Abstract
The serpinopathies are a group of inherited disorders that share as their molecular basis the misfolding and polymerization of serpins, an important class of protease inhibitors. Depending on the identity of the serpin, conditions arising from polymerization include emphysema, thrombosis, and dementia. The structure of serpin polymers is thus of considerable medical interest. Wild-type α1-antitrypsin will form polymers upon incubation at moderate temperatures and has been widely used as a model system for studying serpin polymerization. Using hydrogen/deuterium exchange and mass spectrometry, we have obtained molecular level structural information on the α1-antitrypsin polymer. We found that the flexible reactive center loop becomes strongly protected upon polymerization. We also found significant increases in protection in the center of β-sheet A and in helix F. These results support a model in which linkage between serpins is achieved through insertion of the reactive center loop of one serpin into β-sheet A of another. We have also examined the heat-induced conformational changes preceding polymerization. We found that polymerization is preceded by significant destabilization of β-sheet C. On the basis of our results, we propose a mechanism for polymerization in which β-strand 1C is displaced from the rest of β-sheet C through a binary serpin/serpin interaction. Displacement of strand 1C triggers further conformational changes, including the opening of β-sheet A, and allows for subsequent polymerization.
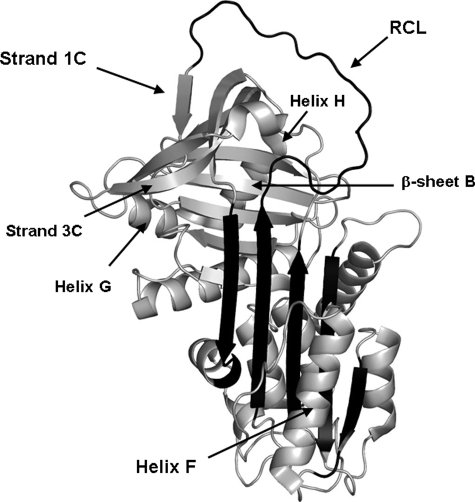
α1-Antitrypsin (α1-AT)2 is a member of the serpin class of protease inhibitors. Serpins inhibit their target proteases via a unique mechanism (Fig. 1) (1). Cleavage of a serpin's reactive center loop (RCL) by a protease triggers a massive conformational change in which the RCL inserts into the central β-sheet A, becoming a sixth strand. This process disrupts the protease active site and traps the acyl-enzyme intermediate, rendering the protease inactive. The active, loop-expelled form of α1-AT is significantly less stable than the loop-inserted form, and as a result of this metastability, the serpin structure is readily disrupted by mutations that result in inappropriate conformational changes. It has been established that a number of serious diseases associated with serpin mutations are caused by the formation and accumulation of serpin polymers (2). The details of the polymerization mechanism are therefore of considerable interest.
FIGURE 1.
Crystal structure of the active metastable form of α1-AT (Protein Data Bank code 1QLP). Helix F, β-strand 1C, and the RCL are indicated. β-Sheet A is shown in black.
Serpin polymerization has been studied extensively by a variety of methods, including circular dichroism, fluorescence spectroscopy, electron microscopy, and x-ray crystallography (3–5). The most generally accepted model of pathological serpin polymerization is one in which the RCL of one serpin anneals between β-strands 3 and 5A of another (1). Two major lines of evidence support this model. The first is that RCL-mimicking peptides have been shown to anneal between strands 3 and 5A and have also been shown to block polymerization in vitro (6). The second comes from fluorescence spectroscopy. Distances between donor-acceptor Förster resonance energy transfer pairs were measured at multiple sites in polymerized α1-AT, and subsequent modeling found that a structure in which the RCL of one molecule anneals between strands 3 and 5A of another was most consistent with the distance constraints (7). In contrast, although several crystal structures of polymerized serpins have been determined, none of them reveal this type of linkage. Linkage mechanisms observed to date in crystal structures include several types of RCL/β-sheet C interactions and an RCL/strand 6A interaction (8–10). Furthermore, electron microscopy has detected a head-to-head disulfide-bonded interaction (11). The structure of pathogenic serpin polymers has thus not been unambiguously resolved, although detailed knowledge of the structure of these polymers is critical for understanding the basis of serpin-linked diseases and for designing effective therapeutics.
Hydrogen/deuterium exchange measured by mass spectrometry (HXMS) has proven to be a powerful method for probing the structure and dynamics of proteins and protein assemblies that are not accessible to other techniques, and several recent studies have employed HXMS to gain insight into the structural organization of amyloid fibrils (12–14). HXMS provides information on dynamics and solvent accessibility throughout the entire structure of a protein and does not require the attachment of exogenous labels (15). In this work, we employed HXMS to probe the local dynamics and solvent accessibility of polymeric wild-type α1-AT and to examine the structural changes that occur in monomeric α1-AT prior to polymerization. Our results reveal the structure of polymerized α1-AT at a new level of detail and suggest a mechanism for polymerization.
MATERIALS AND METHODS
Expression and Purification of Wild-Type α1-AT—Wild-type α1-AT was expressed and purified as described previously (16). The activity assay on purified α1-AT was done as described previously (16). For all samples, the stoichiometry of inhibition was determined to be 1.0.
Native Gel and Silver Staining—Each monomeric and polymeric sample (500 ng each) was loaded onto a native gel (imidazole/HEPES (pH 7.4)) and run at 15 mA in electrophoresis running buffer containing 43 mm imidazole and 35 mm HEPES (pH 7.4). During the run, the electrophoresis apparatus was immersed in ice to prevent sample denaturation and polymer dissociation. Proteins were visualized by silver staining.
Electron Microscopy—A solution containing polymerized α1-AT was placed on a carbon-coated electron microscopy grid and visualized by negative staining.
Peptide Mapping by High Performance Liquid Chromatography (HPLC)-Tandem Mass Spectrometry—Peptide mapping experiments were performed as described previously (16) using an LCQ-DECA mass spectrometer (Thermo Electron Corp.). Sequencing of each peptic peptide was carried out using the search algorithm SEQUEST (Thermo Electron Corp.) and by manual inspection.
Hydrogen/Deuterium Exchange of Monomeric Wild-type α1-AT and Polymers—Hydrogen/deuterium (H/D) exchange of monomeric wild-type α1-AT at 25 °C was performed by 10-fold dilution of the sample with 10 mm sodium phosphate (pD 7.8), 50 mm NaCl deuterium buffer. The exchange reaction was allowed to proceed for different time periods, and the reaction was quenched by dropping the pH by the addition of cold 100 mm NaH2PO4 (pH 2.4) buffer containing 2 m guanidine hydrochloride. The quenched sample was immediately frozen in a dry ice/ethanol bath. The final protein concentration was 0.1 μm. Similarly, H/D exchange of monomeric wild-type α1-AT (0.1 mg/ml) at 45 °C was carried out by adding prewarmed 10 mm sodium phosphate (pD 7.8), 50 mm NaCl deuterium buffer. Each sample was incubated in the deuterium buffer for a different time period, and the reaction was quenched as described above. Deuterium labeling times at 45 °C were 10, 20, 30, 50, 60, 80, 90, 100, 200, 300, 400, 500, 700, 1000, 2000, 3000, and 5000 s. The final protein concentration was 0.1 μm. Temperature dependence of H/D exchange between 25 °C and 45 °C samples was corrected using the HXpep software package, and accordingly, 25 °C samples were incubated in the deuterium buffer for six times longer than the corresponding 45 °C samples. For pulse labeling experiments, α1-AT was preincubated in H2O at either 25 or 45 °C and then pulsed with 10-fold dilution into D2O at the same temperature, incubated for 10 (at 45 °C) or 60 (at 25 °C) s, and quenched.
Wild-type α1-AT polymer was prepared by incubating the protein (0.1 mg/ml) in 10 mm sodium phosphate (pH 7.8), 50 mm NaCl at 45 °C for 4 days. The presence of the polymer was confirmed by electron microscopy and a native gradient gel. The incubated sample was aliquotted and added to prewarmed 10 mm sodium phosphate (pD 7.8), 50 mm NaCl deuterium buffer. The exchange reaction was quenched at different time points by the addition of cold 100 mm NaH2PO4 (pH 2.4) containing 2 m guanidine hydrochloride. The sample was immediately frozen in a dry ice/ethanol bath. The final protein concentration was 0.1 μm.
Isotope Analysis by HPLC-Electrospray Ionization Mass Spectrometry—Deuterated protein was digested with porcine pepsin and analyzed by HPLC-mass spectrometry as described previously (16). Briefly, the frozen sample was thawed and digested with 5 μg of porcine pepsin on ice for 5 min. Peptic peptides were separated using a micropeptide trap and a C18 HPLC column and eluted with an acetonitrile gradient (2 to 45% over 12 min) at a flow rate of 50 μl/min. The centroid mass of each peptide was determined using the MagTran software package. Because we are concerned with differences in deuterium uptake rather than absolute values, the percent exchange for each peptide was calculated directly without a correction for back-exchange. The percent exchange was plotted against deuteration time, and a trend line was drawn using ORIGIN (Origin-Lab). Peptide identification was as described previously (16).
RESULTS
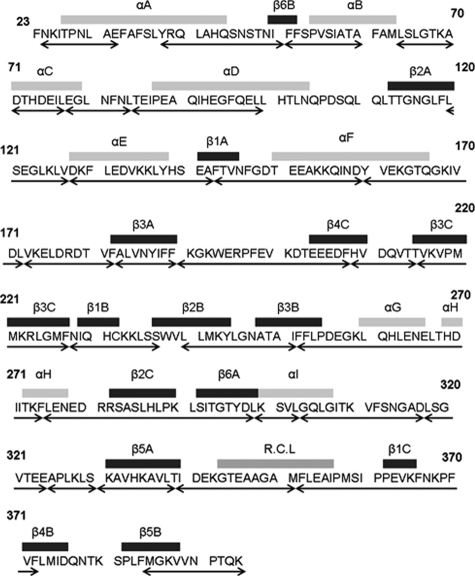
Structure and Dynamics of the Monomeric Intermediate Preceding Polymerization—Amide hydrogens that are solvent-exposed and not hydrogen-bonded will rapidly exchange with deuterium when a protein is incubated in D2O. Hydrogens that are protected from exchange by local structure can exchange if this structure is transiently broken by fluctuations. Hydrogen exchange thus provides a measure of both solvent exposure and conformational flexibility in proteins (17). When coupled with pepsin digestion, HXMS experiments quantify region-specific deuterium uptake (15). The peptides analyzed in this study encompass 71% of the entire amino acid sequence of wild-type antitrypsin covering the most secondary structural regions (Fig. 2). HXMS experiments were performed to monitor the conformational change from the native form to a polymerization-prone intermediate form at 45 °C and pH 7.8.
FIGURE 2.
Sequence of α1-AT. Elements of the secondary structure are indicated above the sequence. Below, the peptides used in this study are indicated by double-headed arrows.
According to a previous study, the active form of α1-AT, upon incubation at 45 °C, undergoes a conformational change to a partially disrupted structure in which β-strands 3 and 5A are separated (3). This conclusion was based on changes seen in intrinsic tryptophan fluorescence and changes in the CD spectrum during the first 4000 s of incubation. This initial conformational change was concentration-independent, but further incubation led to the concentration-dependent formation of polymers. Deuterium labeling was performed at 45 °C, with deuteration times ranging from 10 to 5000 s. Under our experimental conditions, α1-AT at 45 °C retained its monomeric form during the deuterium labeling time (Fig. 3A). Deuterium uptake of monomeric α1-AT at 25 and 45 °C was monitored. To account for the intrinsic temperature dependence of H/D exchange, shorter labeling times were employed at 45 °C relative to 25 °C (the temperature dependence and the resulting incubation time differences were calculated using the program HXpep) to achieve comparable “labeling strength” at both temperatures. Deuterium-labeled α1-AT was digested with pepsin, and the percent exchange for each peptide was determined.
FIGURE 3.
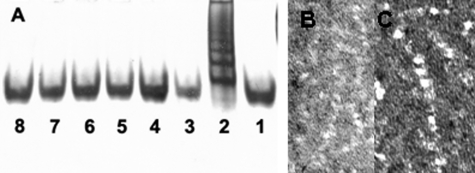
A, native gel. Lane 1, α1-AT after 6 h of incubation at 25 °C; lane 2, polymericα1-AT formed after 4 days of incubation at 45 °C; lanes 3–8, α1-AT at 45 °C at 50, 100, 500, 1000, 3000, and 5000 s, respectively. B, electron microscopic image of α1-AT polymers formed after 4 days of incubation at 45 °C. C, enlarged view.
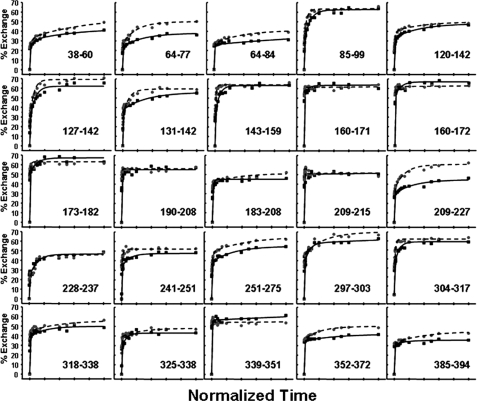
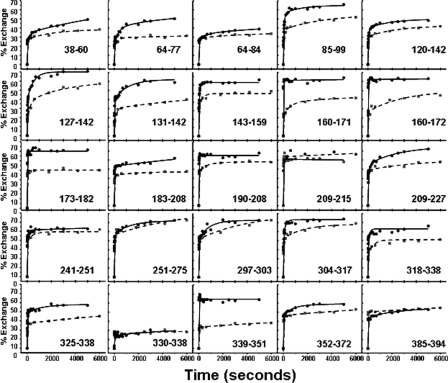
Fig. 4 shows exchange versus time at 25 and 45 °C for 25 peptides covering most structural regions of α1-AT. As mentioned above, it has been proposed that serpin polymerization is preceded by the accumulation of a species in which β-strands 3 and 5A are separated (3). This proposed separation will break a number of backbone hydrogen bonds and expose the amide hydrogens to solvent. Amide hydrogen atoms in these regions should therefore readily undergo exchange. However, in a peptide derived from residues 325–338 that encompasses β-strand 5A, we observed, within experimental error, no significant difference in exchange at 25 and 45 °C (Fig. 4).
FIGURE 4.
Exchange versus time for 25 peptic fragments derived from α1-AT. The percent exchange at 25 and 45 °C is shown by solid and dashed lines, respectively. Samples at the two temperatures were incubated for different times as described under “Materials and Methods.” Time points on the x axis were normalized for ease of comparison.
The largest increase in deuterium uptake was observed in β-strand 3C (residues 209–227). At the longest incubation times, the intermediate at 45 °C showed a percent exchange increase of 16% relative to α1-AT at 25 °C. β-Strand 3C formed amide hydrogen bonds with β-strand 2C (residues 282–290) and β-strand 4C (residues 204–209). Peptide 190–208, which covers the majority of β-strand 4C, showed no increase in exchange at 45 °C, indicating that it is primarily strand 3C/2C interactions that are destabilized by heating. Regrettably, we were unable to identify a peptide covering β-strand 2C and so have no reporter for the dynamics of that region. Helix C also showed a substantial increase in deuterium uptake at longer incubation times (14%). In addition, much of β-sheet B, β-strand 1C, helices G and H, and much of helix A showed smaller but still significant increases in exchange at the longer time points (increases of between 7 and 9%). Of particular interest is the peptide spanning residues 352–372 that contains β-strand 1C. This region showed an 8% increase in exchange at 45 °C. Although this increase is modest, this peptide contains very few residues involved in secondary structure (see Fig. 2), and thus, the observed increase in exchange is consistent with substantial destabilization of those secondary structure elements contained in this region (all of strand 1C and two residues of strand 4B).
Because α1-AT was in D2O during the incubation at 45 °C, the observed increases in deuterium uptake could be due to either increased local fluctuations or local unfolding. To determine which mechanism was responsible for the observed increases in exchange, α1-AT was incubated (at 45 or 25 °C) for 5000 s in H2O buffer and then briefly pulse-labeled in D2O buffer. This type of pulse labeling experiment is well established as a means for detecting local and global protein unfolding (18). When corrected for temperature, we observed no significant difference in deuterium uptake at 25 and 45 °C (Fig. 5). This finding indicates that α1-AT does not adopt a partially unfolded conformation during the initial incubation phase at 45 °C. The observed increases in deuterium uptake are therefore due to decreased stability and increased flexibility in local regions of α1-AT.
FIGURE 5.
Peptide 209–227 preincubated for 5000 s and then pulse-labeled at 25 °C (upper) and 45 °C (lower).
Structure and Stability of Polymeric α1-AT—After the initial conformational change (during the ∼4000-s unimolecular phase), subsequent incubation at 45 °C resulted in polymerization, with the process reaching completion in ∼48 h. We found that incubation of α1-AT at 45 °C for 4 days at pH 7.8 resulted in a “ladder” pattern on a native gel, which is characteristic of serpin polymers (Fig. 3A). Additionally, electron microscopy revealed “bead” polymers (Fig. 3, B and C) similar to those observed in previous studies. α1-AT polymers were labeled with deuterium for different time periods to investigate the stability and solvent accessibility of monomers within the polymers in a region-specific manner. Because dissociation of the polymer occurs much slower than our deuterium labeling time (as a previous study demonstrated (19)), dilution of the polymeric sample into D2O does not affect the structural integrity of the polymer within our experimental time scale.
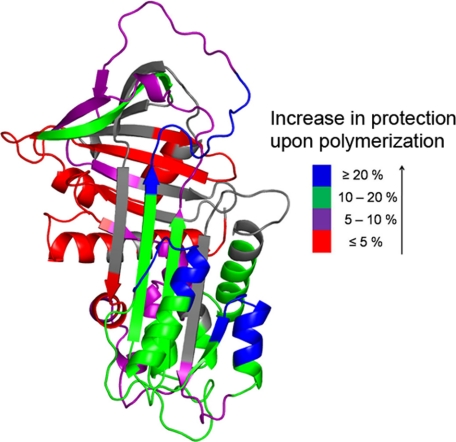
Fig. 6 shows the results of deuterium labeling for both the monomer and polymer at 45 °C. Results for 25 peptides are shown. Regions of α1-AT that become shielded from solvent upon polymerization should show reduced exchange compared with the monomer, and Fig. 6 indeed shows a large decrease in exchange in the polymer. The extent of reduction in percent exchange in the polymer relative to the monomer is color-coded and mapped onto the crystal structure of α1-antitrypsin in Fig. 7. Most regions in the polymeric form have deuterium uptakes comparable to those of the intermediate. However, there are several regions with notable reductions in the polymer compared with the intermediate.
FIGURE 6.
Exchange versus time for monomeric and polymeric α1-AT at 45 °C. Exchange curves are shown in by solid lines for the monomer and dashed lines for the polymer.
FIGURE 7.
Differences in hydrogen exchange (at 5000 s) between monomeric and polymeric α1-AT mapped onto the three-dimensional structure (Protein Data Bank code 1QLP). Darker colors represent larger decreases in observed exchange (i.e. increased protection) in the polymer compared with the monomer.
If the polymer is formed by the loop/sheet A polymerization mechanism, peptides derived from the RCL and the center of β-sheet A should show significant reductions in exchange. Indeed, a specific region within the RCL had a large reduction in the percent exchange in the polymeric form (Fig. 6). This region consists of residues 339–351 and showed a 34% reduction in exchange relative to the monomer (at 10 s of incubation in D2O). The other half of the RCL, residues 352–372, had only a 9% decrease in percent exchange in the polymer (see “Discussion”). Although the results did not indicate the opening of β-sheet A in the monomeric intermediate, a large reduction in percent exchange was observed in a peptide derived from residues 325–338 (16% reduction) containing β-strand 5A. These reductions in percent exchange in residues 339–351 (the RCL) and 325–338 (β-strand 5A) suggest that these two regions form the annealing site. In addition, a noticeable decrease in the percent exchange was observed in regions nearby the annealing site including β-strands 1A and 3A and helices E and F. A significant decrease in exchange was also observed in β-strand 3C (17% reduction). At 45 °C, this region is highly unstable in the monomer, and this reduction may reflect restabilization induced by polymerization.
DISCUSSION
The current proposed mechanism of serpin polymerization suggests that polymerization is preceded by the accumulation of a partially unfolded form (often referred to as M*) in which β-strands 3 and 5A are separated and that this separation readily allows the annealing of the RCL of a second serpin to form a sixth strand. Following this initial dimerization, subsequent RCL/sheet A linkages with additional serpins lead to the formation of polymers (2, 3). There are two chief pieces of evidence that support the proposed structure of M*. The first is the crystal structure of the δ-conformation of antichymotrypsin, which reveals partial RCL insertion and partial separation of strands 3 and 5A (20). The second is the change in tryptophan fluorescence that is observed during the initial unimolecular phase that precedes polymerization when α1-AT is incubated at 45 °C. It is believed that the change in fluorescence is due mainly to changes in the environment of Trp194, which is buried and located in the breach region at the top of sheet A.
The separation of β-strands 3 and 5A that is hypothesized for M* would break five hydrogen bonds and expose the amide hydrogens involved to solvent. This should result in significant increases in deuterium uptake in peptides covering stands 3 and 5A, and we did not observe such increases, even after 24 h of incubation at 45 °C. This lack of increased deuterium uptake in strands 3 and 5A during the first 5000 s of incubation indicates that there is no accumulation of the proposed M* intermediate during this period. Although the hypothesized structure of M* is based in part on the structure of the δ-conformation of α1-antichymotrypsin, it should be noted that the RCL of antichymotrypsin is five residues longer than that of α1-AT, allowing it to partially insert into β-sheet A without displacing strand 1C. In the case of α1-AT with its shortened RCL, it is not clear that a partially inserted conformation would be stable.
We did observe minor increases in flexibility throughout much of the structure and significant increases in flexibility in β-sheet C and helix C. Trp194 is close in space to several residues from region 215–227, which shows the largest increase in exchange upon heating. In particular, the side chain of Trp194 is very nearly in van der Waals contact with the side chain of Met221. Possibly the increase in the flexibility of the local environment is responsible for the changes seen in the fluorescence of Trp194 at 45 °C. Furthermore, increased exchange in residues 64–77 indicates moderate destabilization of helix C, which may account for the very small change in the CD signal previously seen during the first ∼5000 s of incubation at 45 °C (3).
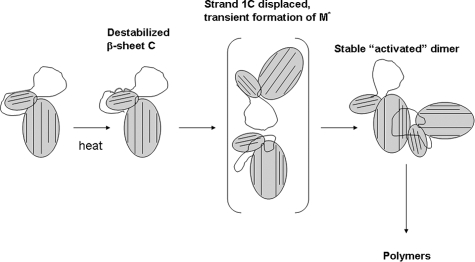
Many studies, including the current H/D exchange study, support a model of polymerization in which the RCL of one serpin inserts between β-strands 3 and 5A of another serpin. If no detectable unfolding occurs during the period preceding polymerization, then how does RCL/sheet A polymerization take place? The increased flexibility seen in β-sheet C at 45 °C suggests a mechanism. Several studies have shown the importance of strand 1C displacement for polymerization. In particular, an engineered disulfide bond between strands 1 and 2C was sufficient to completely block polymerization (21). Increased flexibility in β-sheet C, including strand 1C, at 45 °C indicates that interactions between strand 1 and the rest of sheet C are weakened upon mild heating. With these weakened interactions, strand 1C will be more easily displaced from the rest of sheet C. Thus, at 45 °C, α1-AT initially populates not the M* intermediate, but a species in which β-sheet C is significantly destabilized (during the first ∼5000 s). Following this, there is a lengthy lag phase prior to the appearance of polymers. If strand 1 were simply removed from sheet C by thermal fluctuations, it is not clear why the resulting species would not quickly proceed to the latent form, which is incapable of forming polymers. We suggest instead that strand 1C is displaced by an interaction with another serpin (Fig. 8). This displacement of strand 1C is the initial event that triggers the opening of sheet A and the formation of the M* intermediate. This intermediate could then quickly proceed to form a dimer with the nearby second serpin, which will go on to form polymers. Zhou and Carrell (22) have recently demonstrated that it is dimers, rather than monomers, that initiate and propagate serpin polymerization. They proposed a structure of the “activated,” polymerization-promoting dimer that requires the complete removal of strand 1C from β-sheet C in one of the two monomers (22).
FIGURE 8.
Proposed mechanism for the heat-induced polymerization of α1-AT.
Polymerization results in changes in deuterium uptake throughout much of the structure of α1-AT. We distinguish between two types of changes. Amide hydrogens that are solvent-exposed and not hydrogen-bonded will exchange completely within ∼10 s under our labeling conditions, and therefore, differences in deuterium uptake at the earliest time points can be attributed primarily to changes in solvent exposure upon polymerization. Changes in the rate of deuterium uptake over longer time periods can be attributed to changes in stability/flexibility. The largest change in solvent accessibility upon polymerization is seen in the N-terminal portion of the RCL. Peptide 339–351, which includes RCL residues P14–P8, shows a large decrease in exchange at 10 s in the polymer relative to the monomer. Less dramatic changes are seen in the C-terminal portion of the RCL. This would seem to conflict with the observation that peptides corresponding to P7–P3 can anneal between the lower portions of β-strands 3 and 5A and that this is sufficient to block serpin polymerization in vitro, suggesting that these residues, located in the C-terminal half of the RCL, play a critical role in polymer formation (we note, however, that in the case of wild-type α1-AT, the peptide FLEAIG, corresponding to RCL residues P7–P2, is not an effective blocker of polymerization) (23). However, the peptic fragment that includes these residues (fragment 352–372) is quite large and contains, in addition to part of the RCL, both β-strand 1C and a portion of strand 4B. Interpretation of the results for this peptide is therefore not straightforward. As mentioned previously, considerable evidence indicates that polymerization requires the removal of strand 1 from β-sheet C. If this is the case, the changes in deuterium uptake in peptide 352–372 will reflect the net result of both losing protection in β-strand 1C and gaining protection due to the insertion of the RCL into β-sheet A. The relatively small changes in deuterium uptake seen in this region may be due to these two opposing factors. Although our results are not conclusive regarding the C-terminal half of the RCL, they very strongly demonstrate that the N-terminal half is involved in newly formed interactions in the polymer.
The other region in which large differences in deuterium uptake are seen at the earliest incubation times consists of residues 160–172. This region comprises the C-terminal half of helix F and part of the loop connecting it to β-sheet A. Both this study and a previous study (16) found that this region shows very little protection from exchange in the monomer, indicating that the structure in this region is at best marginally stable. In the polymer, exchange at 10 s is reduced by ∼36%. This increase in protection could indicate either that part of this region is buried in an intermolecular interface or that interactions in the polymer stabilize the C-terminal portion of helix F. Protein engineering has demonstrated that destabilizing mutations in the C-terminal (but not N-terminal) half of helix F accelerates polymerization (24). The dramatic reduction in deuterium uptake in the polymer relative to the monomer strongly supports an important role for helix F in polymerization.
In addition to changes in solvent accessibility, H/D exchange also reveals changes in conformational stability/flexibility upon polymerization. Significant decreases in the rates of deuterium uptake are seen in β-strands 3 and 5A, which is consistent with this region being stabilized by the annealing of the RCL of another serpin to form strand 4A. Helices D and E, β-strands 1A and 3C, and the N-terminal portion of helix F are also substantially stabilized in the polymer. Most likely the large increase in stability seen in the C-terminal portion of helix F propagates to the N-terminal portion, explaining the decreased rates of exchange seen there. Helix E is very close to the C-terminal portion of helix F and is also in van der Waals contact with helix D. The stabilization of the top of helix F might result in stabilizing contacts with helix E, which are in turn propagated to helix D. Although β-strand 3C is clearly stabilized upon polymer formation, the mechanism for this stabilization is unclear.
Unlike wild-type α1-AT, the Z-mutant spontaneously forms polymers at 37 °C (25). Presumably the Glu342 → Lys mutation, which destroys a salt bridge, introduces an unfavorable Lys/Lys electrostatic interaction at the top of β-sheet A and promotes facile separation of strands 3 and 5A. The Z-mutant may therefore populate a monomeric strand-separated structure rather than require an interaction with a second serpin to induce strand separation. As the polymerization of wild-type α1-AT is frequently used as a model for the polymerization of pathogenic mutant serpins, potential differences in the polymerization mechanisms of wild-type and Z-mutant α1-AT require investigation.
In addition to crystal structures that suggest multiple possible linkage mechanisms, there is evidence from polymerization kinetic studies indicating that different serpins may form structurally distinct polymers. It was shown that when preformed polymers of the serpin α1-antichymotrypsin were added to a solution of monomers, the rate of polymerization was increased through a “seeding” mechanism similar to that seen for amyloid proteins (26). However, preformed polymers of other serpins, including α1-AT and antithrombin, did not seed polymerization of antichymotrypsin. This serpin-specific seeding suggests that the polymer structures, and possibly the polymerization mechanisms, of different serpins may differ in significant ways. Comparison of the extensive data on wild-type α1-AT polymerization with not just α1-AT mutants but also with other serpins will be important in establishing the commonalities and differences in serpin polymerization.
The serpinopathies are similar to amyloid diseases in that protein conformational change and subsequent polymerization are strongly implicated as an underlying cause of pathology. It is therefore instructive to compare our H/D exchange results on polymerized α1-AT with H/D exchange studies of amyloid fibrils. At the level of overall morphology, it has already been observed that serpin polymers differ from amyloids. Serpins generally adopt short polymers with a bead-like structure rather than the long fibrils formed by amyloids. We found that there are also differences at the microscopic level. Amyloid fibrils have generally been found to contain a core region showing extreme protection from H/D exchange (>95% of amide hydrogens in these regions remain unexchanged even after several days), indicating the almost complete exclusion of solvent from the core (27). Furthermore, some amyloid proteins, such as human prion protein, undergo a complete alteration in their tertiary and secondary structures upon fibrilization (14). In contrast, the monomers in polymeric α1-AT retain a large degree of conformational flexibility. In fact, excepting the annealing site and helices E and F, the distribution of relative flexibility in the structure is similar to that seen in the monomeric form. These observations are consistent with a model in which serpins do not undergo a major change in their tertiary fold upon polymerization, instead remaining largely native-like apart from local changes required for the formation of loop/sheet linkages.
Protein fibrilization/polymerization is often described in terms of the accumulation of a partially unfolded aggregationprone intermediate arising from destabilization of the native structure, followed by interprotein interactions leading to the formation of amyloid fibrils or polymers. Although we do not detect the accumulation of a partially unfolded structure, the increased rates of H/D exchange seen at 45 °C, particularly in β-sheet C, do indicate that the native structure of α1-AT is destabilized by moderate temperatures and that this destabilization (presumably by facilitating the facile displacement of stand 1C) is an important step in the polymerization process. Factors other than temperature, mutations in particular, can also destabilize the native conformation. This is clearly reflected in the fact that many mutant serpins readily form polymers at physiological temperatures. Now that H/D exchange has elucidated the details of wild-type α1-AT polymerization, comparative studies on the polymerization of pathological mutants such as Glu342 → Lys (the Z-mutant) can be pursued.
Both mild denaturant and mild temperature increases have been shown to induce the polymerization of α1-AT (28). Our current and previous H/D exchange results indicate that these two perturbations have dramatically different effects on α1-AT structure: mild denaturant causes a transition to a molten globule state with a solvent-accessible core (29), whereas mild temperature increases result in increased structural fluctuations but not local or global unfolding. Therefore, subtle changes in the environment allow α1-AT to explore a wide range of different structural states. For a full understanding, serpin polymerization must be considered in the context of a complex energy landscape in which folding, stability, dynamics, and polymerization are intertwined.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1-HL085469. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: α1-AT, α1-antitrypsin; RCL, reactive center loop; HXMS, hydrogen/deuterium exchange measured by mass spectrometry; HPLC, high performance liquid chromatography; H/D, hydrogen/deuterium.
References
- 1.Gettins, P. G. (2002) Chem. Rev. 102 4751-4804 [DOI] [PubMed] [Google Scholar]
- 2.Lomas, D. A., and Carrell, R. W. (2002) Nat. Rev. Genet. 3 759-768 [DOI] [PubMed] [Google Scholar]
- 3.Dafforn, T. R., Mahadeva, R., Elliott, P. R., Sivasothy, P., and Lomas, D. A. (1999) J. Biol. Chem. 274 9548-9555 [DOI] [PubMed] [Google Scholar]
- 4.Dunstone, M. A., Dai, W. W., Whisstock, J. C., Rossjohn, J., Pike, R. N., Feil, S. C., Le Bonniec, B. F., Parker, M. W., and Bottomley, S. P. (2000) Protein Sci. 9 417-420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lomas, D. A., Elliott, P. R., Sidhar, S. K., Foreman, R. C., Finch, J. T., Cox, D. W., Whisstock, J. C., and Carrell, R. W. (1995) J. Biol. Chem. 270 16864-16870 [DOI] [PubMed] [Google Scholar]
- 6.Zhou, A. W., Stein, P. E., Huntington, J. A., and Carrell, R. W. (2003) J. Biol. Chem. 278 15116-15122 [DOI] [PubMed] [Google Scholar]
- 7.Sivasothy, P., Dafforn, T. R., Gettins, P. G. W., and Lomas, D. A. (2000) J. Biol. Chem. 275 33663-33668 [DOI] [PubMed] [Google Scholar]
- 8.Sharp, A. M., Stein, P. E., Pannu, N. S., Carrell, R. W., Berkenpas, M. B., Ginsburg, D., Lawrence, D. A., and Read, R. J. (1999) Structure (Camb.) 7 111-118 [DOI] [PubMed] [Google Scholar]
- 9.Carrell, R. W., Stein, P. E., Wardell, M. R., and Fermi, G. (1994) Structure (Camb.) 2 257-270 [DOI] [PubMed] [Google Scholar]
- 10.Zhang, Q. W., Law, R. H. P., Bottomley, S. P., Whisstock, J. C., and Buckle, A. M. (2008) J. Mol. Biol. 376 1348-1359 [DOI] [PubMed] [Google Scholar]
- 11.Marszal, E., Danino, D., and Shrake, A. (2003) J. Biol. Chem. 278 19611-19618 [DOI] [PubMed] [Google Scholar]
- 12.Redeker, V., Halgand, F., Le Caer, J. P., Bousset, L., Laprevote, O., and Melki, R. (2007) J. Mol. Biol. 369 1113-1125 [DOI] [PubMed] [Google Scholar]
- 13.Eghiaian, F., Daubenfeld, T., Quenet, Y., van Audenhaege, M., Bouin, A. P., van der Rest, G., Grosclaude, J., and Rezaei, H. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7414-7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu, X. J., Wintrode, P. L., and Surewicz, W. K. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1510-1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wales, T. E., and Engen, J. R. (2006) Mass Spectrom. Rev. 25 158-170 [DOI] [PubMed] [Google Scholar]
- 16.Tsutsui, Y., Liu, L., Gershenson, A., and Wintrode, P. L. (2006) Biochemistry 45 6561-6569 [DOI] [PubMed] [Google Scholar]
- 17.Englander, S. W., Mayne, L., Bai, Y., and Sosnick, T. R. (1997) Protein Sci. 6 1101-1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng, Y. Z., and Smith, D. L. (1999) Anal. Biochem. 276 150-160 [DOI] [PubMed] [Google Scholar]
- 19.Purkayastha, P., Klemke, J. W., Lavender, S., Oyola, R., Cooperman, B. S., and Gai, F. (2005) Biochemistry 44 2642-2649 [DOI] [PubMed] [Google Scholar]
- 20.Gooptu, B., Hazes, B., Chang, W. S., Dafforn, T. R., Carrell, R. W., Read, R. J., and Lomas, D. A. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 67-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang, W. S. W., Whisstock, J. C., Hopkins, P. C. R., Lesk, A. M., Carrell, R. W., and Wardell, M. R. (1997) Protein Sci. 6 89-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou, A., and Carrell, R. W. (2008) J. Mol. Biol. 375 36-42 [DOI] [PubMed] [Google Scholar]
- 23.Zhou, A., Stein, P. E., Huntington, J. A., Sivasothy, P., Lomas, D. A., and Carrell, R. W. (2004) J. Mol. Biol. 342 931-941 [DOI] [PubMed] [Google Scholar]
- 24.Cabrita, L. D., Dai, W., and Bottomley, S. P. (2004) Biochemistry 43 9834-9839 [DOI] [PubMed] [Google Scholar]
- 25.Lomas, D. A., Evans, D. L., Stone, S. R., Chang, W. S. W., and Carrell, R. W. (1993) Biochemistry 32 500-508 [DOI] [PubMed] [Google Scholar]
- 26.Crowther, D. C., Serpell, L. C., Dafforn, T. R., Gooptu, B., and Lomas, D. A. (2003) Biochemistry 42 2355-2363 [DOI] [PubMed] [Google Scholar]
- 27.Kheterpal, I., Cook, K. D., and Wetzel, R. (2006) Methods Enzymol. 413 140-166 [DOI] [PubMed] [Google Scholar]
- 28.Koloczek, H., Guz, A., and Kaszychi, P. (1996) J. Protein Chem. 15 447-454 [DOI] [PubMed] [Google Scholar]
- 29.Tsutsui, Y., and Wintrode, P. L. (2007) J. Mol. Biol. 371 245-255 [DOI] [PubMed] [Google Scholar]