Abstract
An image-based phenotypic screen was developed to identify small molecule regulators of intracellular traffic. Using this screen we found that AG1478, a previously known inhibitor of epidermal growth factor receptor, had epidermal growth factor receptor-independent activity in inducing the disassembly of the Golgi in human cells. Similar to brefeldin A (BFA), a known disrupter of the Golgi, AG1478 inhibits the activity of small GTPase ADP-ribosylation factor. Unlike BFA, AG1478 exhibits low cytotoxicity and selectively targets the cis-Golgi without affecting endosomal compartment. We show that AG1478 inhibits GBF1, a large nucleotide exchange factor for the ADP-ribosylation factor, in a Sec7 domain-dependent manner and mimics the phenotype of a GBF1 mutant that has an inactive mutation. The treatment with AG1478 leads to the recruitment of GBF1 to the vesicular-tubular clusters adjacent to the endoplasmic reticulum exit sites, a step only transiently observed previously in the presence of BFA. We propose that the treatment with AG1478 delineates a membrane trafficking intermediate step that depends upon the Sec7 domain.
The Golgi apparatus, an intracellular membrane-bound structure organized as a series of stacked cisternae and tubules, plays an important role in packaging and transporting macro-molecules. The cisternae stack can be divided into five functional regions: the cis-Golgi network, cis-Golgi, medial Golgi, trans-Golgi, and trans-Golgi network. Newly synthesized proteins and lipids are delivered by the coat protein complex II (COPII)4 vesicles from the endoplasmic reticulum (ER) via the vesicular-tubular clusters (VTCs) to the cis-Golgi network and subsequently progress through the stack to the trans-Golgi network, where they are packaged and sent to the cell surface, secretory vesicles, or late endosomal compartments (1, 2). p58 (also called ERGIC53), a cargo receptor involved in transporting proteins from the ER to the Golgi complex, has been used as a marker for the VTC structure and the cis-Golgi (3, 4).
The integrity and functions of the Golgi depend critically on the small membrane-bound vesicles that shuttle macromolecules in and out of the Golgi stacks (5). The ADP-ribosylation factor (ARF), a small GTPase of the Ras superfamily, plays a major role in driving Golgi membrane trafficking by recruiting coatomer (COPI) to Golgi membranes in the exocytotic pathway. The cellular activity of ARFs is stimulated by the Sec7 family of guanine nucleotide exchange factors (GEFs), which promote the exchange of inactive GDP-bound forms to active GTP-bound forms (6, 7). GBF1, the only known GEF localized to the cis-Golgi, plays an important role in mediating protein trafficking between the ER and the cis-Golgi (8, 9).
Brefeldin A (BFA), a lactone isolated from fungi, interferes with anterograde transport from the endoplasmic reticulum to the Golgi apparatus by binding to the ARF1-GDP-Sec7 complex to inhibit the Sec7 guanine nucleotide exchange activity, which in turn prevents the activation of ARF (10, 11). The treatment with BFA causes rapid but reversible dispersal of the Golgi apparatus leading to the mixing of the cis-Golgi with the ER and vesicularization of the trans-Golgi network (12, 13). BFA has pleiotropic effects on intracellular organelles other than the Golgi. For example, BFA induces extensive formation of membrane tubules from endosome compartment (14-16). BFA is highly cytotoxic and causes rapid cell death. It is not clear whether its toxicity is related to its effects on Golgi dispersal (13, 17, 18).
In an effort to discover novel small molecules as tools to study membrane trafficking, we developed an image-based assay for compounds that can induce disassembly of the Golgi. In this manuscript, we show that tyrphostin AG1478 inhibits the nucleotide exchange of ARF1 in a manner dependent upon the Sec7 domain of GBF1.
EXPERIMENTAL PROCEDURES
Plasmids, Antibodies, and Cell Reagents—AG1478, BFA, nocodazole, bafilomycin A1, EGF, H89, and 2-deoxy-d-glucose were from Sigma-Aldrich. The EGFR inhibitors were from ICCB Known Bioactives Library. Aluminum fluoride was prepared by (50 μm AlCl3 + 30 mm NaF) before use (19). Polyclonal rabbit anti-sec13 antibody was a kind gift from Wanjin Hong (Institute of Molecular Biology, Singapore). Monoclonal anti-GM130, anti-GBF1, anti-p230, anti-adaptin γ, and anti-adaptin δ antibodies were purchased from BD Biosciences. Monoclonal anti-α tubulin, anti-Myc, and polyclonal rabbit anti-calnexin antibodies were from Sigma-Aldrich. Monoclonal anti-phospho-EGFR, monoclonal anti-phospho-ERK, and polyclonal rabbit anti-EGFR antibodies were from Cell Signaling Technology. Alexa Fluor 546-conjugated transferrin and LysoTracker red DND-99 were purchased from Molecular Probes. Polyclonal rabbit anti-GFP antibody and monoclonal anti-LAMP3 was from Santa Cruz Biotechnology. Polyclonal rabbit anti-β-COP antibody was from Affinity Bioreagents. Secondary antibodies conjugated with Texas Red or fluorescein isothiocyanate were from Rockland Immunochemicals. p58-YFP, GalT-CFP, and ARF1-GFP were kind gifts from Jennifer Lippincott-Schwartz (National Institutes of Health). GBF1-GFP and E794K-GFP were kindly provided by Elizabeth Sztul (University of Alabama, Birmingham). Sec13-GFP, ERGIC53-Myc, and GST-VHS-GAT-GGA3 were kinds gifts from Benjamin S. Glick (University of Chicago), Hans-Peter Hauri (University of Basel), and Paul A. Randazzo (National Institutes of Health). The different deleted constructs of GBF1-1 or GBF1-2 were digested from GBF1-GFP. The constructs were verified by sequencing.
Cell Culture and Transfection—The cells used for the experiments were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin/streptomycin, and 2 mm l-glutamine (Invitrogen) at 37 °C, 5% CO2. Transfection of H4 cells was done using Lipofectamine 2000 reagent (Invitrogen). A stable cell line expressing p58-YFP was established in the presence of 1 mg/ml G418 (Invitrogen).
Phenotypic Screen—H4-p58 cells were plated in 96-well plates at 3,000 cells/100 μl/well and cultured in a 5% CO2 incubator overnight. The compounds were diluted by sampling 100 μl of 5 mg/ml or 0.5 mg/ml of stocks dissolved in DMSO from the ICCB Known Bioactives Library of 480 compounds and transferred into wells in duplication. The images were visually examined using fluorescence microscopy (IX81; Olympus) after incubating with the compounds for 4, 8, and 24 h. DMSO (0.1%) and BFA (5 μm) were used as negative or positive control, respectively.
Immunofluorescence Microscopy and Image Analysis—Cells grown on coverslips were prepared and washed with phosphate-buffered saline for three times and fixed in 3.8% paraformaldehyde (Sigma) for 20 min at room temperature. After being blocked with 1% bovine serum albumin in TBST for 30 min, the cells were incubated with primary antibody diluted in 1% bovine serum albumin/TBST for 2 h or overnight at 4 °C. Coverslips were washed with TBST for three times and incubated with secondary antibody diluted in 1% bovine serum albumin/TBST for 1 h at room temperature. The coverslips were again washed with TBST three times, stained with 4,6-diamidino-2-phenylindole (1 μg/ml; Sigma), and mounted on slides with fluorescent mounting medium (DAKO). The cells were imaged using a fluorescent microscopy (IX81; Olympus) with a 100× oil 1.35 NA objective and Image-Pro Express software (Media Cybernetics). Confocal images (see Fig. 8) were acquired using an LSM 510 (Carl Zeiss) with a 100× oil 1.35 NA objective, and a single focal plane (0.9 um) was analyzed. For Fig. 1, the analysis for Golgi dispersal course was performed using CellProfiler software. The analysis pipeline used to quantify images by CellProfiler was, briefly, as follows: after correcting the illumination pattern across the field of view for each channel, the nuclei were identified based on the DNA-stained images. Then a ring 36 pixels around each nucleus was defined as a proxy for the cytoplasm. Within each cell, the Golgi were identified based on the GM130-stained image. Over 300 features were measured for each cell in each image; the most reliable measure of the change in Golgi localization was found to be a texture measure of the GM130 staining within each cell (sum average, at a scale of 5, nucleus + cytoplasm together). Example processed images are shown in supplemental Fig. S3.
FIGURE 8.
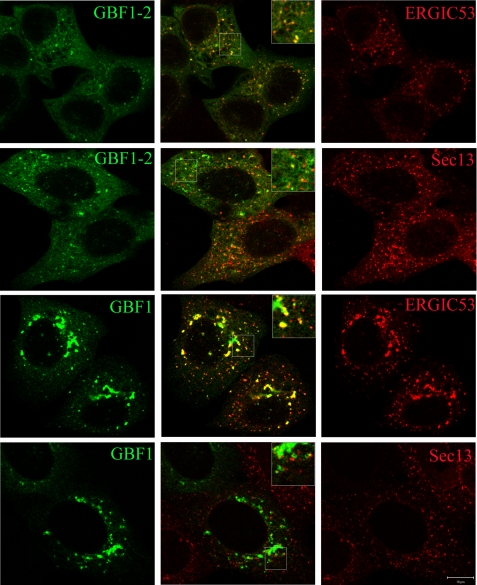
AG1478 causes accumulation of GBF1-2 and GBF1 spots on VTCs. H4 cells were transfected with GBF1-2-GFP, GBF1-GFP (green) alone or with ERGIC53-Myc for 24 h and then treated with 14 μm AG1478 for 1 h. The fixed cells were stained with an antibody against Myc or sec13 (red) as indicated. The middle row shows overlapping images. Single-slice confocal images are shown. The insets show a higher magnification of the boxed areas of the merged images. Bar, 10 μm.
FIGURE 1.
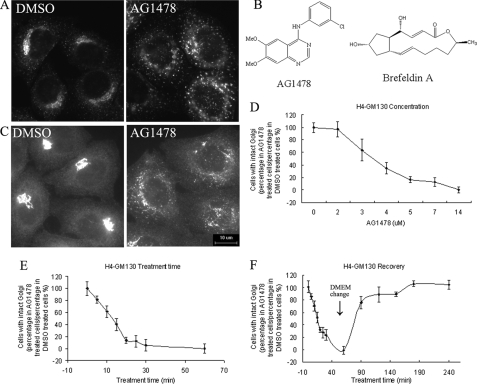
AG1478 treatment causes Golgi fragmentation in H4 cells. H4-p58 cells (A) or H4 cells (C) were treated with DMSO or 14 μm AG1478 as indicated for 1 h. H4 cells were fixed and stained with anti-GM130 antibody. B, chemical structures of AG1478 and BFA. D-F, H4 cells were treated with DMSO or AG1478 for different concentrations, different time, and then stained with an antibody against GM130. The images were analyzed by CellProfiler software. Bar, 10 μm.
EGFR siRNA—For transient inhibition of EGFR mRNA production, the small interfering RNA (siRNA) targeting the EGFR was synthesized (20): 5′-CTCTGGAGGAAAAGAAAGT-3′ and 5′-ACTTTCTTTTCCTCCAGAG-3′. H4 cells were co-transfected EGFR siRNA or negative control siRNA with pEGFP-N1 plasmid at 10:1 ratio by Lipofectamine 2000 reagent (Invitrogen). After 24 h, the cells were treated with compounds and analyzed by Western blotting for EGFR siRNA effect. The experiments were repeated twice with consistent results.
ARF1-GTP Pulldown Assay—The pulldown assay to assess the activity of ARF1 by a GST-VHS-GAT-GGA3 protein were performed as described previously (21, 22). The stable H4-ARF1-GFP cells were transiently transfected with GBF1-GFP for 24 h or pEGFP-C1 or vector alone and then treated with compounds. The cells were lysed with pulldown buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 2 mm MgCl2, 1% Triton X-100, and protease inhibitors). The cell lysates were cleared by glutathione-Sepharose beads and incubated with beads containing 50 μg of GST-VHS-GAT-GGA3 protein for 2 h at 4 °C. The bound proteins were removed and analyzed by Western blotting using anti-GFP antibody.
Cell Fractionation—The cell fractionation assay was performed as described previously (23). H4 cells were grown on 100-cm dishes for 24 h and treated with compounds. The cells were washed with phosphate-buffered saline and lysed with ice-cold homogenization buffer (50 mm HEPES, pH 7.5, 100 mm KCl, 1 mm MgCl2, 1 mm dithiothreitol, and protease inhibitors) by passaging 10 times through a 22-gauge needle and centrifuged at 1,000 × g for 15 min at 4 °C to remove the unbroken cells. The postnuclear supernatant was centrifuged at 100,000 × g for 60 min at 4 °C to obtain cytosol and membrane fraction. The supernatant was used as a cytosol fraction. The pellet was rinsed with homogenization buffer, dissolved in radioimmune precipitation assay buffer and used as membrane fraction. Fractions containing same volume were analyzed by 6% SDS-PAGE and transferred to nitrocellulose membrane. The membrane was detected by anti-GBF1, anti-calnexin, and antitubulin antibodies, respectively. Calnexin and tubulin were considered as standards for membrane and cytosol fraction.
RESULTS
An Image-based Phenotypic Screen for Small Molecules That Disrupt the Golgi—We established a H4 human glioblastoma cell line stably expressing a p58-YFP fusion protein (H4-p58-YFP). p58-YFP normally appears in peripheral punctate structures close to the cis-Golgi (Fig. 1A). Using H4-p58-YFP cells, we developed an image-based assay using fluorescent microscopy and screened the ICCB Known Bioactive Library for compounds that may have effects on Golgi morphology (Biomol catalogue number 2840). H4-p58-YFP cells were plated in 96-well plates for 24 h and treated with different compounds individually at concentration of 3∼12 μm. 0.1% DMSO was used as a negative control. After incubating with the compounds for 4, 8, and 24 h, the images were visually examined using fluorescence microscopy. The screen was carried out in duplicates. Most of the compounds were found to have no effect on Golgi morphology. Twenty compounds were isolated as Golgi-disrupting agents. These include some well known Golgi disrupting reagents such as brefeldin A, bafilomycin A1, and monensin (data not shown). Here we focus on tyrphostin AG1478, the most potent novel Golgi dispersing compound identified from our screen. It has a structure distinct from that of BFA (Fig. 1B).
AG1478 Is a Potent and Reversible Disruptor of the Golgi—The treatment of H4 cells with AG1478 caused p58-YFP to redistribute from its normal perinuclear compact localization to a dispersed localization throughout the cytosol in a manner very similar to that of BFA (Fig. 1A). The Golgi dispersing effect of AG1478 in the parental H4 cells that do not express p58-YFP was confirmed by indirect immunofluorescence using an antibody against GM130, a Golgi matrix protein (24). GM130 was redistributed to peripheral punctate structures throughout the cytoplasm after the treatment of AG1478 (Fig. 1C). We quantified the Golgi dispersing effect of AG1478 using the image analysis software, CellProfiler (25, 26). We found that AG1478 can rapidly disperse the cis-Golgi protein GM130 with a half-time of ∼10 min and an EC50 of 3.35 μm (Fig. 1, D and E). The effect of AG1478 on the Golgi was fully reversible, because the normal Golgi structure reappeared in the perinuclear area after the cells were cultured in medium without AG1478 for 3 h (Fig. 1F).
Unlike nocodazole or bafilomycin A1, which disperse the Golgi through their effects on the microtubules and intracellular pH, AG1478 had no apparent effect on the cytoskeleton or intracellular pH. The treatment of AG1478 for 1 h also did not have any obvious effect on the intracellular ATP levels (supplemental Fig. S1). Furthermore, the ability of AG1478 to disrupt the Golgi was not associated with significant cytotoxicity; incubation of H4 cells in the presence of 14 μm AG1478 for 24 h had no detectable effect on cell survival. After 48 h, a loss of 25% of viability was observed that could be rescued by a caspase inhibitor z-VAD-fmk. In contrast, the treatment of H4 cells with 5 μm BFA for 24 or 48 h led to a significant loss of cell viability that could not be blocked by z-VAD-fmk (supplemental Fig. S2). From these results, we conclude that dispersing Golgi per se is not affecting the viability of H4 cells. Therefore, the cytotoxicity of BFA may not be directly linked to its ability to disrupt the Golgi.
The Effect of AG1478 on the Golgi Is Independent of EGFR—AG1478 was shown to be a highly potent inhibitor of the EGF receptor tyrosine kinase (27, 28). Indeed, the treatment of H4 cells with 250 nm AG1478 was sufficient to block EGFR phosphorylation, which is much lower than that of the EC50 required to disperse the Golgi (Fig. 2A). To further determine whether the Golgi dispersing effect of AG1478 was associated with its inhibitory activity for EGFR, we treated H4 cells with multiple different EGFR inhibitors individually (AG213, Lavendustin A, and RG-14620), inhibitors for other RTPKs (AG-370, AG-879, and AG-825), as well as genistein, which is a general inhibitor of tyrosine protein kinase. None of these inhibitors demonstrated a similar Golgi dispersing effect even at very high concentrations (50-100 μm) and after prolonged treatment time (2-4 h) (data not show). Thus, the Golgi dispersing ability of AG1478 is not shared by the other known tyrosine kinase inhibitors tested.
FIGURE 2.
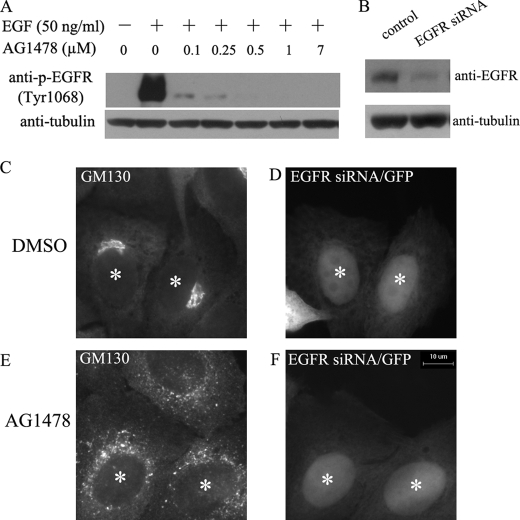
The effect of AG1478 on Golgi is independent of EGFR. A, H4 cells were pretreated with AG1478 for 30 min, then treated with 50 ng/ml EGF for 5 min and analyzed for anti-phospho-EGFR (tyr1068) or tubulin by immunoblot. B, control Western blot for the effect of EGFR siRNA. Anti-tubulin was used as a loading control. C-F, H4 cells were co-transfected pEGFP-N1 plasmid with EGFR siRNA at 1:10 ratio for 24 h and then treated with DMSO or 14 μm AG1478 as indicated for 1 h. H4 cells expressing EGFP siRNA (D and F) were stained with anti-GM130 antibody (C and E). GFP-positive transfected cells are indicated by white asterisks. Bar, 10 μm.
To formally exclude the involvement of EGFR, we used siRNA to decrease the expression of EGFR. Expression of EGFR siRNAs for 24 h significantly decreased the cellular levels of EGFR (Fig. 2B), but reducing the expression of EGFR had no effect on the Golgi structure per se nor on the Golgi dispersing effect of AG1478 (Fig. 2, C-F). From these results, we conclude that AG1478 has a distinct activity in inducing Golgi dispersing that is independent from its activity toward EGFR.
Rodent Cells Are Resistant to the Golgi Dispersal by AG1478 but Not BFA—To characterize the Golgi dispersing effect of AG1478, we examined whether the Golgi structures in multiple cell lines are sensitive to AG1478. AG1478 showed a similar Golgi dispersing effect in different human cell lines tested, including HeLa, HepG2, A549, and Hs-578Bst fibroblast cells (supplemental Table S1). Surprisingly, AG1478 could not disrupt the Golgi structures in multiple rodent cell lines tested, including two rat cell lines, Rat2 and NRK, and a mouse cell line, NIH3T3. The Golgi complex marked by anti-GM130 antibody in Rat2 cells remained intact even after the treatment with AG1478 at concentrations as high as 140 μm (50 μg/ml) prolonged incubation (Fig. 3A). In contrast, Rat2 cells were fully sensitive to the Golgi dispersing effect of BFA (Fig. 3A).
FIGURE 3.
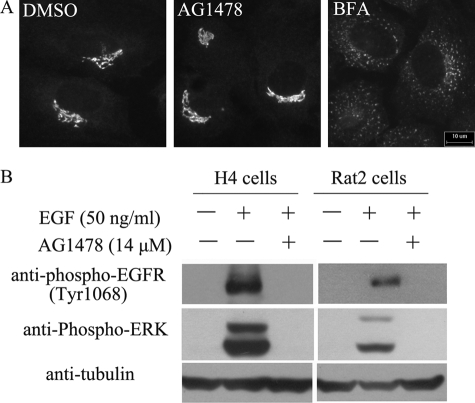
Rat2 cells are resistant to AG1478 but not BFA. A, Rat2 cells were treated with DMSO, 140 μm AG1478, or 5 μm BFA as indicated for 4 h, stained with anti-GM130 antibody. B, H4 cells or Rat2 cells were pretreated with 14 μm AG1478 for 30 min or not, then stimulated by 50 ng/ml EGF for 5 min, and analyzed by Western blotting using anti-phospho-EGFR (tyr1068), anti-phospho-ERK, and anti-tubulin antibody. Bar, 10 μm.
To further ascertain the resistance of rodent cell lines to AG1478, we evaluated other markers of Golgi such as GalT, ERGIC53/p58, ARF1, β-COP, and GBF1 in AG1478-treated Rat2 cells. None of these proteins was redistributed from its normal perinuclear compact localization (data not shown). We also examined the effect of AG1478 in PtK1 and Madin-Darby canine kidney cells, which were kangaroo rat and dog epithelial cell lines, respectively, and reported to be resistant to BFA (29, 30). The Madin-Darby canine kidney and PtK1 cells were also resistant to the effect of AG1478 in dispersing the Golgi. On the other hand, AG1478 inhibited the EGFR signaling pathway in both human and rodent cells (Fig. 3B), supporting the conclusion that the effect of AG1478 for dispersing the Golgi is independent of its activity on EGFR. Taken together, these results suggest that AG1478 targets a molecular entity that exhibits species specificity for its Golgi dispersing effect.
Differential Effects of AG1478 on the Proteins Associated the cis-Golgi and trans-Golgi—To understand the mechanism by which AG1478 disperses the Golgi, we compared the activity of AG1478 with that of BFA for their effects on the distribution of Golgi-associated proteins. COPI has been shown to be the most important coat protein in facilitating retrograde intracellular transport from the Golgi complex to the ER. Recruitment of COPI to the membrane requires the activation of ARF1 (31, 32). In control cells, most of COPI and ARF1 are localized on the cis-Golgi membrane. The addition of AG1478 caused a rapid release of COPI and ARF1 into the cytoplasm (Fig. 4, A and B). The treatment with AG1478 also resulted in the redistribution of GBF1 from the Golgi region to a dispersed localization throughout the cytoplasm (Fig. 4C), an effect similar to that of BFA (33).
FIGURE 4.
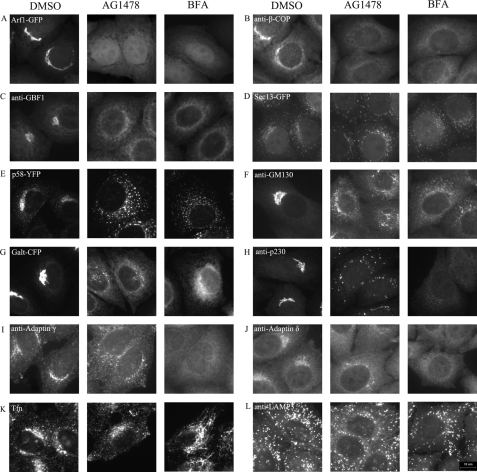
Effects of AG1478 on the distribution of intracellular Golgi-associated proteins. H4 cells expressing ARF1-GFP (A), Sec13-GFP (D), p58-YFP (E), and GalT-CFP (G) were incubated with DMSO, 14 μm AG1478, 5 μm BFA for 1 h, or H4 cells were incubated with DMSO, 14 μm AG1478, 5 μm BFA for 1 h and then stained with antibodies against β-COP (B), GBF1 (C), GM130 (F), p230 (H), adaptin γ (I), adaptin δ (J), and LAMP3 (L). K, H4 cells incubated with Alexa 568-labeled transferrin for 5 min and then treated with DMSO, 14 μm AG1478, 5 μm BFA for 1 h. Bar, 10 μm.
Sec13 is a component of the COPII coat that mediates vesicle budding from ER exit site (ERES) (34, 35). The treatment with AG1478 had no effect on the distribution of Sec13-GFP, which is localized to the punctate ERES, suggesting that the treatment of AG1478 does not perturb the COPII-dependent ER export machinery (Fig. 4D). The treatment with AG1478, however, caused the redistribution of p58/ERGIC53, a marker for the VTCs, from its normal compact localization to a dispersed cytoplasmic localization (Fig. 4E). GM130, a Golgi matrix protein, was also redistributed to the peripheral punctate structures after treatment by AG1478, similar to that of BFA (Fig. 4F). The peripheral punctate structures of p58/ERGIC53 and GM130 were similar to the “Golgi remnants” after BFA treatment (36, 37). GalT, a Golgi-resident enzyme, was found in both the ER and some punctate structures after the treatment with AG1478 (Fig. 4G). Taken together, the effects of AG1478 on the proteins localized in the cis-Golgi are very similar to that of BFA.
We also compared the effects of AG1478 and BFA on proteins associated with the trans-Golgi. After incubation of H4 cells with either compound for 1 h, 5 μm BFA completely dispersed the trans-Golgi network marker p230 from the perinuclear region into the cytosol, whereas after AG1478 treatment p230 remained as small punctate structures with partial redistribution to the cytoplasm (Fig. 4H) (38). Longer treatment with AG1478 (2-3 h) still did not completely disperse the punctate structures (data not shown). The effects of AG1478 on adaptin γ and δ, which are markers for AP-1 and AP-2, were similar to that of p230 (Fig. 4, I and J). Taken together, these results suggest that the cis-Golgi is more sensitive to AG1478 than the trans-Golgi.
The treatment of BFA is known to cause extensive formation of membrane tubules from endosomes (15, 39). Using Alexa 568-labeled transferrin, we analyzed the effect of AG1478 on the formation of tubular endosomes. BFA induced enlargement and tubulation of transferrinpositive endosomes, but AG1478 had no effect (Fig. 4K), suggesting that AG1478 has no effect on early/recycling endosomes as BFA. In addition, the treatment with AG1478 had no effect on the distribution of lysosomes systems, as marked by LAMP3, similar to that of BFA (Fig. 4L). Because of these similarities and differences, we conclude that AG1478 might target a similar target as BFA in dispersing the Golgi, but AG1478 might be more selective because it does not affect the endosomal compartment.
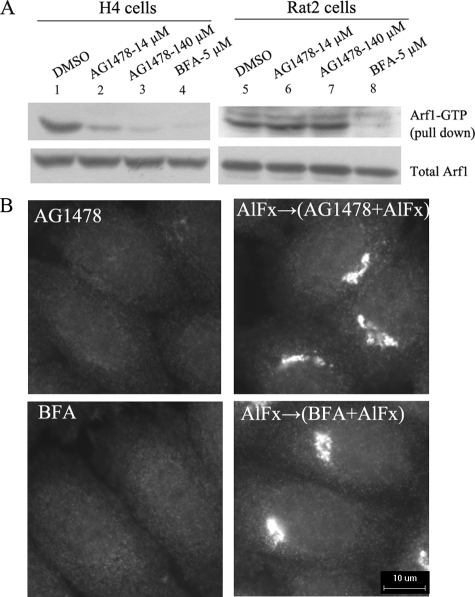
AG1478 Blocks ARF1 Activity in Human Cells—Our data described above suggest that in AG1478-treated cells, ARF1 might reside in an inactive state in the cytosol, similar to that of BFA-treated cells. To measure the ARF1 activity in AG1478-treated cells, we used an ARF1-GTP pull-down assay that monitors the amount of ARF1-GTP using a GST-GGA3-GAT domain fusion protein (21, 22). The GAT domain of GGA3, an effector of ARF1, preferentially binds the active ARF1-GTP over the inactive ARF1-GDP. Treatment with AG1478 and BFA for 1 h dramatically reduced the amount of ARF1-GTP compared with that of control cells (Fig. 5A). On the other hand, although the treatment of BFA reduced the amount of active ARF1 in Rat2 cells as predicted, incubation of Rat2 cells with AG1478 had no effect on the levels of active ARF1 even if in the presence of high concentration of AG1478 (140 μm). The species specificity of AG1478 on the human and rodent cells in reducing the amount of active ARF1 is consistent with its Golgi dispersing activity as described above and supports our hypothesis that AG1478 causes Golgi disassembly by blocking ARF1 activation in human cells.
FIGURE 5.
ARF1 inactivity causes Golgi disassembly induced by AG1478 on H4 cells. A, ARF1-GTP pulldown assay. H4 cells or Rat2 cells were transfected transiently with ARF1-GFP for 24 h, treated with DMSO (lanes 1 and 5), 14 μm AG1478 (lanes 2 and 6), 140 μm AG1478 (lanes 3 and 7), and 5 μm BFA (lanes 4 and 8) for 1 h before lysis. The upper panels show the amount of GTP bound ARF1-GTP, and the lower panels show the total amount of ARF1-GTP in cells. B, H4 cells were pretreated with control or 50 μm AlCl3 and 30 mm NaF for 10 min as indicated and then treated with 14 μm AG1478 or 1 μm BFA as indicated together for 30 min. The cells were stained with an antibody against β-COP. Bar, 10 μm.
Aluminum fluoride has been shown to activate the GDP-bound α subunit of heterotrimeric G-protein and trigger the conversion to the GTP-bound state. By stabilizing a subset of ARF1 on the Golgi membrane, aluminum fluoride was shown to slow the dissociation of ARF1 from the Golgi membrane in the presence of BFA (40-42). Consistent with our hypothesis that AG1478 inhibits the activation of ARF1, we found that the treatment with aluminum fluoride significantly inhibited the Golgi dispersing effect of AG1478 (Fig. 5B).
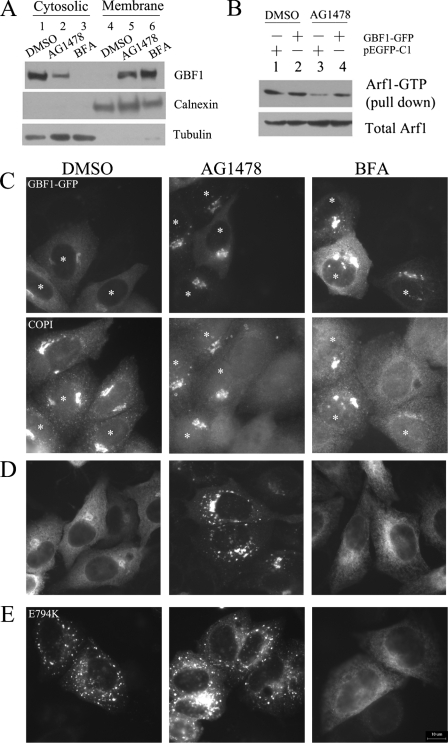
The Effects of AG1478 on GBF1—Because GBF1 has been shown to be stabilized on the VTCs and Golgi membrane in BFA-treated cells, we performed subcellular fractionation experiment in H4 cells treated with AG1478 and BFA. In control cells, most of the endogenous GBF1 was in the cytoplasmic fractions. As previously observed (23, 33), the treatment of BFA caused a dramatic redistribution of GBF1 from cytoplasmic to membrane fraction (Fig. 6A). The treatment of AG1478 also led to a similar increase of GBF1 in the membrane fractions and a corresponding reduction in the cytoplasmic fractions, indicating that AG1478 may also lead to stabilization of GBF1 on membranes (Fig. 6A).
FIGURE 6.
The effect of AG1478 on GBF1. A, H4 cells were treated with DMSO (lanes 1 and 4), 14 μm AG1478 (lanes 2 and 5), 5 μm BFA (lanes 3 and 6) for 1 h, homogenized, and subjected to differential centrifugation. Equivalent amounts of the cytosolic and membrane fractions were resolved by SDS-PAGE and Western blotted with anti-GBF1, anti-calnexin, and anti-tubulin. B, H4 cells were transfected with ARF1-GFP together with GBF1-GFP or pEGFP-C1 for 24 h and then treated with DMSO, 14 μm AG1478 for 30 min. The cell lysates were analyzed by ARF1 pulldown assay. C, H4 cells were transfected transiently with GBF1-GFP plasmid for 24 h and treated with DMSO, 14 μm AG1478, or 1 μm BFA for 30 min. Cells expressing GBF1-GFP are indicated by white asterisks. The cells were stained with an antibody against β-COP. D, H4 cells were transfected transiently with GBF1-GFP plasmid for 24 h and treated with DMSO, 14 μm AG1478, or 5 μm BFA for 1 h. E, H4 cells were transfected transiently with E794K-GFP plasmid for 24 h and treated with DMSO, 14 μm AG1478, or 5 μm BFA for 1 h. Bar, 10 μm.
We further determined the effect of overexpressing GBF1 on the Golgi dispersing effect of AG1478. Overexpression of GBF1 has been shown to reduce the sensitivity of cells to BFA (43). GBF1-GFP was localized to both cytoplasm and the Golgi in H4 cells that were transiently transfected with an expression construct of GBF1-GFP (Fig. 6C) (33, 44). Expression of GBF1 at low levels had no effect on the localization of COPI to the cis-Golgi. The treatment with AG1478 (14 μm) for 30 min led to the rapid release of COPI into cytosol in control H4 cells but was ineffective in GBF1-GFP expressing H4 cells. Thus, overexpression of GBF1 prevents the release of COPI induced by AG1478, similar to the effect seen with BFA. Consistent with a protective effect by GBF1, increased amounts of ARF1-GTP were found in cells overexpressing GBF1 compared with control cells in the presence of AG1478 (Fig. 6B). These observations indicate that overexpression of GBF1 also confers resistance to AG1478-induced Golgi disassembly.
Prolonged treatment of cells with 5 μm BFA led to the redistribution of GBF1 into the ER (Fig. 6D) (33, 43). Interestingly, the treatment with 14 μm AG1478 for 1 h led to the appearance of GBF1-GFP in smaller punctate structures throughout the cell and larger bright clusters around the nucleus, which persisted even after longer treatment (4 h) at high concentration (42 μm) (Fig. 6D). Thus, AG1478, in contrast to BFA, is unable to localize GBF1-GFP into the reticular network. The small punctate structures labeled by GBF1-GFP in the presence of AG1478 resemble the structures formed by the catalytically inactive GBF1 mutant E794K (Fig. 6E) (44).
E794K mutation, which caused a change reversal at the edge of a hydrophobic catalytic guanine nucleotide exchange center, has been shown to completely abolish the nucleotide exchange activity and stabilize the interaction between the mutant GEF and ARF1-GDP (45, 46). The expression of E794K GBF1 causes the complete disassembly of the Golgi, similar to that of BFA-treated cells, whereas E794K GBF1 itself is known to be localized to the VTCs adjacent to the ERES (44). The treatment of BFA caused redistribution of E794K GBF1 from the VTCs adjacent to ERES into the ER (23, 44). In striking contrast, the punctate dots of E794K GBF1-GFP were resistant to the effect of AG1478 (Fig. 6E), suggesting that different from that of BFA, AG1478 is unable to redistribute E794K GBF1 from the VTCs adjacent to the ERES into the ER. Taken together, we conclude that the treatment of AG1478 induces wild type GBF1-GFP to phenocopy the E794K GBF1 mutant. Furthermore, the interaction of AG1478 with GBF1 most likely requires the catalytic activation of the Sec7 domain in GBF1. On the other hand, the catalytic activity of the Sec7 domain in GBF1 is not required for BFA to induce the redistribution of E794K mutant from the VTCs adjacent to the ERES into the ER.
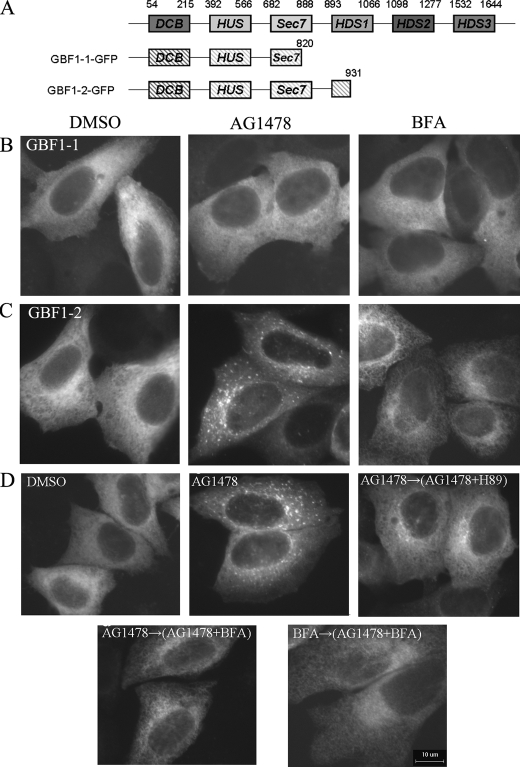
The Requirement of the Sec7 Domains of GBF1 for the Activity of AG1478—In addition to the Sec7 domain, GBF1 contains two noncatalytic domains (DCB and HUS) at its N terminus and three additional noncatalytic domains of its C terminus (HDS1, HDS2, and HDS3) (Fig. 7A) (47, 48). To gain more insight into the domains of GBF1 that are responsible for the effect of AG1478, we made two deletion constructs from GBF1-GFP plasmid without the C-terminal domain. The GBF1-1-GFP construct contains the DCB, HUS, and parts of the Sec7 domain, whereas the GBF1-2-GFP comprises the DCB, HUS, and the entire Sec7 domain. Both GBF1-1-GFP and GBF1-2-GFP were localized to the cytosol in a homogeneous manner and did not induce Golgi disassembly (data not shown). The treatment with BFA or AG1478 did not cause any significant change in GBF1-1-GFP distribution (Fig. 7B). Interestingly, the treatment with AG1478 induced GBF1-2-GFP to form peripheral punctate structures, similar to those seen with the E794K mutant (Fig. 6E). On the other hand, the addition of BFA redistributed GBF1-2 into a reticular network, most likely the ER, without any obvious punctate structure (Fig. 7C). Because GBF1-2-GFP but not GBF1-1-GFP contains an intact Sec7 domain, this result is consistent with the requirement of the Sec7 domain in interaction with AG1478. The requirement of an intact Sec7 domain suggests that AG1478 targets the Sec7 domain directly or indirectly; alternatively, the deletion could indirectly affect the targeting of AG1478 by affecting the overall structure of GBF1. These possibilities need to be examined in the future.
FIGURE 7.
The effect of AG1478 on the GBF1-1 protein. A, schematic representation of different deletion constructs from GBF1-GFP. B and C, H4 cells were transfected with GBF1-1-GFP or GBF1-2-GFP for 24 h and then treated with DMSO, 14 μm AG1478, or 5 μm BFA as indicated for 1 h. D, H4 cells were transfected with GBF1-2-GFP for 24 h and treated with DMSO; treated with 14 μm AG1478 for 1 h; pretreated with 14 μm AG1478 for 1 h and then treated with 100 μm H89 together with AG1478 for 10 min; treated with 5 μm BFA together with AG1478 for 1 h; or pretreated with 5 μm BFA for 1 h and then treated with 14 μm AG1478 together with BFA for 1 h. Bar, 10 μm.
To characterize the peripheral punctate structures to which GBF1-2 localizes in the presence of AG1478, we used small molecules that are known to have effects on the Golgi. H89, a cAMP-dependent protein kinase inhibitor, was shown to block COPII recruitment to ER membrane. The “Golgi remnants” localized to VTCs adjacent to ERES induced by BFA could be redistributed to ER after combined treatment with BFA and H89 (49, 50). Treatment with H89 and AG1478 together in H4 cells redistributed the peripheral punctate structures of GBF1-2-GFP to the ER, suggesting that GBF1-2 punctates formed upon AG1478 treatment may also be localized to VTCs adjacent to ERES (Fig. 7D).
Because the treatment of BFA led to the redistribution of GBF1-2-GFP into a reticular network, we tested whether BFA can further redistribute the punctate structures formed by GBF1-2-GFP upon AG1478 treatment. As shown in Fig. 7D, the addition of BFA after AG1478 treatment led to the redistribution of the punctate structures of GBF1-2-GFP into a reticular network, likely corresponding to the ER. On the other hand, the addition of AG1478 after the treatment of BFA was not able to induce GBF1-2-GFP to form punctate structures. Thus, the effect of BFA is dominant over that of AG1478.
The VTCs adjacent to the ERES can be differentiated by the presence of ERGIC53 and COPII blank marker (44, 51). To further characterize the peripheral punctate structures of GBF1-2-GFP and GBF1-GFP induced by AG1478, we examined their distribution relative to that of ERGIC53 and Sec13, which are markers for the VTCs and the ERES, respectively. The punctate spots of GBF1-2-GFP and GBF1-GFP induced by AG1478 overlapped almost completely with ERGIC53 but only partially with that of Sec13 (Fig. 8). Together, these results confirm that AG1478 treatment causes the accumulation of GBF1 and GBF1-2 on the VTCs adjacent to the ERES, whereas the treatment of BFA is known to cause the complete redistribution of GBF1 into the ER network.
DISCUSSION
We screened a small molecule library to identify new molecules that affect the Golgi for reagents as tools to study intracellular membrane trafficking. We identified a novel activity of known compound, AG1478, in down-regulation of the ARF1 activity, which in turn leads to the dispersal of the Golgi structure. The effect of AG1478 on the cis-Golgi is more selective than that of BFA, and AG1478 does not affect the endosomal systems as BFA does. We show that the Golgi dispersing effect of AG1478 can be partially suppressed by the overexpression of GBF1, a large GEF that promotes the GDP to GTP exchange of ARF1. This effect is similar to that of BFA. Interestingly, however, in contrast to BFA, AG1478 was unable to disperse overexpressed GBF1 from the peripheral VTCs adjacent to the ERES into the ER. Instead, in the presence of AG1478, wild type GBF1-GFP forms punctate structures on the VTCs adjacent to the ERES, a phenotype similar to that with the E794K GBF1-GFP mutant. We conclude that AG1478 is likely to target a molecular entity that regulates the Sec7 nucleotide exchange activity of GBF1 in human cells but not rodent, canine, or kangaroo rat cells.
The ARF family of GTPases plays a central role in maintaining Golgi structure and function. Six ARF isoforms (ARF1-6) in mammals can be classified into three classes based on their primary sequences; however, ARF2 is lost in the human genome. ARF isoforms exhibit similar biochemical activities in regulating membrane traffic (52, 53). ARF1, the best characterized ARF, localizes primarily to the Golgi complex and regulates several types of coat proteins. Our results demonstrate that Golgi disassembly induced by AG1478 is caused by inhibition of ARF1 activation. We show that ARF1 exists in an inactive state localized in the cytosol of AG1478 treated cells, indicating a failure in the recruitment of ARF1 to membranes to facilitate COPI transport. The amount of active ARF1-GTP production in H4 human cells was dramatically decreased after AG1478 treatment.
The selective effect of AG1478 on the cis-Golgi associated proteins led us to first examine the role of GBF1, which is the only GEF localized at the cis-Golgi. GBF1 is a high molecular weight GEF that cycles rapidly on and off the Golgi membrane. The mobility of GBF1 is associated with its exchange activity because an inactive mutant E794K cycles slower and stabilizes on membrane longer than wild type GBF1 (23). Sztul et al. (23) propose that GBF1 is stabilized on membrane when in a complex with ARF1-GDP, and the catalytic activity of GBF1 is required for its dissociation from ARF and membrane. Because the treatment of AG1478 led to an increased association of GBF1 with the membrane, we propose that AG1478 also inhibits the catalytic activity of GBF1, and in the presence of AG1478, GBF1 is also stabilized on the membrane in complex with ARF1-GDP.
GBF1 is localized to the peripheral VTCs and the juxtanuclear early Golgi in normal mammalian cells (33, 44). However, the anti-GBF1 antibody that we used does not recognize the endogenous GBF1 in peripheral puncta on the VTCs in control cells (Fig. 4C) as demonstrated elegantly by the affinity purified anti-GBF1 antibody by the Melançon laboratory (33) and the Sztul laboratory (44). AG1478 cannot disperse the transfected wild type GBF1 from the VTCs (Fig. 6D), which mimics the behavior of the E794K GBF1 mutant. On the other hand, the treatment of BFA can disperse E794K from the VTCs into the ER (44). Thus, the treatment of AG1478, but not that of BFA, provides a phenocopy of the Sec7 mutant GBF1. An time-lapsed imaging analysis showed that the dispersal of GBF1-GFP induced by BFA occurs in two steps: a transient accumulation of GBF1-GFP in both peripheral puncta and juxtanuclear Golgi area and a subsequent dispersal into the ER (33). Thus, the treatment of AG1478 might arrest the GBF1-GFP at the first step. Because GBF1 knockdown also blocks cargo in peripheral VTCs structures (51), it has been proposed that inactive GBF1 is localized to the VTCs adjacent to the ERES. We propose that AG1478 selectively targets the Sec7 activity of GBF1 and traps GBF1 on VTCs adjacent to the ERES, whereas BFA inhibits not only the nucleotide exchange activity of GBF1 but also an additional step that is independent of Sec7 activity and leads to the dispersal of GBF1 into the ER. For example, BFA might works with multiple GEFs, each affecting a different compartment including the compartment to which E794K targets, thereby changing its distribution. On the other hand, the effect of AG1478 might be more selective. Therefore, AG1478 might provide a useful tool in defining an intermediate vesicular trafficking step that can be only transiently observed using BFA. The low cytotoxicity of AG1478 also suggests that the cytotoxicity of BFA might not be directly related to its inhibitory effect on the Sec7 nucleotide exchange activity. Because BFA has been proposed to inhibit additional targets to affect the endosomal system (14-16), whereas AG1478 does not have such side effects, it is possible that the inhibitory effect of BFA on its additional target(s) involved in the endosomal trafficking leads to its cytotoxicity.
The selectivity of AG1478 for human cells is also interesting. One might speculate that such selectivity is caused by species difference in the target of AG1478. Alternatively, the presence of ARF2 isoform in rodent cells but not human cells might explain the resistance of rodent cells to AG1478. To test the latter possibility, we have used siRNA to decrease expression of ARF2 in Rat2 cells. However, the expression of siRNA for ARF2 did not sensitize the rodent cells to AG1478 (data not shown). We have also considered the possibility that GBF1 is the direct target of AG1478. Because the Sec7 domains of human and mouse GBF1 proteins are highly similar with 96% identity at the amino acid level, we made an expression construct expressing a h(m)GBF1-2 fusion protein that contains the Sec7 domain of mouse GBF1 with other domains of human GBF1. AG1478 treatment redistributed this h(m)GBF1-2 to the VTCs adjacent to the ERES in H4 cells, similar to the effect seen with the wild type (data not shown), suggesting that differences in the Sec7 domains of GBF1 between mouse and human cannot account for the resistance in rodent cells.
In conclusion, we have identified a novel Golgi dispersing molecule AG1478. We show that similar to BFA, AG1478 inhibits the activity of ARF1. AG1478 exhibits a number of interesting properties distinct from that of BFA. First, AG1478 exhibits low cytotoxicity, whereas BFA is highly toxic. Second, the Golgi dispersing effect of AG1478 is highly species specific, whereas BFA can affect both human and rodent cells. Third, AG1478 preferentially targets the cis-Golgi compartment, whereas BFA targets both the cis- and trans-Golgi. Fourth, AG1478 redistributes GBF1 to the peripheral VTCs adjacent to that of ERES but not into the ER, mimicking the phenotype of the E794K GBF1 mutant, whereas BFA can disperse the E794K mutant into the ER network. Because the interaction of AG1478 with GBF1 depends upon a functional Sec7 domain, we propose that AG1478 provides an interesting new tool to define the functional role of nucleotide exchange factors.
Supplementary Material
Acknowledgments
We are grateful to Dr. Tom Rapoport (Harvard Medical School) and Drs. Lippincott-Schwartz (National Institutes of Health) for critical reading and editing of this manuscript. We thank Renxiao Wang (Shanghai Institute of Organic Chemistry) for model analysis and helpful discussion. We express our deep appreciation for the kind gifts of plasmids and antibodies from Drs. Lippincott-Schwartz (National Institutes of Health), Elizabeth Sztul (University of Alabama, Birmingham), Benjamin S. Glick (University of Chicago), Hans-Peter Hauri (University of Basel), Paul A. Randazzo (National Institutes of Health), and Wanjin Hong (Institute of Molecular Biology, Singapore).
This work was supported, in whole or in part, by National Institutes of Health Grant R37 AG12859 (to J. Y.). This work was also supported by National Natural Science Foundation of China Grant 20321202 and Chinese Academy of Science Grant KGCX2-SW-209 (to D. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1-S3.
Footnotes
The abbreviations used are: COP, coat protein complex; ARF, ADP-ribosylation factor; BFA, brefeldin A; DMSO, dimethyl sulfoxide; EGF, epidermal growth factor; EGFR, EGF receptor; ER, endoplasmic reticulum; ERES, ER exit sites; ERGIC, ER-Golgi intermediate compartment; GBF1, Golgi-specific brefeldin A resistance factor 1; GEF, guanine nucleotide exchange factor; GFP, green fluorescent protein; VTCs, vesicular tubular clusters structure; ERK, extracellular signal-regulated kinase; siRNA, small interfering RNA; GST, glutathione S-transferase; z, benzyloxycarbonyl; fmk, fluoromethyl ketone; GalT, galactosyltransferase; DCB, dimerization/cyclophilin binding region; HUS, homology upstream of Sec7; HDS, homology downstream of Sec7.
References
- 1.Rothman, J. E. (1994) Nature 372 55-63 [DOI] [PubMed] [Google Scholar]
- 2.Schekman, R. a. O. L. (1996) Science 271 1526-1533 [DOI] [PubMed] [Google Scholar]
- 3.Hauri, H. P., Kappeler, F., Andersson, H., and Appenzeller, C. (2000) J. Cell Sci. 113 587-596 [DOI] [PubMed] [Google Scholar]
- 4.Ward, T. H., Polishchuk, R. S., Caplan, S., Hirschberg, K., and Lippincott-Schwartz, J. (2001) J. Cell Biol. 155 557-570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altan-Bonnet, N., Sougrat, R., and Lippincott-Schwartz, J. (2004) Curr. Opin. Cell Biol. 16 364-372 [DOI] [PubMed] [Google Scholar]
- 6.D'Souza-Schorey, C., and Chavrier, P. (2006) Nat. Rev. Mol. Cell. Biol. 7 347-358 [DOI] [PubMed] [Google Scholar]
- 7.Donaldson, J. G., Honda, A., and Weigert, R. (2005) Biochim. Biophys. Acta 1744 364-373 [DOI] [PubMed] [Google Scholar]
- 8.Claude, A., Zhao, B. P., Kuziemsky, C. E., Dahan, S., Berger, S. J., Yan, J. P., Armold, A. D., Sullivan, E. M., and Melançon, P. (1999) J. Cell Biol. 146 71-84 [PMC free article] [PubMed] [Google Scholar]
- 9.Kawamoto, K., Yoshida, Y., Tamaki, H., Torii, S., Shinotsuka, C., Yamashina, S., and Nakayama, K. (2002) Traffic 3 483-495 [DOI] [PubMed] [Google Scholar]
- 10.Renault, L., Guibert, B., and Cherfils, J. (2003) Nature 426 525-530 [DOI] [PubMed] [Google Scholar]
- 11.Mossessova, E., Corpina, R. A., and Goldberg. J. (2003) Mol. cell 12 1403-1411 [DOI] [PubMed] [Google Scholar]
- 12.Tsai, S. C., Adamik, R., Haun, R. S., Moss, J., and Vaughan, M. (1993) J. Biol. Chem. 268 10820-10825 [PubMed] [Google Scholar]
- 13.Dinter, A., and Berger, E. G. (1998) Histochem. Cell Biol. 109 571-590 [DOI] [PubMed] [Google Scholar]
- 14.Lippincott-Schwartz, J., Yuan, L., Tipper, C., Amherdt, M., Orci, L., and Klausner, R. D. (1991) Cell 67 601-616 [DOI] [PubMed] [Google Scholar]
- 15.Wood, S. A., Park, J. E., and Brown, W. J. (1991) Cell 67 591-600 [DOI] [PubMed] [Google Scholar]
- 16.Hunziker, W., Whitney, J. A., and Mellman, I. (1991) Cell 67 617-627 [DOI] [PubMed] [Google Scholar]
- 17.Guo, H., Tittle, T. V., Allen, H., and Maziarz, R. T. (1998) Exp. Cell Res. 245 57-68 [DOI] [PubMed] [Google Scholar]
- 18.Citterio, C., Vichi, A., Pacheco-Rodriguez, G., Aponte, A. M., Moss, J., and Vaughan, M. (2008) Proc. Natl. Acad. Sci. U. S. A 105 2877-2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brewer, C. B., and Roth, M. G. (1995) J. Cell Sci. 108 789-796 [DOI] [PubMed] [Google Scholar]
- 20.Smith, K., Gunaratnam, L., Morley, M., Franovic, A., Mekhail, K., and Lee, S. (2005) Cancer Res. 65 5221-5230 [DOI] [PubMed] [Google Scholar]
- 21.Shinotsuka, C., Yoshida, Y., Kawamoto, K., Takatsu, H., and Nakayama, K. (2002) J. Biol. Chem. 277 9468-9473 [DOI] [PubMed] [Google Scholar]
- 22.Yoon, H. Y., Bonifacino, J. S., and Randazzo, P. A. (2005) Methods Enzymol. 404 316-332 [DOI] [PubMed] [Google Scholar]
- 23.Szul, T., Garcia-Mata, R., Brandon, E., Shestopal, S., Alvarez, C., and Sztul, E. (2005) Traffic 6 374-385 [DOI] [PubMed] [Google Scholar]
- 24.Nakamura, N., Rabouille, C., Watson, R., Nilsson, T., Hui, N., Slusarewicz, P., Kreis, T. E., and Warren, G. (1995) J. Cell Biol. 131 1715-1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carpenter, A. E., Jones, T. R., Lamprecht, M. R., Clarke, C., Kang, I. H., Friman, O., Guertin, D. A., Chang, J. H., Lindquist, R. A., Moffat, J., Golland, P., and Sabatini, D. M. (2006) Genome Biol. 7 R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamprecht MR, S. D., and Carpenter, A. E. (2007) BioTechniques 42 71-75 [DOI] [PubMed] [Google Scholar]
- 27.Daub, H., Weiss, F. U., Wallasch, C., and Ullrich, A. (1996) Nature 379 557-560 [DOI] [PubMed] [Google Scholar]
- 28.Levitzki, A., and Gazit, A. (1995) Science 267 1782-1788 [DOI] [PubMed] [Google Scholar]
- 29.Low, S. H., Wong, S. H., Tang, B. L., Tan, P., Subramaniam, V. N., and Hong, W. (1991) J. Biol. Chem. 266 17729-17732 [PubMed] [Google Scholar]
- 30.Ktistakis, N. T., Roth, M. G., and Bloom, G. S. (1991) J. Cell Biol. 113 1009-1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donaldson, J. G., Kahn, R. A., and Lippincott-Schwartz, J., and Klausner, R. D. (1991) Science 254 1197-1199 [DOI] [PubMed] [Google Scholar]
- 32.Rabouille, C., and Klumperman, J. (2005) Nat. Rev. Mol. Cell. Biol. 6 812-817 [DOI] [PubMed] [Google Scholar]
- 33.Zhao, X., Claude, A., Chun, J., Shields, D. J., Presley, J. F., and Melançon, P. (2006) J. Cell Sci. 119 3743-3753 [DOI] [PubMed] [Google Scholar]
- 34.Hammond, A. T., and Glick, B. S. (2000) Mol. Biol. Cell 11 3013-3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang, B. L., Peter, F., Krijnse-Locker, J., Low, S. H., Griffiths, G., and Hong, W. (1997) Mol. Cell. Biol. 17 256-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orci, L., Perrelet, A., Ravazzola, M., Wieland, F. T., Schekman, R., and Rothman, J. E. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 11089-11093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seemann, J., Jokitalo, E., Pypaert, M., and Warren, G. (2000) Nature 407 1022-1026 [DOI] [PubMed] [Google Scholar]
- 38.Gleeson, P. A., Anderson, T. J., Stow, J. L., Griffiths, G., Toh, B. H., and Matheson, F. (1996) J. Cell Sci. 109 2811-2821 [DOI] [PubMed] [Google Scholar]
- 39.Stephen, G. M., Lucinda, C., and Hsiao-Ping, H. M. (1992) J. Cell Biol. 118 267-2831629235 [Google Scholar]
- 40.Rittinger, K., Walker, P. A., Eccleston, J. F., Smerdon, S. J., and Gamblin, S. J. (1997) Nature 389 758-762 [DOI] [PubMed] [Google Scholar]
- 41.Finazzi, D., Cassel, D., Donaldson, J. G., and Klausner, R. D. (1994) J. Biol. Chem. 269 13325-13330 [PubMed] [Google Scholar]
- 42.Feng, Y., Yu, S., Lasell, T. K., Jadhav, A. P., Macia, E., Chardin, P., Melancon, P., Roth, M., Mitchison, T., and Kirchhausen, T. (2003) Proc. Natl. Acad. Sci. (U. S. A) 100 6469-6474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niu, T. K., Pfeifer, A. C., Lippincott-Schwartz, J., and Jackson, C. L. (2005) Mol. Biol. Cell 16 1213-1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.García-Mata, R., Szul, T., Alvarez, C., and Sztul, E. (2003) Mol. Biol. Cell 14 2250-2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cherfils, J., Ménétrey, J., Mathieu, M., Le Bras, G., Robineau, S., Béraud-Dufour, S., Antonny, B., and Chardin, P. (1998) Nature 392 101-105 [DOI] [PubMed] [Google Scholar]
- 46.Mossessova, E., Gulbis, J. M., and Goldberg, J. (1998) Cell 92 415-423 [DOI] [PubMed] [Google Scholar]
- 47.Mouratou, B., Biou, V., Joubert, A., Cohen, J., Shields, D. J., Geldner, N., Jürgens, G., Melançon, P., and Cherfils, J. (2005) BMC Genomics 6 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramaen, O., Joubert, A., Simister, P., Belgareh-Touzé, N., Olivares-Sanchez, M. C., Zeeh, J. C., Chantalat, S., Golinelli-Cohen, M. P., Jackson, C. L., Biou, V., and Cherfils, J. (2007) J. Biol. Chem. 282 28834-28842 [DOI] [PubMed] [Google Scholar]
- 49.Lee, T. H., and Linstedt, A. D. (2000) Mol. Biol. Cell 11 2577-2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puri, S., and Linstedt, A. D. (2003) Mol. Biol. Cell 14 5011-5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manolea, F., Claude, A., Chun, J., Rosas, J., and Melançon, P. (2008) Mol. Biol. Cell 19 523-535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chun, J., Shapovalova, Z., Dejgaard, S. Y., Presley, J. F., and Melançon, P. (2008) Mol. Biol. Cell. 19 3488-3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Volpicelli-Daley, L. A., Li, Y., Zhang, C. J., and Kahn, R. A. (2005) Mol. Biol. Cell 16 4495-4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.