Abstract
Primary hyperoxaluria type I is a severe kidney stone disease caused by mutations in the protein alanine:glyoxylate aminotransferase (AGT). Many patients have mutations in AGT that are not deleterious alone but act synergistically with a common minor allele polymorphic variant to impair protein folding, dimerization, or localization. Although studies suggest that the minor allele variant itself is destabilized, no direct stability studies have been carried out. In this report, we analyze AGT function and stability using three approaches. First, we describe a yeast complementation growth assay for AGT, in which we show that human AGT can substitute for function of yeast Agx1 and that mutations associated with disease in humans show reduced growth in yeast. The reduced growth of minor allele mutants reflects reduced protein levels, indicating that these proteins are less stable than wild-type AGT in yeast. We further examine stability of AGT alleles in vitro using two direct methods, a mass spectrometry-based technique (stability of unpurified proteins from rates of H/D exchange) and differential scanning fluorimetry. We also examine the effect of known ligands pyridoxal 5′-phosphate and aminooxyacetic acid on stability. Our work establishes that the minor allele is destabilized and that pyridoxal 5′-phosphate and aminooxyacetic acid binding significantly stabilizes both alleles. To our knowledge, this is the first work that directly measures relative stabilities of AGT variants and ligand complexes. Because previous studies suggest that stabilizing compounds (i.e. pharmacological chaperones) may be effective for treatment of primary hyperoxaluria, we propose that the methods described here can be used in high throughput screens for compounds that stabilize AGT mutants.
Deficiencies in the enzyme alanine:glyoxylate aminotransferase (AGT)3 cause primary hyperoxaluria type I (PH1), a severe autosomal recessive kidney stone disease (1, 2). In humans, AGT is responsible for conversion of glyoxylate to glycine in the liver. Without functional AGT, glyoxylate builds up and is converted to calcium oxalate, which is deposited in the kidneys and can lead to kidney stones and renal failure. In many patients, deficiency of AGT results from one or two amino acid changes that decrease the stability of this enzyme. As a result, AGT may be degraded, become improperly localized, or form nonfunctional aggregates (1, 2).
Over 50 different mutations of AGT and two polymorphic variants have been identified (1, 3). The two allelic forms consist of a “wild-type” major allele, AGTma, and a minor allele, AGTmi. The minor allele is present in ∼20% of European and North American populations and contains P11L and I340M substitutions in the amino acid sequence and a 74-bp duplication in intron 1 (4, 5). The P11L and I340M substitutions, particularly P11L, have several biochemical effects, including decreasing catalytic activity and slowing the dimerization rate, but by themselves are not disease-causing (4, 5). The minor allele of AGT is deleterious when combined with certain additional mutations, leading to impairments in the stability, localization, and/or dimerization of the enzyme. Interacting mutations include I244T, found in ∼9% of PH1 patients. This mutation is not disease-causing itself but leads to protein aggregation and disease when combined with the minor allele polymorphism (6). The mutation G170R, found in ∼30% of PH1 patients, is also not by itself deleterious but when combined with the minor allele is thought to reduce stability, delay dimerization, and result in protein mislocalization (5).
Previous studies have characterized wild-type AGT and PH1-associated variants biochemically and in cell culture (5–10). The minor allele itself appears to have a particularly complex phenotype; studies suggest that it has reduced catalytic activity (5, 8) and reduced stability (5, 7) and that the P11L mutation generates a cryptic mitochondrial targeting signal that affects protein trafficking (4, 10, 11). Although the trafficking differences between the major and minor allele proteins are well established (2), there is only indirect evidence supporting that the minor allele is destabilized, and some of these experiments are difficult to interpret because they assessed the minor allele in combination with other mutations or characterized proteins containing only one of the two minor allele mutations. Evidence for minor allele destabilization includes in vitro transcription/translation studies demonstrating that AGTmi is more aggregation-prone than AGTma (7) and protease protection and overexpression results showing that variants containing the P11L mutation are more susceptible to proteolytic digestion (6) and prone to aggregation (5).
Studies have suggested that certain PH1-associated forms of AGT may be suitable for rescue by pharmacological chaperones, small molecules that promote folding of a protein in an active conformation. For example, the mistrafficking of the G170R/minor allele mutant mentioned above could be rescued by the addition of betaine, a nonspecific chemical chaperone, as well as by lowering temperature (9), supporting the idea that increasing the stability of unstable variants can alleviate protein dysfunction. Molecules that provide even a modest increase in protein stability may therapeutically benefit PH1 patients with partially functional AGT variants. One chemical that has been used in pharmacological treatment of PH1 is pyridoxine, which appears to improve outcome for certain subtypes of PH1 (1, 12) through an unknown mechanism. Pyridoxine is broken down in the cell to form pyridoxal 5′-phosphate (PLP), a cofactor for AGT. Several in vitro studies suggest that PLP stabilizes AGT; PLP was found to increase the solubility of a variant (AGTmi-I244T) expressed in COS cells (6), to improve the yield and increase dimerization of in vitro transcribed/translated protein (7), to protect AGT mutants from proteasomal degradation (7), and to protect wild-type AGT from heat inactivation (13). None of these studies, however, incorporated direct stability measurements, and the issue is further complicated because attempts to observe an effect of PLP in vivo (similar to the effect seen with betaine, for example) have failed (9).
In this work, we use in vivo and in vitro approaches to evaluate the activity and stability of wild-type AGT and PH1-associated variants. First, we demonstrate the utility of a yeast-based complementation assay, which provides a rapid and facile means of evaluating activity of wild-type and mutant AGT proteins in vivo. We show that human AGT can substitute for the yeast enzyme and that variants of human AGT associated with disease, including the minor allele, show reduced protein levels and reduced growth in yeast. We further explore differences of AGT variants in vitro, with the aim of directly characterizing the stability of major and minor alleles in the presence and absence of ligand complexes. These studies include thermal denaturation experiments utilizing catalytic activity and fluorescence spectroscopy as well as chemical denaturation experiments using a hydrogen/deuterium (H/D) exchange- and mass spectrometry-based approach termed SUPREX. Our results show that the minor allele polymorphism substitutions destabilize the enzyme and that both wild-type and minor allele proteins are significantly stabilized in the presence of ligands PLP and aminooxyacetic acid (AOA).
EXPERIMENTAL PROCEDURES
Strains and Chemicals—Yeast strain YCG-Fr (MATa ura3-1 trp1-1 ade2-1 his3-11,-15 leu2-3,-112 can1-100 shm1::HIS3 shm2::LEU2 gly1::URA3 AGX1::kanMX4) was a gift from Dr. Peter Stahmann (Lausitz University of Applied Sciences, Senftenberg, Germany). Strain W303–1A (MATa ade2-1 can1-100 his3-11,15 leu2-3112 trp1-1 ura3-1) was used to express HA-tagged proteins for Western blotting. Major and minor allele AGT plasmids were a gift from Dr. Marion Coulter-Mackie (University of British Columbia, Vancouver, Canada). Chemicals, including pyridoxal 5′-phosphate and aminooxyacetic acid, were from Sigma.
Yeast Complementation Experiments—AGTma and AGTmi clones for expression in the YCG-Fr strain were cloned by homologous recombination into the plasmid p416GPD. Forward and reverse oligonucleotides were designed that added sequences to AGT clones that match the p416GPD vector at the insert site and used for PCR (Phusion; New England Biolabs). To generate mutations, 32-bp forward and reverse oligonucleotides were generated with the mutation. These oligonucleotides were used with the above mentioned forward and reverse primers to generate two overlapping fragments, using either AGTma or AGTmi as a template. The two fragments were transformed with p416GPD (cut with BamHI and EcoRI) and cloned by homologous recombination in yeast. For production of HA-tagged proteins, 2× HA tags were amplified from a construct that contained the sequence GMRYPYDVPDYAGYPYDVPDYASGR. During PCR amplification, the sequences were added to allow homologous recombination with p416GPD-AGT constructs cut with EcoRI and HindIII enzymes, such that the HA tag would be inserted at the C terminus of the AGT protein. All of the clones were verified by sequencing.
For growth assays, yeast were grown overnight in SD –Ura/–Gly medium containing 2% glucose and supplemented with 10 mm glycine. The following morning, the strains were diluted in the same medium and grown to log phase. The strains were washed in water and diluted to A600 0.05 in SD –Ura/–Gly with 3% ethanol/no glycine for assays. Growth was carried out at the indicated temperatures in either 96-well plates or 13-mm culture tubes. A600 measurements were taken after indicated times. Fig. 3A and Table 1 report the average of three A600 measurements taken at a single time point, at which AGTma was in log phase with A600 between 0.6 and 1.
FIGURE 3.
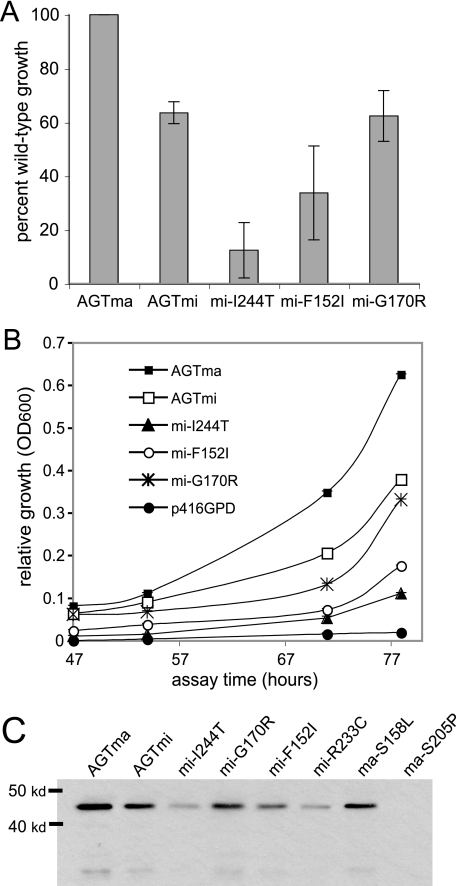
Disease-associated human AGT variants show reduced growth in yeast. A, relative growths of AGTma and variants expressed in YCG-Fr cells averaged over several experiments. The growths were collected from ≥3 independent assays and are expressed as percentages of wild type. B, growth of yeast expressing AGT variants in medium containing 3% ethanol from 47 to 78 h in strain YCG-Fr at 30 °C. The solid circles indicate growth of empty vector p416GPD. C, Western blot analysis of HA-tagged AGT variants expressed in yeast.
TABLE 1.
Activities of AGT variants and percentage of growth in yeast assay
| Variant | In vitro activity (reference (s)) | Liver activity (reference) | Yeast complementationa |
|---|---|---|---|
| % | % | % | |
| AGTma | 100 | 100 | 100 |
| AGTmi | 46-50 (5) | NA | 63.5 ± 4.1 |
| AGTmi | 62-84 (7, 8, 20) | NA | 63.5 ± 4.1 |
| AGTmi G170R | 40-57 (7, 8) | 23 (range 0-72) (20) | 62.4 ± 9.5 |
| AGTmi I244T | 8-26 (6) | 11 (range 0-36) (20) | 12.4 ± 10.3 |
| AGTmi R233C | 0 (20) | 6 (20) | 0.29 ± 0.4 |
| AGTma R233C | 14 (20) | NA | 52.3 ± 10.7 |
| AGTmi F1521 | NA | 12 (20) | 33.7 ± 17.4 |
| AGTma S205P | NA | 1 (21) | 6.0 ± 1.0 |
| AGTma S158L | 0 (20) | 6 (20) | 2.8 ± 0.3 |
The values are the averages and standard deviation of three replicate A600 measurements at 78-90 h of growth, with A600 of AGTma between 0.6 and 1.0.
For Western blotting, HA-tagged proteins were expressed in strain YCG-Fr in the presence of glycine and grown to log phase. Equal amounts of total protein were blotted onto a nitrocellulose membrane, and labeling was performed using an anti-HA monoclonal antibody (HA.11; Covance). The secondary antibody used was an IRDye 680 goat anti-mouse IgG (Li-COR), and the proteins were visualized using an Odyssey infrared imaging system (Li-COR).
Protein Purification—To generate C-terminal His6 tag clones for protein expression, NdeI and XhoI sites were added to AGTma or AGTmi using PCR (Phusion). PCR products were digested with NdeI and XhoI and ligated into pET-30a digested with NdeI and XhoI. The clones were verified by sequencing and were transformed into BL21(DE3) Escherichia coli (Novagen). For purification, the bacteria containing the AGT clones were grown to 0.8 A600 and then induced for 6 h at room temperature with 1 mm isopropyl-β-d-thiogalactopyranoside to produce His-tagged proteins. The cells were harvested by centrifugation and frozen for later use. For purification, the cells were lysed in Y-PER (Pierce) and purified on a nickel-nitrilotriacetic acid column using the Y-PER His6 fusion protein purification kit (Pierce) according to the manufacturer's protocol. Following purification, protein concentration was determined by Bradford assay, and the samples were stored at –80 °C in aliquots in elution buffer (50 mm Tris, 300 mm NaCl, 200 mm imidazole, 10% glycerol, pH 6.8) or storage buffer (50 mm Tris, 300 mm NaCl, 10% glycerol, pH 6.8).
Determination of AGT Enzyme Activity—AGT activity was determined as described (14), using a two-step reaction. Initial reactions contained 100 mm potassium phosphate (K2HPO4 + KH2PO4), pH 8.0, 10 mm glyoxylate, 150 μm PLP, and 150 mm alanine in a total of 200 μl. The samples were incubated 30 min at 37 °C, and the reaction was stopped by trichloroacetic acid precipitation using 50 μl of 50% trichloroacetic acid. For thermal inactivation experiments (see Fig. 4), the purified proteins were preincubated for 1 h at specified temperatures in dilution buffer (300 mm NaCl, 50 mm Tris, pH 6.8). Following preincubation, the samples were placed on ice for 10 min and then assayed for AGT activity as usual.
FIGURE 4.
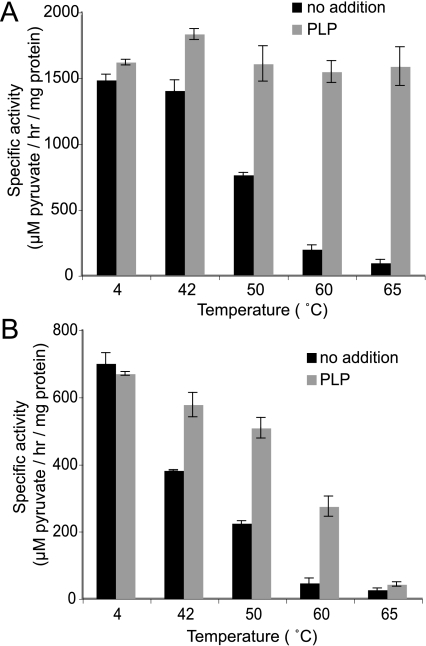
Thermal inactivation analysis of AGTma (A) or AGTmi (B). Purified proteins were incubated in the presence or absence of 50 μm PLP and subjected to heat treatment for 1 h at the specified temperature. Following heat treatment, the samples were cooled on ice, adjusted such that they contained equivalent amounts of PLP, and assayed for AGT activity.
SUPREX Analysis of Purified AGT—SUPREX experiments were performed using a series of deuterated H/D exchange buffers (20 mm phosphate in D2O, pD 7.4) containing 0–7.6 m deuterated guanidine hydrochloride. The concentration of guanidine hydrochloride in each H/D exchange buffer was determined with a Bausch & Lomb refractometer as described (15). Prior to SUPREX analysis, AGT samples (13 μm) were preincubated for 1 h at room temperature in phosphate-buffered saline (pH 7.4) in the presence or absence of ligands (1.3 mm). In some cases, AGTma samples were equilibrated overnight at 4 °C, but this did not result in an observable change in the SUPREX results. Hydrogen exchange reactions were initiated by adding 7 μl of each exchange buffer to 3 μl of protein solution, resulting in a final protein concentration of 4 μm and final ligand concentrations of 400 μm. The H/D exchange reactions were carried out at room temperature for the specified times and quenched with 1 μl of 10% trifluoroacetic acid. The protein was concentrated and desalted using C4 or C18 ZipTips (Millipore). The protein samples were eluted from the ZipTips directly onto a MALDI target using the MALDI matrix solution, which consisted of saturated sinapinic acid in 0.1% trifluoroacetic acid, 50–75% acetonitrile, and 25–50% water. The matrix solution also contained aldolase as an internal standard for the MALDI measurements.
After H/D exchange, the protein masses were measured using a Bruker
Ultraflex II TOF/TOF mass spectrometer. The mass of the protein after it was
subjected to H/D exchange at each concentration of denaturant was extracted
from the MALDI mass spectra using a MATLAB program that performed a 19-point
floating average smoothing of the data, a one-point mass calibration using the
ion signal from the internal mass standard, and a center of mass determination
for the ion signal of the protein. Mass determinations from 10 replicate MALDI
mass spectra were averaged to obtain an average molecular mass of the
deuterated protein in each buffer. An average change in mass caused by the
uptake of deuterons (i.e. Δmass) was calculated at each
concentration of guanidine hydrochloride by subtracting the mass of the
protonated protein from the average molecular mass of the deuterated protein.
SUPREX curves were constructed by plotting Δmass versus
[guanidine hydrochloride], and the transition midpoint of each curve
( ) was extracted by
fitting the data to a sigmoidal equation using IGOR Pro software
(Wavemetrics). The concentration of guanidine hydrochloride in the plot was
adjusted to reflect the final concentration after mixing the exchange buffer
with the protein solution.
) was extracted by
fitting the data to a sigmoidal equation using IGOR Pro software
(Wavemetrics). The concentration of guanidine hydrochloride in the plot was
adjusted to reflect the final concentration after mixing the exchange buffer
with the protein solution.
Differential Scanning Fluorimetry—Purified AGTma or AGTmi proteins (1–5 μm) were diluted in SYPRO buffer containing 10 mm HEPES, 150 mm NaCl, and 5× SYPRO orange (Molecular Probes). The proteins were subjected to a ramp of 1 °C/min in a Lightcycler 2.0 real time PCR thermal cycler (Roche Applied Science) at a temperature gradient from 35 to 95 °C. The graphs were generated using IGOR Pro software. Calculation of Tm was carried out by determining the maximum of the first derivative of each transition using the Lightcycler 2.0 software.
RESULTS
Expression of AGT Disease Variants in Yeast—Yeast alanine: glyoxylate aminotransferase (AGX1) is involved in the glyoxylate pathway and is important for glycine synthesis under growth on nonfermentous carbon sources. Although AGX1 is not essential for viability, yeast lacking AGX1 in a shm1, shm2, and gly1 background are unable to grow on medium that contains ethanol unless exogenous glycine is provided (16). Plasmid-expressed yeast Agx1 protein can restore ethanol-dependent growth to an agx1-deficient strain, YCG-Fr (16). The Agx1 enzyme has 23% identity to human AGT, with residues that are conserved among all AGTs, such as a common pyridoxal 5′-phosphate-binding site (Fig. 1). Agx1 in yeast has been proposed to be mitochondrially localized based on green fluorescent protein fusion analysis (17), although this localization may not be exclusive. In humans, the AGT enzyme is peroxisomal, but in other mammals the enzyme can be peroxisomal, mitochondrial, or both (18, 19).
FIGURE 1.
Lineup of human AGT protein (AGXT) with Saccharomyces cerevisiae AGX1. The shaded areas indicate homology. Residues involved in binding PLP are marked with asterisks.
We examined whether expression of plasmid-derived human AGT could complement growth of yeast lacking AGX1 in the YCG-Fr strain (16). This strain is unable to grow on glycine-free medium, because it cannot synthesize glycine endogenously (16). When streaked on plates containing ethanol, YCG-Fr cells expressing Agx1 (p416ADH-Agx1) are able to grow, because the Agx1 enzyme converts glyoxylate to glycine (Fig. 2). Expression of human AGT (p416GPD-hAGT) in YCG-Fr also allowed growth on ethanol plates, indicating that the human enzyme is able to substitute for the yeast protein (Fig. 2). High level expression of human AGT was required for complementation, because yeast expressing human AGT under control of a weaker ADH promoter showed no growth (data not shown).
FIGURE 2.
Human AGT protein complements a yeast AGX1 deficiency. Shown is a Ura–/3% ethanol plate containing YCG-Fr yeast lacking Agx1, expressing either empty vector control (p416GPD), yeast Agx1 (p416ADH-AGX1), or human AGT (p416GPD-hAGT).
We examined whether the minor allele polymorphisms P11L and I340M would confer reduced growth to YCG-Fr yeast expressing AGT. We also examined the effects on growth of the three most common mutations that cause disease when in combination with the minor allele: I244T, G170R, and F152I. In Fig. 3A, we show the growth of yeast expressing mutant versions of AGT. In Fig. 3B, we show the relative growths of yeast expressing wild-type and mutant proteins after 47–78 h at 30 °C. Growth of yeast expressing wild-type AGT was highest, followed by AGTmi and AGTmi-G170R at about 45–70% of wild-type growth. The reduced growth seen by AGTmi is consistent with reported measurements showing that this allele has ∼50–80% of wild-type activity (5, 8). There was no significant difference between growth of yeast expressing AGTmi and AGTmi-G170R, indicating that the G170R mutation does not significantly affect function of AGT in yeast. In contrast, growth of yeast expressing AGTmi-F152I was on average ∼33% of wild-type growth, whereas those expressing AGTmi-I244T showed about 12% of wild-type growth. Yeast transformed with the control plasmid p416GPD showed no significant growth after ∼90 h.
We examined variants of AGT in the yeast assay and compared these growths with reported activities from bacterially expressed proteins and from human liver (Table 1). The results from the yeast assays are generally consistent with the reported activities. One exception to this was the reported in vitro activity for AGTma-R233C, which was found to show greatly reduced catalytic activity (14% of wild type) when expressed in bacteria (20). The mutation R233C causes disease when combined with the minor allele mutations P11L and I340M but is not disease-causing in the major allele background. We examined R233C in both minor and major allele backgrounds in yeast. Yeast expressing AGTmi-R233C were unable to grow at any temperature, indicating the severity of this mutation. Yeast expressing AGTma-R233C showed reduced growth compared with wild type but not to the same degree as was reported from bacterial protein expression. To explore this discrepancy, we expressed and purified the AGTma-R233C mutant from bacteria. In our hands, we find that the AGTma-R233C mutant shows 72–88% of wild-type activity (data not shown), which is more consistent with the yeast results. We also examined two additional mutations in the major allele background, S205P and S158L. These mutations are predicted to be quite severe, because they cause disease in the major allele background. The S205P replacement is predicted to alter hydrogen bonding of the central β-sheet (21, 22), whereas Ser158 helps to orient the PLP cofactor in the enzyme active site (22). Using the yeast assay, we found the growth of the S205P variant in yeast to be 6% of wild-type and S158L to be 2.8% of wild-type (Table 1), correlating well with the reported severity of these mutations.
The reduced growth of the AGT mutants expressed in yeast could be due to a reduction in protein levels and/or a reduction in protein activity. In Fig. 3C, we show immunoblots of HA-tagged AGT proteins expressed in yeast. For wild-type and all of the minor allele mutants, the protein levels correlated well with yeast growth, with wild-type AGTma showing the highest levels, followed by AGTmi and AGTmi-G170R, AGTmi-F152I, AGTmi-I244T, and AGTmi-R233C. These results indicate that the different levels of yeast growth seen with these AGTmi mutants reflect different steady-state levels of protein in the yeast. We also examined levels of protein containing the S205P or S158L mutation, which conferred severely reduced growth to yeast (Table 1). We were unable to detect a Western band with S205P, a mutation that is predicted to significantly disrupt AGT structure, indicating that this mutant also greatly destabilizes AGT in yeast. This protein was previously examined in vitro (7), where it was found to have an extremely short half-life. S158L, a mutation that affects PLP binding in the active site, was not predicted to cause a large decrease in stability, and the protein correspondingly showed a significant band on immunoblots. However, the levels of S158L were reduced ∼50% compared with wild-type, indicating that the mutation affects stability as well as activity.
We examined whether the addition of PLP would stabilize AGT mutants and increase growth of yeast. Yeast expressing AGTmi or AGTmi-I244T were incubated in the presence or absence of up to 30 μm PLP, but we were unable to observe an effect of PLP using this approach. Our results are consistent with another study that also failed to observe an effect of PLP on AGT expressed in cell culture (9). PLP is a cofactor for many proteins in yeast, and yeast contain a TPN1 gene encoding a vitamin B6 (pyridoxine) transporter (23). The transporter is regulated by the extracellular levels of pyridoxine, such that a decrease in PLP levels results in an increase in the levels of Tpn1 (23). In addition to regulating intracellular levels of PLP, yeast are able to synthesize PLP endogenously. It is possible that our inability to observe a stabilizing effect with exogenous PLP in the cell may be due to the ability of yeast to strongly buffer PLP.
In Vitro Studies—Because our yeast experiments indicated a difference in stability of wild-type and minor allele forms of AGT (as observed by a difference in steady-state protein levels), we examined these differences in more detail in vitro using recombinant protein. Major and minor allele forms of AGT were cloned into the vector pET-30a to generate a C-terminal His6 tag, and proteins were expressed and purified from bacteria. To lyse and solubilize the bacterial extracts, we used Y-PER (Pierce), which we had found useful for solubilizing AGT protein. Approximately 50% of the expressed AGT was soluble after lysis using Y-PER (data not shown). C-terminal His6-tagged AGTma and AGTmi proteins consistently showed ∼2-fold increased activity when compared with a previous study using N-terminal protein fusions (8). AGTma showed an average specific activity of 1639 ± 145 μmol/h/mg, whereas AGTmi protein activity was on average ∼50% that of AGTma (specific activity 853 ± 249 μmol/h/mg). A previous study of purified AGT with a C-terminal His6 tag also reported activity of AGTmi as ∼50% of AGTma, although the total specific activity was higher (5).
We used thermal inactivation to explore whether the difference we observed in activity between AGTma and AGTmi was due to a difference in stability of the two variants (Fig. 4). In these studies, we also examined protection by PLP. Although previous studies, including our yeast experiments, were unable to demonstrate protection of AGT by PLP in vivo, indirect effects of PLP on stability have been observed in vitro by several groups (6, 7, 13). Purified proteins were preincubated for 1 h at the indicated temperatures in the presence or absence of 50 μm PLP. Following preincubation, PLP was added such that all samples contained an equal amount of PLP, and AGT activity was assayed. AGTma (Fig. 4A) is inactivated after a 1-h incubation at 65 °C in the absence of ligand but is significantly protected by the addition of PLP. In contrast, the activity of AGTmi is less thermally stable than AGTma, showing reduced activity at 42 °C and complete inactivation after incubation at 60 °C in the absence of added ligand (Fig. 4B). AGTmi was also significantly stabilized by PLP, although not to the same degree as AGTma.
The thermal inactivation studies suggested that both AGTma and AGTmi are protected by the presence of PLP and supported previous studies that suggest AGTma is more stable than AGTmi. Because these studies rely on activity as an indirect indication of stability, we sought to validate these results by directly examining the thermodynamic stabilities of AGT using SUPREX, a mass spectrometry-based technique, and differential scanning fluorimetry (DSF), a fluorescence-based method that examines protein unfolding during heat denaturation.
Mass Spectrometry-based Analysis of AGT Stability Using
SUPREX—The SUPREX technique
(24–27)
is designed to evaluate the thermodynamic stability of proteins in solution
and can provide quantitative stability measurements of both unliganded and
ligand-bound protein. SUPREX is performed by diluting a protein into a series
of H/D exchange buffers containing increasing concentrations of a chemical
denaturant. At higher concentrations of denaturant, a larger population of the
protein is unfolded, leading to an increase in the uptake of deuterons. This
increase in deuteration leads to an increase in mass, which can be measured
using MALDI-TOF mass spectrometry. The mass increase (i.e.
Δmass) is plotted versus denaturant concentration, and the
midpoint of the resulting SUPREX curve transition
( ) reflects the stability
of the protein. Because ligand binding results in protein stabilization, a
greater amount of chemical denaturant is required to unfold a ligand-bound
protein. Thus, upon ligand binding, the midpoint of the SUPREX curve
transition moves toward higher concentrations of denaturant. Unlike
conventional protein denaturation curves, the midpoint of the SUPREX curve
transition is dependent on H/D exchange time. Thus, in the case of very stable
proteins or complexes, a longer H/D exchange time can be employed to move the
transition midpoint to lower denaturant concentrations.
) reflects the stability
of the protein. Because ligand binding results in protein stabilization, a
greater amount of chemical denaturant is required to unfold a ligand-bound
protein. Thus, upon ligand binding, the midpoint of the SUPREX curve
transition moves toward higher concentrations of denaturant. Unlike
conventional protein denaturation curves, the midpoint of the SUPREX curve
transition is dependent on H/D exchange time. Thus, in the case of very stable
proteins or complexes, a longer H/D exchange time can be employed to move the
transition midpoint to lower denaturant concentrations.
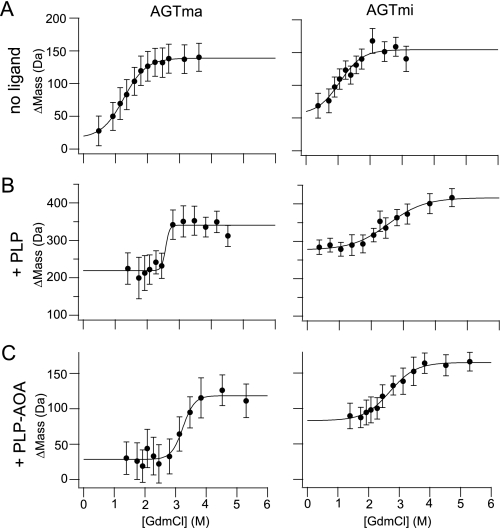
Typical SUPREX data obtained on AGTma and AGTmi are shown in
Fig. 5A, whereas the
average  values obtained
from three to four replicate measurements are shown in
Table 2. The average
values obtained
from three to four replicate measurements are shown in
Table 2. The average
 value for AGTma was
consistently greater than the average
value for AGTma was
consistently greater than the average
 value for AGTmi,
indicating that AGTma is more thermodynamically stable than AGTmi. SUPREX was
also used to detect binding between AGT and two of its known ligands, PLP and
AOA. As shown in Fig. 5 (B and
C), PLP binding resulted in an increase in thermodynamic
stability for AGTma and AGTmi, with further stabilization when both PLP and
AOA were added. The Δmass values for AGT in the presence of PLP were
higher than the Δmass values for AGT without added ligand because of a
covalent linkage between AGT and PLP. (Noncovalent complexes dissociate in the
MALDI matrix solution, but covalent bonds generally persist throughout the
sample preparation protocol.) The Δmass values for AGT-PLP-AOA were
similar to those for AGT without added ligand because of the reaction between
PLP and AOA, which breaks the Schiff base linkage between AGT and PLP
(28).
value for AGTmi,
indicating that AGTma is more thermodynamically stable than AGTmi. SUPREX was
also used to detect binding between AGT and two of its known ligands, PLP and
AOA. As shown in Fig. 5 (B and
C), PLP binding resulted in an increase in thermodynamic
stability for AGTma and AGTmi, with further stabilization when both PLP and
AOA were added. The Δmass values for AGT in the presence of PLP were
higher than the Δmass values for AGT without added ligand because of a
covalent linkage between AGT and PLP. (Noncovalent complexes dissociate in the
MALDI matrix solution, but covalent bonds generally persist throughout the
sample preparation protocol.) The Δmass values for AGT-PLP-AOA were
similar to those for AGT without added ligand because of the reaction between
PLP and AOA, which breaks the Schiff base linkage between AGT and PLP
(28).
FIGURE 5.
SUPREX analysis of AGTma and AGTmi. AGTma and AGTmi (4 μm, with C-terminal His6 tags) were analyzed in the absence of added ligand (5 min of exchange time) (A), in the presence of 400 μm PLP (5 min of exchange time) (B), and in the presence of 400 μm PLP and 400 μm AOA (1 h of exchange time) (C). Graphs show changes in mass at increasing concentrations of guanidine hydrochloride (GdmCl).
TABLE 2.
 values for AGTma
and AGTmi
values for AGTma
and AGTmi
AGTma [guanidine
hydrochloride]a [guanidine
hydrochloride]a
|
AGTmi [guanidine
hydrochloride]a [guanidine
hydrochloride]a
|
|
|---|---|---|
| m | m | |
| AGTb | 1.4 ± 0.1 | 1.0 ± 0.1 |
| AGT + PLPb | 2.7 ± 0.1 | 2.4 ± 0.2 |
| AGT + PLP + AOAc | 3.2 ± 0.1 | 2.7 ± 0.1 |
The values are the averages and standard deviation obtained from three to four replicate measurements.
Exchange time, 5 min.
Exchange time, 1 h.
SUPREX analysis of AGTma and AGTmi in the presence of both PLP and AOA
(PLP-AOA) revealed an increase in the average
 values over those seen
with PLP alone. Because of this considerable stabilization effect, we were
unable to observe a SUPREX transition using a 5-min H/D exchange time, as was
used with PLP only, and found it necessary to increase the H/D exchange time
to 1 h. As mentioned above, by varying the exchange time, the transition
midpoint of the SUPREX curve can be moved into an experimentally accessible
guanidine hydrochloride concentration range. Because PLP and AOA are known to
react with each other (28,
29), we also evaluated whether
AOA is capable of binding AGT in the absence of added PLP. We were unable to
detect a shift in the SUPREX transition midpoint for AGTma when AOA was added
in the absence of additional PLP (data not shown), suggesting that AOA either
cannot bind to AGTma without PLP or that it binds too weakly to produce a
detectable shift in the transition midpoint.
values over those seen
with PLP alone. Because of this considerable stabilization effect, we were
unable to observe a SUPREX transition using a 5-min H/D exchange time, as was
used with PLP only, and found it necessary to increase the H/D exchange time
to 1 h. As mentioned above, by varying the exchange time, the transition
midpoint of the SUPREX curve can be moved into an experimentally accessible
guanidine hydrochloride concentration range. Because PLP and AOA are known to
react with each other (28,
29), we also evaluated whether
AOA is capable of binding AGT in the absence of added PLP. We were unable to
detect a shift in the SUPREX transition midpoint for AGTma when AOA was added
in the absence of additional PLP (data not shown), suggesting that AOA either
cannot bind to AGTma without PLP or that it binds too weakly to produce a
detectable shift in the transition midpoint.
Analysis of AGT Stability Using DSF—We also examined the stability of major and minor alleles of AGT using DSF, a fluorescence method that can be used to monitor solution phase protein stability and ligand-induced changes in stability (30–32). The technique involves subjecting a protein to heat denaturation under continuous fluorescence monitoring in the presence of an environmentally sensitive fluorescent dye. The dyes used, such as SYPRO orange, are quenched in aqueous solutions but show increased emission in nonpolar environments, binding to hydrophobic sites exposed during protein unfolding. As a given protein is exposed to increasing temperature, it undergoes denaturation, leading to an increase in fluorescence. The temperature at which this transition occurs is related to the protein stability. Because unfolding of a protein results in an increase in fluorescence, effects on protein unfolding can be separated from protein aggregation, which is seen as a general decrease in the base-line fluorescence as the temperature increases. For mutant proteins that are very aggregation-prone, this decrease in fluorescence can be quite pronounced but can be overcome by reducing the protein concentration in the assay.4
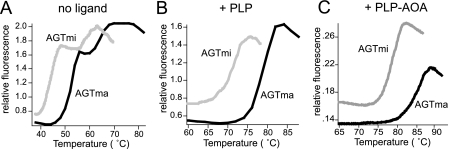
We used DSF to determine melting curves for AGTma and AGTmi. Fig. 6A shows fluorescence changes of the dye SYPRO orange in the presence of 5 μm AGTma or AGTmi over a temperature ramp from 37 to 95 °C. The temperature at which the concentration of folded protein is equivalent to unfolded protein is termed the melting temperature, Tm. A two-step transition was observed for AGTma, with inflection points (i.e. Tm values) at 54 and 66 °C. AGTmi showed a similar two-step transition curve, but both transitions were shifted to the left, with inflection points at 45 and 61 °C. These results are consistent with the SUPREX results, which also show that AGTmi is less stable than AGTma.
FIGURE 6.
Differential scanning fluorimetry of AGTma and AGTmi. Fluorescence changes of 5 μm solutions of AGTma or AGTmi in the presence of SYPRO orange were analyzed in the absence of added ligand (A), in the presence of 70 μm PLP (B), and in the presence of 70 μm PLP and 200 μm AOA (C).
For protein-ligand interactions, ligand binding results in an overall increase in protein stability. In DSF experiments this typically corresponds to an increase in the Tm in the presence of ligand. As shown in Fig. 6 (B and C), we observed shifts in the DSF transitions of AGTma and AGTmi in the presence of ligands PLP and PLP-AOA. The addition of 70 μm PLP causes the Tm values of both AGTma and AGTmi to increase by more than 20 °C, raising the Tm to 78 °C for AGTma and 73 °C for AGTmi. In the presence of both PLP and AOA, the shift in Tm is even greater, to 86 °C for AGTma and 78 °C for AGTmi. With AOA alone, only a small shift in stability (4 °C for AGTma) for the second of the two transitions was detected (data not shown). These DSF results indicate that ligands PLP and AOA exert significant stabilizing effects on AGTma and AGTmi.
DISCUSSION
In this study, we use three separate approaches to analyze the activity and stability of wild-type and PH1-associated AGT variants. First, we developed a cell-based assay for AGT that links activity to yeast growth. Our results show that human AGT can substitute for yeast AGX1 and that mutations associated with disease in humans cause a reduction in growth of yeast. The yeast assay is comparable with other methods of assaying AGT, in that for the mutant proteins that we characterized, growth in yeast roughly parallels activity of the same proteins assayed from human liver (Table 1). For patients affected by PH1, diagnosis can be difficult if DNA sequencing does not reveal one of several known mutations associated with the disease. Thus, straightforward methods to quantify activity are essential. Current methods either require liver biopsy, which has added risk for the patient, or recombinant expression of the mutant allele. The latter method is time-consuming and laborious, requiring harvesting of cells and/or purification, determination of protein levels, and biochemical assay of activity. The yeast assay is convenient because it does not require protein purification or biochemical activity determination. In addition, new mutations can be quickly cloned in a high throughput manner using homologous recombination, a specific advantage of yeast. Finally, the yeast assay is also amenable to high throughput analysis, which we discuss in more detail below.
Upon expressing wild-type and mutant forms of AGT in yeast, we found that disease-associated variants showed reduced growth. We examined variants by Western blotting and found that in most cases the reduced growth could be attributed to reduced steady-state protein levels. One exception was AGTma-S158L, containing a mutation in an amino acid involved in orienting PLP in the active site, which retained significant protein levels despite reduced yeast growth. Significantly, we found that minor allele AGT, a variant that is not disease-causing, showed ∼50% of wild-type activity and yielded ∼50% of wild-type levels of protein in yeast. This finding suggested that this allele is significantly destabilized in cells, which is in agreement with previous studies showing that AGTmi is more prone to aggregation than AGTma when expressed in bacteria (7). To further explore the relationship between activity and stability, we expressed the major and minor AGT alleles in bacteria and directly assayed for stability differences using several different methodologies. Our results, the first such direct studies of AGT stability to be carried out, clearly show that minor allele AGT is significantly destabilized when compared with the major allele protein. We hypothesize that the reduction in yeast growth and reduction in in vitro activity of the minor allele AGT variant is largely due to destabilization.
Although bacterially expressed AGTmi has been observed to be more aggregation-prone than AGTma (7), we did not observe significant aggregation in our stability studies. Protein aggregation would be expected to reduce the cooperativity of the SUPREX and DSF transitions. An aggregated species that dominated our sample would thus greatly reduce or eliminate the denaturant dependence to the Δmass values in the transition regions of the SUPREX curves and the temperature dependence of the fluorescence signals in the DSF curves. For severely aggregated proteins these assays may be unable to differentiate between severe destabilization and aggregation, but such aggregation issues can also be alleviated by decreasing the protein concentration in the assays. The observation of cooperative curve transitions in our studies (see Figs. 5 and 6) indicates that it is unlikely that such aggregated species dominate.
We directly measured the stability of AGTma and AGTmi in the presence of known ligands PLP and AOA. Pyridoxine, which is broken down to form PLP in cells, has long been known to improve the outcome in patients with certain forms of PH1 (1, 12), but it has been unclear how this effect is mediated. Such treatment improves the disease in certain patients with residual AGT enzymatic activity but has no effect on patients that lack enzyme activity, leading researchers to hypothesize that PLP may be acting to enhance protein folding or stability. However, studies that have examined the rescue of mislocalized AGT variants in cell culture have failed to see an effect with PLP (9). In vitro studies have provided indirect evidence of AGT stabilization with PLP (6, 7, 13), but no direct stability analysis has been undertaken.
In the studies described here, we demonstrate ligand-induced stability changes in the presence of PLP. Using an activity-based readout, we show that both AGTma and AGTmi are significantly protected against thermal inactivation by the presence of PLP (Fig. 4) and that AGTma is stabilized to a greater degree than AGTmi. The reduced degree of stabilization for AGTmi may reflect a reduced affinity for PLP, as has been shown previously (8). Our results with His-tagged AGTma in this experiment are similar to those from a recent report using untagged purified AGTma (13), indicating that the C-terminal His6 tag does not have a significant effect on stability. We also detected large stability increases of AGT in the presence of PLP using two direct approaches, SUPREX and DSF. We were unable to observe a difference in degree of PLP stabilization between AGTma and AGTmi using these methods, which we suspect is because the structure-based assays are less sensitive than the activity-based assay. PLP-induced stabilization has been observed previously for many, but not all, PLP-binding enzymes. For example, rat liver mitochondrial aspartate aminotransferase, O-acetylserine sulfhydrylase, glutamate decarboxylase, sheep liver serine hydroxymethyltransferase, and tryptophan synthase were found to be stabilized by PLP (33–38), but no stabilization has been observed for serine hydroxymethyltransferase (39) or dopa decarboxylase (40).
We were unable to examine thermal inactivation of AGT in the presence of PLP-AOA because AOA is an inhibitor of AGT activity. However, because DSF and SUPREX do not rely on activity measurements as an indication of stability, we used these direct methods to examine stability changes of the AGT-PLP complex upon binding of AOA. Results from these approaches show an increase in the stability of AGTma-PLP and AGTmi-PLP upon AOA binding. Although AOA is well known as an inhibitor of many PLP-binding enzymes, to our knowledge, this is the first report of AOA-induced stabilization of AGT-PLP.
Unstable proteins that retain partial function, such as certain AGT mutant subtypes, represent promising targets for drug intervention. These proteins may benefit from pharmacological chaperones, which are compounds that specifically bind and stabilize a native protein conformation. The strong stabilizing effect of AOA on AGT-PLP suggests that an AGT inhibitor could be useful as a pharmacological chaperone. Unfortunately, AOA inhibits a variety of PLP-binding enzymes (41), and this nonspecificity prevents it from being viable as a pharmacological chaperone. Despite their inhibition of protein activity, enzyme inhibitors show promise as pharmacological chaperones, because they can promote proper folding of the active conformation of a protein. Once the active site domain has adopted the proper structure, exchange of the inhibitor for the substrate (which is most likely at a higher concentration than the inhibitor because of the deficiency in protein activity) occurs more readily. For example, unstable variants of lysosomal α-galactosidase A, associated with Fabry disease, were rescued by addition of a competitive inhibitor, 1-deoxygalactonorijimycin (42, 43). In other studies, increased protein levels were seen following treatment of cells expressing mutant acid β-glucosidase (defective in Gaucher disease) or a V2 vasopressin receptor mutant (associated with nephrogenic diabetes insipidus) with active site inhibitors (44, 45). These observations, along with our results with AGT-PLP-AOA, suggest that an AGT-specific inhibitor could be therapeutically useful for PH1, particularly when used in conjunction with PLP.
In the studies described here, we found a clear difference in stability between AGTma and AGTmi. The minor allele acts synergistically with a number of other mutations to cause ∼30–50% of PH1 disease. We speculate that pharmacological chaperones that target this variant, even in the absence of other mutations, may be therapeutically beneficial. Previously, it has been speculated that the minor allele, although not itself disease-causing, could play a role in idiopathic calcium oxalate kidney stone disease, a much more common disease (2). Although this link remains to be established, our studies suggest that a large percentage of the population that is homozygous for the minor allele (∼4% of European and North American (2)) may, depending on their diet, benefit from treatment with PLP or novel stabilizing compounds.
To effectively identify new pharmacological chaperones for AGT, efficient high throughput assays are required. Each of the methodologies that we have developed with AGT (the yeast complementation, SUPREX, and DSF assays) can be used in a high throughput manner to detect ligand binding in screens for pharmacological chaperones. The yeast complementation assay is adaptable to a 96-well format and is straightforward, inexpensive, and requires little set-up time. A major advantage of this assay is that it not only allows identification of compounds that directly bind AGT but also allows for the identification of compounds that act indirectly, such as those that may stimulate protein chaperones. Previous high throughput compound library screens with yeast growth assays have successfully targeted a variety of proteins, such as polyglutamine-containing proteins (46, 47), Sir2 (48), and HSP90 ATPase (49).
The in vitro assays we describe with AGT are also adaptable to high throughput screening. SUPREX can be scaled up for high throughput assays using a single-point protocol (50), which involves collecting data at a single denaturant concentration rather than multiple concentrations and detecting binding events according to the magnitude of the Δmass values. Important advantages of this assay are the lack of requirement for purified protein, because protein mass changes can be evaluated in unpurified extracts, and the ability to detect ligand-binding events at any location in the protein. The DSF assay also has advantages that have made it attractive for use in identifying stabilizing ligands for protein crystallization (51, 52) and in drug discovery (53–55). The method is easily adaptable to a high throughput format, can be carried out using a conventional real time PCR machine, requires small amounts of protein and ligand, and moreover is rapid and easy to set up. Together, these approaches should provide powerful platforms on which to initiate screening studies for stabilizing molecules for AGT.
Acknowledgments
We thank Dr. K.-Peter Stahmann for providing the YCG-Fr yeast strain and Dr. Marion Coulter-Mackie for providing major and minor AGT protein constructs that we adapted for use in this study.
This work was supported, in whole or in part, by National Institutes of Health Grant R21 DK075291-01A1 (to C. L. T.). This work was also supported by a grant from the Oxalosis and Hyperoxaluria Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: AGT, alanine:glyoxylate aminotransferase; PH1, primary hyperoxaluria type I; SUPREX, stability of unpurified proteins from rates of H/D exchange; PLP, pyridoxal 5′-phosphate; H/D, hydrogen/deuterium; AOA, aminooxyacetic acid; HA, hemagglutinin; MALDI, matrix-assisted laser desorption ionization; TOF, time-of-flight; DSF, differential scanning fluorimetry.
A. M. C. Pittman and C. L. Tucker, unpublished data.
References
- 1.Danpure, C. J. (2005) Am. J. Nephrol. 25 303–310 [DOI] [PubMed] [Google Scholar]
- 2.Danpure, C. J. (2006) Biochim. Biophys. Acta 1763 1776–1784 [DOI] [PubMed] [Google Scholar]
- 3.Coulter-Mackie, M. B., and Rumsby, G. (2004) Mol. Genet. Metab. 83 38–46 [DOI] [PubMed] [Google Scholar]
- 4.Purdue, P. E., Takada, Y., and Danpure, C. J. (1990) J. Cell Biol. 111 2341–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lumb, M. J., and Danpure, C. J. (2000) J. Biol. Chem. 275 36415–36422 [DOI] [PubMed] [Google Scholar]
- 6.Santana, A., Salido, E., Torres, A., and Shapiro, L. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7277–7282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coulter-Mackie, M. B., and Lian, Q. (2006) Mol. Genet. Metab. 89 349–359 [DOI] [PubMed] [Google Scholar]
- 8.Coulter-Mackie, M. B., Lian, Q., and Wong, S. G. (2005) Protein Expr. Purif. 41 18–26 [DOI] [PubMed] [Google Scholar]
- 9.Lumb, M. J., Birdsey, G. M., and Danpure, C. J. (2003) Biochem. J. 374 79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lumb, M. J., Drake, A. F., and Danpure, C. J. (1999) J. Biol. Chem. 274 20587–20596 [DOI] [PubMed] [Google Scholar]
- 11.Purdue, P. E., Allsop, J., Isaya, G., Rosenberg, L. E., and Danpure, C. J. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 10900–10904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monico, C. G., Olson, J. B., and Milliner, D. S. (2005) Am. J. Nephrol. 25 183–188 [DOI] [PubMed] [Google Scholar]
- 13.Cellini, B., Montioli, R., Bianconi, S., Lopez-Alonso, J. P., and Voltattorni, C. B. (2008) Protein Pept. Lett. 15 153–159 [DOI] [PubMed] [Google Scholar]
- 14.Rumsby, G., Weir, T., and Samuell, C. T. (1997) Ann. Clin. Biochem. 34 400–404 [DOI] [PubMed] [Google Scholar]
- 15.Nozaki, Y. (1972) Methods Enzymol. 26 43–50 [DOI] [PubMed] [Google Scholar]
- 16.Schlosser, T., Gatgens, C., Weber, U., and Stahmann, K. P. (2004) Yeast 21 63–73 [DOI] [PubMed] [Google Scholar]
- 17.Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., and O'Shea, E. K. (2003) Nature 425 686–691 [DOI] [PubMed] [Google Scholar]
- 18.Danpure, C. J., Fryer, P., Jennings, P. R., Allsop, J., Griffiths, S., and Cunningham, A. (1994) Eur. J. Cell Biol. 64 295–313 [PubMed] [Google Scholar]
- 19.Danpure, C. J., Guttridge, K. M., Fryer, P., Jennings, P. R., Allsop, J., and Purdue, P. E. (1990) J. Cell Sci. 97 669–678 [DOI] [PubMed] [Google Scholar]
- 20.Williams, E., and Rumsby, G. (2007) Clin. Chem. 53 1216–1221 [DOI] [PubMed] [Google Scholar]
- 21.Nishiyama, K., Funai, T., Katafuchi, R., Hattori, F., Onoyama, K., and Ichiyama, A. (1991) Biochem. Biophys. Res. Commun. 176 1093–1099 [DOI] [PubMed] [Google Scholar]
- 22.Zhang, X., Roe, S. M., Hou, Y., Bartlam, M., Rao, Z., Pearl, L. H., and Danpure, C. J. (2003) J. Mol. Biol. 331 643–652 [DOI] [PubMed] [Google Scholar]
- 23.Stolz, J., and Vielreicher, M. (2003) J. Biol. Chem. 278 18990–18996 [DOI] [PubMed] [Google Scholar]
- 24.Ghaemmaghami, S., Fitzgerald, M. C., and Oas, T. G. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 8296–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powell, K. D., and Fitzgerald, M. C. (2003) Biochemistry 42 4962–4970 [DOI] [PubMed] [Google Scholar]
- 26.Powell, K. D., Ghaemmaghami, S., Wang, M. Z., Ma, L., Oas, T. G., and Fitzgerald, M. C. (2002) J. Am. Chem. Soc. 124 10256–10257 [DOI] [PubMed] [Google Scholar]
- 27.Powell, K. D., Wales, T. E., and Fitzgerald, M. C. (2002) Protein Sci. 11 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, W., Peterson, P. E., Carter, R. J., Zhou, X., Langston, J. A., Fisher, A. J., and Toney, M. D. (2004) Biochemistry 43 10896–10905 [DOI] [PubMed] [Google Scholar]
- 29.Beeler, T., and Churchich, J. E. (1976) J. Biol. Chem. 251 5267–5271 [PubMed] [Google Scholar]
- 30.Epps, D. E., Sarver, R. W., Rogers, J. M., Herberg, J. T., and Tomich, P. K. (2001) Anal. Biochem. 292 40–50 [DOI] [PubMed] [Google Scholar]
- 31.Pantoliano, M. W., Petrella, E. C., Kwasnoski, J. D., Lobanov, V. S., Myslik, J., Graf, E., Carver, T., Asel, E., Springer, B. A., Lane, P., and Salemme, F. R. (2001) J. Biomol. Screen. 6 429–440 [DOI] [PubMed] [Google Scholar]
- 32.Poklar, N., Lah, J., Salobir, M., Macek, P., and Vesnaver, G. (1997) Biochemistry 36 14345–14352 [DOI] [PubMed] [Google Scholar]
- 33.Seifert, T., Bartholmes, P., and Jaenicke, R. (1985) Biochemistry 24 339–345 [DOI] [PubMed] [Google Scholar]
- 34.Wu, T. H., Oses-Prieto, J. A., Iriarte, A., and Martinez-Carrion, M. (2003) Biochim. Biophys. Acta 1647 315–320 [DOI] [PubMed] [Google Scholar]
- 35.Bettati, S., Benci, S., Campanini, B., Raboni, S., Chirico, G., Beretta, S., Schnackerz, K. D., Hazlett, T. L., Gratton, E., and Mozzarelli, A. (2000) J. Biol. Chem. 275 40244–40251 [DOI] [PubMed] [Google Scholar]
- 36.Chen, C. H., Wu, S. J., and Martin, D. L. (1998) Arch. Biochem. Biophys. 349 175–182 [DOI] [PubMed] [Google Scholar]
- 37.Venkatesha, B., Udgaonkar, J. B., Rao, N. A., and Savithri, H. S. (1998) Biochim. Biophys. Acta 1384 141–152 [DOI] [PubMed] [Google Scholar]
- 38.Zetina, C. R., and Goldberg, M. E. (1980) J. Biol. Chem. 255 4381–4385 [PubMed] [Google Scholar]
- 39.Cai, K., Schirch, D., and Schirch, V. (1995) J. Biol. Chem. 270 19294–19299 [DOI] [PubMed] [Google Scholar]
- 40.Dominici, P., Moore, P. S., and Borri Voltattorni, C. (1993) Biochem. J. 295 493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zollner, H. (1989) Handbook of Enzyme Inhibitors, p. 239, VCH, Weinheim, Germany
- 42.Yam, G. H., Zuber, C., and Roth, J. (2005) FASEB J. 19 12–18 [DOI] [PubMed] [Google Scholar]
- 43.Fan, J. Q., Ishii, S., Asano, N., and Suzuki, Y. (1999) Nat. Med. 5 112–115 [DOI] [PubMed] [Google Scholar]
- 44.Fan, J. Q. (2003) Trends Pharmacol. Sci. 24 355–360 [DOI] [PubMed] [Google Scholar]
- 45.Morello, J. P., Salahpour, A., Laperriere, A., Bernier, V., Arthus, M. F., Lonergan, M., Petaja-Repo, U., Angers, S., Morin, D., Bichet, D. G., and Bouvier, M. (2000) J. Clin. Investig. 105 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ehrnhoefer, D. E., Duennwald, M., Markovic, P., Wacker, J. L., Engemann, S., Roark, M., Legleiter, J., Marsh, J. L., Thompson, L. M., Lindquist, S., Muchowski, P. J., and Wanker, E. E. (2006) Hum. Mol. Genet. 15 2743–2751 [DOI] [PubMed] [Google Scholar]
- 47.Zhang, X., Smith, D. L., Meriin, A. B., Engemann, S., Russel, D. E., Roark, M., Washington, S. L., Maxwell, M. M., Marsh, J. L., Thompson, L. M., Wanker, E. E., Young, A. B., Housman, D. E., Bates, G. P., Sherman, M. Y., and Kazantsev, A. G. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 892–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bedalov, A., Gatbonton, T., Irvine, W. P., Gottschling, D. E., and Simon, J. A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 15113–15118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rowlands, M. G., Newbatt, Y. M., Prodromou, C., Pearl, L. H., Workman, P., and Aherne, W. (2004) Anal. Biochem. 327 176–183 [DOI] [PubMed] [Google Scholar]
- 50.Powell, K. D., and Fitzgerald, M. C. (2004) J. Comb. Chem. 6 262–269 [DOI] [PubMed] [Google Scholar]
- 51.Niesen, F. H., Berglund, H., and Vedadi, M. (2007) Nat. Protoc. 2 2212–2221 [DOI] [PubMed] [Google Scholar]
- 52.Vedadi, M., Niesen, F. H., Allali-Hassani, A., Fedorov, O. Y., Finerty, P. J., Jr., Wasney, G. A., Yeung, R., Arrowsmith, C., Ball, L. J., Berglund, H., Hui, R., Marsden, B. D., Nordlund, P., Sundstrom, M., Weigelt, J., and Edwards, A. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 15835–15840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baum, E. Z., Crespo-Carbone, S. M., Klinger, A., Foleno, B. D., Turchi, I., Macielag, M., and Bush, K. (2007) Antimicrob. Agents Chemother. 51 4420–4426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grasberger, B. L., Lu, T., Schubert, C., Parks, D. J., Carver, T. E., Koblish, H. K., Cummings, M. D., LaFrance, L. V., Milkiewicz, K. L., Calvo, R. R., Maguire, D., Lattanze, J., Franks, C. F., Zhao, S., Ramachandren, K., Bylebyl, G. R., Zhang, M., Manthey, C. L., Petrella, E. C., Pantoliano, M. W., Deckman, I. C., Spurlino, J. C., Maroney, A. C., Tomczuk, B. E., Molloy, C. J., and Bone, R. F. (2005) J. Med. Chem. 48 909–912 [DOI] [PubMed] [Google Scholar]
- 55.Lo, M. C., Aulabaugh, A., Jin, G., Cowling, R., Bard, J., Malamas, M., and Ellestad, G. (2004) Anal. Biochem. 332 153–159 [DOI] [PubMed] [Google Scholar]