrium binding. Cell-free methods suffer from different problems, including the challenge to ensure the proper conformation of the binding domain. It is also possible that our use of short peptide domains minimized interference from neighboring protein domains that would be present in the intact native proteins, but this remains to be tested.
While our own approach is not immune from the issues described above, we can provide a few arguments that our method does indeed provide a reasonable measure of peptide/Cldn-4 binding. First, the binding affinity calculated for Cpe peptides binding to GST-Cldn-4 is among the highest reported for Cpe binding to Cldn-4, and two different versions of the C-terminal peptide of Cpe (Cpe30 and Cpe17) showed similar calculated affinity in the nanomolar range. Second, phage display peptides selected for binding to native Cldn-4 on transfected CHO cells also showed high affinity specific binding to the GST-Cldn-4 fusion protein, again in the nanomolar affinity range, confirmed also in pull-down assays of binding to native protein on transfected CHO cells. That is, peptides selected for binding to the native transmembrane conformation also showed high affinity binding to our recombinant fusion protein analyte, confirming in a parallel system the specific binding interaction among the peptides. Third, the peptides showing similar high affinity binding, whether derived from Cpe or from phage display selection, showed a striking common structural motif. This suggested that a common structural conformation was maintained in all of the binding studies, and this is likely to reflect the natural conformation of the second extracellular domain of Cldn-4 under physiological conditions.
Most previous studies on Cpe binding to Cldn-4 in vitro or in vivo used either the intact Cpe protein, or versions truncated at the N-terminal end, leaving a large polypeptide such as the 136 amino acid long “C-Cpe” protein. Despite that fact that the C-Cpe peptide no longer contains direct cytotoxic activity, a recent report showed that a similar size truncated version of Cpe still retained the ability to form membrane “prepore complexes,” suggesting that this peptide retained the ability to form structures distinct from its ability to simply bind the second extracellular domain of Cldn-4 (18, 19). Some studies also showed binding by the smaller Cpe30 peptide, but our present studies show that Cpe peptides as small as Cpe17 also can bind with high affinity, and even shorter versions, such as a mutated version (MT2) only 12 amino acids long could still bind with high affinity. The shorter peptides did not interfere with TER in Caco-2BBe monolayers, but this may be due to a relative inability of short peptides to form larger secondary complexes. So while larger Cldn-4 binding peptides might be used intentionally for their tight junction effects, the shorter peptides might instead be used for targeting without side effects on epithelial barrier function. Such short peptides could have a practical advantage in applications such as targeting to Cldn-4-expressing tumors (20), or targeted delivery of vaccines to mucosal M cells (21, 22). The small peptides would be much less immunogenic than the larger Cpe fragments such as C-Cpe and would be less likely to induce blocking antibodies that could interfere with subsequent doses.
The short peptides also provide an interesting clue to the minimal structure required for binding to Cldn-4. By comparison among all peptides with positive binding, we identified a common binding motif as shown in Fig. 5. Published studies with mutation of the three tyrosines (11) and the alanine scan (12) were consistent with our findings, but our study is the first to test whether the core sequence was sufficient to mediate binding to Cldn-4 Ecl2. The specific conformation of the binding of short peptides to Cldn-4 Ecl2 could be explained partially by the recently published structure of C-Cpe (10). Three tyrosines near the C-terminal end were localized to a large surface loop held by two flanking β-strands; this relatively stable conformation could still provide some plasticity for docking into the Ecl2 structure. Without the availability of Cldn-4 crystal structure, it is difficult to know how they exactly interact with each other. However, the recently solved structure of GPCR β2AR (β2-adrenergic receptor) (13, 23) may provide some clues. β2AR uses Ecl2 and amino acids from transmembrane helices (III, IV, V, and VII) to form the binding cleft, and its ligand (carazolol) is hydrophobic with multiple aromatic rings. The Ecl2 is composed of an α-helix and a coiled-coil secondary structure; one phenylalanine (Phe-193) in the loop and a downstream tyrosine (Tyr-199) were involved in the direct binding with carazolol by hydrophobic contacts. Cldn-4 Ecl2 shares some similarity with β2AR Ecl2 in amino acid composition, as both contain 2 to 3 tyrosines and phenylalanines. Theoretical predictions suggested that the Cldn-4 Ecl2 forms a α-helix at the N terminus followed by a loop structure (14), which resembles the conformation of β2AR Ecl2. The “FY” in the loop region of Ecl2 could be the direct contact sites for interaction with Tyr or Trp in Cpe30 and other binding peptides.
In sum, this study established a quick and accurate SPR method for measuring the binding kinetics of Cldn-4 ligands and furthermore revealed some important structural constraints in Cldn-4 Ecl2 conformation and the ligand binding motif for efficient interactions.
This work was supported, in whole or in part, by the National Institutes of Health. This work was also supported by a Grand Challenges in Global Health grant from the Bill and Melinda Gates Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: Cldn-4, Claudin-4; Cpe, C. perfringens enterotoxin; C-Cpe, C-terminal domain of Cpe; Ecl2, extracellular loop 2; SPR, surface plasmon resonance; HA, hemagglutinin; TER, trans-epithelial electrical resistance; TM, transmembrane; GFP, green fluorescent protein; GST, glutathione S-transferase; Aa, amino acids.
References
- 1.Katahira, J., Inoue, N., Horiguchi, Y., Matsuda, M., and Sugimoto, N. (1997) J. Cell Biol. 136 1239-1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonoda, N., Furuse, M., Sasaki, H., Yonemura, S., Katahira, J., Horiguchi, Y., and Tsukita, S. (1999) J. Cell Biol. 147 195-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koval, M. (2006) Cell Commun. Adhes. 13 127-138 [DOI] [PubMed] [Google Scholar]
- 4.Fujita, K., Katahira, J., Horiguchi, Y., Sonoda, N., Furuse, M., and Tsukita, S. (2000) FEBS Lett. 476 258-261 [DOI] [PubMed] [Google Scholar]
- 5.Rahner, C., Mitic, L. L., and Anderson, J. M. (2001) Gastroenterology 120 411-422 [DOI] [PubMed] [Google Scholar]
- 6.Kuo, W. L., Lee, L. Y., Wu, C. M., Wang, C. C., Yu, J. S., Liang, Y., Lo, C. H., Huang, K. H., and Hwang, T. L. (2006) Oncol. Rep. 16 729-734 [PubMed] [Google Scholar]
- 7.Lo, D., Tynan, W., Dickerson, J., Scharf, M., Cooper, J., Byrne, D., Brayden, D., Higgins, L., Evans, C., and O'Mahony, D. J. (2004) Int. Immunol. 16 91-99 [DOI] [PubMed] [Google Scholar]
- 8.Oliveira, S. S., and Morgado-Díaz, J. A. (2007) Cell Mol. Life Sci. 64 17-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi, A., Kondoh, M., Masuyama, A., Fujii, M., Mizuguchi, H., Horiguchi, Y., and Watanabe, Y. (2005) J. Control Release 108 56-62 [DOI] [PubMed] [Google Scholar]
- 10.Van Itallie, C. M., Betts, L., Smedley, J. G., 3rd, McClane, B. A., and Anderson, J. M. (2008) J. Biol. Chem. 283 268-274 [DOI] [PubMed] [Google Scholar]
- 11.Ebihara, C., Kondoh, M., Harada, M., Fujii, M., Mizuguchi, H., Tsunoda, S., Horiguchi, Y., Yagi, K., and Watanabe, Y. (2007) Biochem. Pharmacol. 73 824-830 [DOI] [PubMed] [Google Scholar]
- 12.Takahashi, A., Komiya, E., Kakutani, H., Yoshida, T., Fujii, M., Horiguchi, Y., Mizuguchi, H., Tsutsumi, Y., Tsunoda, S., Koizumi, N., Isoda, K., Yagi, K., Watanabe, Y., and Kondoh, M. (2008) Biochem. Pharmacol. 75 1639-1648 [DOI] [PubMed] [Google Scholar]
- 13.Rosenbaum, D. M., Cherezov, V., Hanson, M. A., Rasmussen, S. G., Thian, F. S., Kobilka, T. S., Choi, H. J., Yao, X. J., Weis, W. I., Stevens, R. C., and Kobilka, B. K. (2007) Science 318 1266-1273 [DOI] [PubMed] [Google Scholar]
- 14.Rost, B., Yachdav, G., and Liu, J. (2004) Nucleic Acids Res. 32 W321-W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka, M., Kamata, R., and Sakai, R. (2005) J. Biol. Chem. 280 42375-42382 [DOI] [PubMed] [Google Scholar]
- 16.Van Itallie, C. M., Colegio, O. R., and Anderson, J. M. (2004) J. Membr. Biol. 199 29-38 [DOI] [PubMed] [Google Scholar]
- 17.Kokai-Kun, J. F., and McClane, B. A. (1997) Clin. Infect. Dis. Suppl. 2 S165-7 [DOI] [PubMed] [Google Scholar]
- 18.Medley, J. G., 3rd, Uzal, F. A., and McClane, B. A. (2007) Infect. Immun. 75 2381-2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masuyama, A., Kondoh, M., Seguchi, H., Takahashi, A., Harada, M., Fujii, M., Mizuguchi, H., Horiguchi, Y., and Watanabe, Y. (2005) J. Pharmacol. Exp. Ther. 314 789-795 [DOI] [PubMed] [Google Scholar]
- 20.Morin, P. J. (2005) Cancer Res. 65 9603-9606 [DOI] [PubMed] [Google Scholar]
- 21.Kondoh, M., Masuyama, A., Takahashi, A., Asano, N., Mizuguchi, H., Koizumi, N., Fujii, M., Hayakawa, T., Horiguchi, Y., and Watanbe, Y. (2005) Mol. Pharmacol. 67 749-756 [DOI] [PubMed] [Google Scholar]
- 22.Brayden, D. J., Jepson, M. A., and Baird, A. W. (2005) Drug Discov. Today 10 1145-1157 [DOI] [PubMed] [Google Scholar]
- 23.Cherezov, V., Rosenbaum, D. M., Hanson, M. A., Rasmussen, S. G., Thian, F. S., Kobilka, T. S., Choi, H. J., Kuhn, P., Weis, W. I., Kobilka, B. K., and Stevens, R. C. (2007) Science 318 1258-1265 [DOI] [PMC free article] [PubMed] [Google Scholar]