Abstract
Hepsin, a cell surface protease, is widely reported to be overexpressed in more than 90% of human prostate tumors. Hepsin expression correlates with tumor progression, making it a significant marker and target for prostate cancer. Recently, it was reported that in a prostate cancer mouse model, hepsin up-regulation in tumor tissue promotes progression and metastasis. The underlying mechanisms, however, remain largely uncharacterized. Hepsin transgenic mice displayed reduced laminin-332 (Ln-332) expression in prostate tumors. This is an intriguing cue, since proteolytic processing of extracellular matrix macromolecules, such as Ln-332, is believed to be involved in cancer progression, and Ln-332 expression is lost during human prostate cancer progression. In this study, we provide the first direct evidence that hepsin cleaves Ln-332. Cleavage is specific, since it is both inhibited in a dose-dependent manner by a hepsin inhibitor (Kunitz domain-1) and does not occur when catalytically inactive hepsin is used. By Western blotting and mass spectrometry, we determined that hepsin cleaves the β3 chain of Ln-332. N-terminal sequencing identified the cleavage site at β3 Arg245, in a sequence context (SQLR245↓LQGSCFC) conserved among species and in remarkable agreement with reported consensus target sequences for hepsin activity. In vitro cell migration assays showed that hepsin-cleaved Ln-332 enhanced motility of DU145 prostate cancer cells, which was inhibited by Kunitz domain-1. Further, hepsin-overexpressing LNCaP prostate cancer cells also exhibited increased migration on Ln-332. Direct cleavage of Ln-332 may be one mechanism by which hepsin promotes prostate tumor progression and metastasis, possibly by up-regulating prostate cancer cell motility.
Prostate cancer is the second leading cause of cancer death in men in the United States; according to the American Cancer Society, 186,320 new prostate cancer cases and 28,660 deaths from prostate cancer are projected to occur in 2008 (1). This high rate of mortality is largely due to metastasis of the primary tumor (2). For metastasis to occur, primary tumor cells must breach the basement membrane (BM)3 by degrading extracellular matrix (ECM) molecules to initiate the invasion process (3). Escaped tumor cells interact with neighboring ECM molecules to promote this activity. This interaction sometimes promotes remodeling of the ECM to create a more conducive environment for tumor cell migration and invasion and thereby aids in cancer progression (4). The remodeling of certain ECM molecules has been reported to occur as a direct result of protease processing, which results in increased tumor cell migration and invasion (5–7).
Laminin-332 (Ln-332; previously known as laminin-5), an ECM molecule, is an important component of BM (8). It is a trimeric glycoprotein consisting of disulfide-bonded subunits: α3, β3, and γ2 polypeptide chains (9). The importance of Ln-332 in BM assembly was established by the discovery of the occurrence of a lethal skin blistering disorder, junctional epidermolysis bullosa, which is due to mutation in any of the three chains of Ln-332 (10). Ln-332 also plays an important role in development, wound healing, and tumorigenesis (11). Ln-332 is overexpressed in several tumor types, such as esophageal, cutaneous, oral, laryngeal, colon, tracheal, and cervical cancers (12); however, interestingly, there is loss of Ln-332 in prostate cancer (13–16). All three chains of Ln-332 can be processed by different protease systems (17), sometimes as a means of motility regulation of cells (18).
Proteolytic cleavage of Ln-332 γ2 chain by several matrix metalloproteinases (MMPs) has been reported to increase cell migration; our laboratory previously reported Ln-332 γ2 cleavage by MMP2 and MT1-MMP (19, 20), and others have shown that MMP3, -8, -12, -13, -14, and -20 cleave the γ2 chain of Ln-332 (21). Other enzymes reported to cleave Ln-332 are cathepsin S (22), mTLD (23), BMP-1 (24), and neutrophil elastase (25). The β3 chain of Ln-332, believed to be relatively resistant to proteolytic processing, has also been reported to be processed by both MT1-MMP (26) and matrilysin (27). Another study reported that β3 chain of Ln-332 is cleaved at its N terminus by endogenous proteinase(s) in human keratinocytes and other cell lines (28); however, the authors did not indicate the specific protease involved in the cleavage. Clearly, these studies have established that proteolytic processing of Ln-332 occurs physiologically and can alter cellular behavior, at least in terms of motility.
During the progression of prostate cancer, various genetic and epigenetic changes occur (29). Loss or down-regulation of various tumor suppressor genes (30, 31), as well as up-regulation or overexpression of various genes, has been reported (32–34). One of the genes found to be up-regulated in more than 90% of human prostate cancer cases is the cell surface protease, hepsin (35, 36). Recently, a prostate cancer mouse model demonstrated that hepsin overexpression causes disorganization of BM and promotes prostate cancer progression and metastasis (37). The mechanism involved in this disorganization of BM was not revealed; however, the authors did report that prostate tissues from hepsin-overexpressing mice exhibited weaker immunohistochemical staining of Ln-332 compared with wild type mice. Decreased staining of Ln-332 in hepsin-overexpressing mice led us to hypothesize that Ln-332 is processed by hepsin, which may be a key step in the progression of prostate cancer. Hepsin is a member of the type II transmembrane serine protease family. The physiological function of hepsin remains unclear (38). In vitro studies have identified blood coagulation factors as substrates of hepsin (39). Prohepatocyte growth factor and prourokinase-type plasminogen activator are also substrates for hepsin (40, 41). The role of hepsin in prostate cancer remains of great interest, since it overexpressed in more than 90% of human prostate cancer, and its expression correlates with progression of the disease (42).
In summary, a role for either Ln-332 or hepsin in prostate cancer progression is supported by several studies. However, the roles of these two molecules have been studied separately, and they have not been previously linked to date. In this report, we demonstrate for the first time that hepsin cleaves Ln-332. Using Western blot analysis and mass spectrometry, we identified that cleavage occurs specifically in the β3 chain. Further, N-terminal sequencing identified the hepsin cleavage site between the Arg245 and Leu246 residues in the β3 chain. Cleavage of Ln-332 was inhibited by a known hepsin inhibitor (KD1) and did not occur in the presence of catalytically inactive hepsin, confirming specificity. We also report increased migration of DU145 prostate cancer cells on hepsin-cleaved Ln-332, which was also inhibited in the presence of KD1. Similarly, we show that hepsin-overexpressing LNCaP-34 cells exhibit enhanced motility on Ln-332, compared with low hepsin-expressing LNCaP-17 cells. This study suggests a physiological role for hepsin in proteolytic cleavage of Ln-332 and gives new insight into possible mechanisms for hepsin in prostate cancer progression.
EXPERIMENTAL PROCEDURES
Cell Culture—Prostate cancer cell line DU145 (American Type Culture Collection, Manassas, VA) and 804G bladder squamous cell carcinoma cells (previously described by Falk-Marzillier et al. (43)) were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Gemini, Irvine, CA) and 1% glutamine/penicillin/streptomycin antibiotics (Invitrogen) in an incubator with 5% CO2 at 37 °C. LNCaP-17 (low hepsin-expressing) and LNCaP-34 (hepsin-overexpressing) prostrate cancer cells were created as previously described by Moran et al. (40) and were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 500 μg/ml Geneticin (Invitrogen), 0.5 μg/ml puromycin (Sigma), and 1% glutamine/penicillin/streptomycin and incubated with 5% CO2 at 37 °C.
Purification of Rat Ln-332—Rat Ln-332 was purified from spent medium of 804G bladder squamous cell carcinoma cells. Briefly, 804G cells were cultured in 10% fetal bovine serum containing Dulbecco's modified Eagle's medium in a 150-mm dish. Cells were then washed with PBS twice and cultured in serum-free conditioned medium for 2 days in roller bottles. The serum-free conditioned medium was collected and concentrated by ammonium sulfate at 80% saturation and dialyzed against 20 mm Tris-HCl (pH 7.5), 0.5 m NaCl, 0.005% Brij-35 (TNB buffer). The concentrated serum-free conditioned medium was then used for immunoaffinity chromatography. The Protein A-Sepharose column (0.8 × 4.0 cm; Bio-Rad) chemically conjugated with nonfunctional Ln-332 mouse antibody, TR-1 (44), was equilibrated with TNB buffer at a flow rate of 15 ml/h. The concentrated and dialyzed sample was applied to the TR1 column. The column was then washed with TNB, and absorbed Ln-332 was eluted with 10 ml of 0.05% trifluoroacetic acid, pH 2.5. The eluted fractions were neutralized by 300 μl of 1 m Tris-HCl, pH 8.0, and then 1% CHAPS was added to each fraction.
Cleavage of Ln-332—The cleavage of rat Ln-332 was studied with human recombinant hepsin, consisting of the entire extracellular domain (40). To study the cleavage of Ln-332 by hepsin, purified rat Ln-332 (0.2 μm) was incubated with the recombinant protease domain of hepsin (at both 0.13 and 1.3 μm) and reaction buffer containing 250 mm NaCl and 50 mm Tris (pH 7.5) for 1.5 h at 37 °C. For the time course experiment, Ln-332 (0.8 μm) was incubated with hepsin (5.2 μm) and reaction buffer containing 250 mm NaCl and 50 mm Tris, pH 7.5, for 0, 2, and 6 h at 37 °C. After incubation, hepsin and Ln-332 reaction mixture was electrophoresed on 4–12% precast SDS-polyacrylamide gradient gel under reducing/nonreducing (as indicated) conditions and then stained with SimplyBlue™ Safe Coomassie Blue stain (Invitrogen). A standard marker (identified as M in the figures; Precision Plus™ protein dual color standard; Bio-Rad) was also run for comparison.
Western blot analysis was performed after transferring the untreated and treated protein on a polyvinylidene difluoride membrane (PerkinElmer Life Sciences), from reducing gel. Polyclonal antibody (pAb; 1:500) against the C terminus of Ln-332 β3 chain (sc-20775; H-300; Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)) and secondary anti-rabbit IgG horseradish peroxidase monoclonal antibody (1:5000; GE Healthcare) were used for Western blot. Protein bands were visualized with the ECL+ plus system (PerkinElmer Life Sciences).
Mass Spectrometry—The cleaved product of Ln-332 by hepsin was further identified using mass spectrometry analysis performed by the Mass Spectrometry Research Center at Vanderbilt University (Nashville, TN). After digestion, the proteins in the reaction mixture were separated by SDS-PAGE under nonreducing conditions and visualized using Coomassie Blue stain. The protein bands of interest were excised from the SDS-polyacrylamide gel and then equilibrated in 100 mm NH4HCO3, reduced with 3 mm dithiothreitol in 100 mm NH4HCO3 at 37 °C for 15 min. Alkylation was carried out with iodoacetamide (6 mm in 100 mm NH4HCO3 for 15 min). After destaining, the gel slices were dehydrated with 50% acetonitrile in 50 mm NH4HCO3, followed by 100% acetonitrile. Gel slices were rehydrated with 15 μl of 25 mm NH4HCO3 containing 0.01 μg/μl modified trypsin. Trypsin digestion was performed for 2 h at 37 °C. Peptides were extracted with 60% acetonitrile and 0.1% trifluoroacetic acid, dried by vacuum centrifugation, and reconstituted in 10 μl of 0.1% trifluoroacetic acid. After desalting, peptides were concentrated into 2 μl of 60% acetonitrile with 0.1% trifluoroacetic acid using ZipTipC18 pipette tips. For the preparation of sample for matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), 0.4 μl of the sample was applied to a target plate and overlaid with 0.4 μl of α-cyano-4-hydroxycinnamic acid matrix (10 mg/ml in 60% acetonitrile, 0.1% trifluoroacetic acid). MALDITOF MS and tandem TOF/TOF MS/MS were performed using a Voyager 4700 mass spectrometer (Applied Biosystems, Foster City, CA). TOF/TOF fragmentation spectra were acquired in a data-dependent fashion based on the MALDI-TOF peptide mass map for the protein. Both types of mass spectral data were collectively used to examine the protein data bases to generate statistically significant candidate identification using GPS Explorer software (Applied Biosystems) running the MASCOT data base search algorithm (Matrix Science). Searches were performed against the SWISS PROT and the NCBI databases.
Edman Degradation Sequencing—NH2-terminal sequencing of the polyvinylidene difluoride membrane containing the cleaved protein was carried out on an Applied Biosystems Procise® 494 cLC protein sequencer at the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University (New Haven, CT).
Enzyme Inhibition Assay—We used hepatocyte growth factor activator inhibitor-1-derived Kunitz domain inhibitor (KD1) to inhibit hepsin activity (45). As described above under “Cleavage of Ln-332,” purified rat Ln-332 (0.2 μm) was incubated alone or with recombinant hepsin (1.3 μm) and reaction buffer with or without KD1 inhibitor (5.6 or 11.2 μm) at 37 °C. The reaction mixtures were electrophoresed on 4–12% precast gradient gel under reducing conditions and then stained with Coomassie Blue.
Inactivation of Hepsin Enzymatic Activity by Glu-Gly-Arg Chloromethyl Ketone (EGR-cmk)—Purified recombinant hepsin as described (40) was incubated either with 10-fold molar excess of an irreversible covalent inhibitor, EGR-cmk (Hematological Technologies, Essex Junction, VT) or buffer containing 50 mm Tris, pH 8.0, 150 mm NaCl (for Ctrl-hepsin) for 3 h at room temperature. EGR-cmk-inactivated hepsin (EGR-hepsin) complex was separated from unreacted EGR-cmk by size exclusion chromatography using a Superdex S-200 column (GE Healthcare Inc.) with a buffer containing 50 mm Tris, pH 8.0, 150 mm NaCl. Ctrl-hepsin was also subjected to size exclusion chromatographic purification as described above. Relative to Ctrl-hepsin, EGR-hepsin had lost >99% of its catalytic activity, as assessed by the rate of substrate hydrolysis with a small synthetic substrate, S2366 (Diapharma, West Chester, OH).
EGR-hepsin Assay—EGR-hepsin was used to further examine the specificity of Ln-332 cleavage by hepsin. As with earlier assays, 0.2 μm purified rat Ln-332 was incubated with reaction buffer alone or with recombinant EGR-hepsin (1.3 μm), Ctrl-hepsin (1.3 μm), or the recombinant hepsin (1.3 μm) used for the initial cleavage reactions at 37 °C. The reaction mixtures were electrophoresed on 4–12% precast gradient gel under reducing conditions and then stained with Coomassie Blue.
Transwell Migration Assays—Cell migration assays were performed using 8.0-μm pore size Transwell™ permeable supports (Corning Costar, Lowell, MA). The undersides of the filters were coated with either untreated or hepsin-treated rat Ln-332 (1 μg/ml), Ln-332 coincubated with hepsin and KD1, PBS, hepsin (1.3 μm), or KD1 (5.6 μm) overnight at 4 °C. Transwells were then blocked with 5% milk in PBS with 0.2% Tween 20 for 1 h. DU145 or LNCaP cells were trypsinized, resuspended in serum-free medium, and washed twice with serum-free medium, and cells (20,000 or 50,000, respectively) were seeded in the upper chamber of inserts. After 5 h (DU145) or 24 h (LNCaP) incubation in 5% CO2 at 37 °C, cells remaining on the upper filter were scraped off gently using a cotton swab, and the inserts were gently washed with PBS. Those cells that migrated to the lower chamber were fixed with 400 μl of fixation solution (Hema-3® stain kit, catalog number 122-911; Fisher) for 10 min, stained with 400 μl of staining solution for 20 min, and imaged with a Zeiss LSM-510 inverted microscope (Carl Zeiss). Five representative images (×10 magnification) were randomly captured for each insert and used to manually count the number of cells present. Results are presented as mean number of cells per field ± S.D. Student's t tests (α = 0.05) were performed on final data to test significance of effects.
Cell-mediated Cleavage of Ln-332—LNCaP-17 (low hepsin-expressing) or LNCaP-34 (hepsin-overexpressing) cells (9.0 × 104) in 100 μl of RPMI 1640 were incubated with either 100 μg/ml Ln-332 or PBS for 12 h at 37 °C. After 12 h, Ln-332 solution with medium and cells was collected and centrifuged for 5 min at 15,000 rpm, and supernatant was collected. SDS-PAGE analysis was performed under reducing conditions, and protein was transferred to a nitrocellulose membrane. A pAb against the C terminus of Ln-332 β3 chain (1:200; sc-20775; H-300; Santa Cruz Biotechnology), and the secondary anti-rabbit IgG horseradish peroxidase antibody (1:5000) was used for visualization in Western blot. Protein bands were visualized with the ECL+ plus system (PerkinElmer Life Sciences).
RESULTS
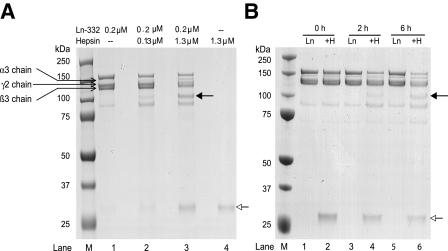
Processing of Ln-332 by Hepsin—To determine whether hepsin cleaves Ln-332, purified rat Ln-332 was incubated with recombinant hepsin at different concentrations in reaction buffer for 1.5 h with an enzyme/substrate (i.e. hepsin/Ln-332) molar ratio of 1.0:1.5 and 1.0:0.15 (latter ratio used in all follow-up experiments), as described under “Experimental Procedures.” After incubation, protein samples were separated under reducing conditions by SDS-PAGE and stained with Coomassie Blue (Fig. 1A). We observed a unique band at ∼100 kDa in the lanes containing Ln-332 incubated with hepsin at both concentrations tested (solid arrow; lanes 2 and 3) but not in untreated samples (lane 1). The intensity of the cleaved band correlated with increasing hepsin concentration (lane 3). Additionally, lanes 2 and 3 produced a unique band at ∼30 kDa (open arrow), which corresponds to the protease domain of hepsin. Lane 4, which includes hepsin alone, also revealed this ∼30 kDa band. Ln-332, both in the absence and presence of hepsin, was also electrophoresed under nonreducing conditions (supplemental Fig. 1). Under both reducing and nonreducing conditions, Ln-332 appeared proteolytically degraded by hepsin on SDS-PAGE; however, under nonreducing conditions, the bands appeared less defined, which was due to the absence of the reducing agent (dithiothreitol). This electrophoretic behavior under nonreducing conditions is consistent with the presence of disulfide bonds between the Ln-332 chains, as mentioned in the Introduction.
FIGURE 1.
SDS-PAGE analysis of hepsin cleavage of purified rat laminin-332. A, purified rat Ln-332 (0.2 μm) was incubated alone or with the recombinant extracellular domain of hepsin for 1.5 h at 37 °C, electrophoresed on 4–12% gradient gel under reducing conditions, and stained with Coomassie Blue. After incubation of Ln-332 alone, the gel included bands identified as the α3 (190 kDa), β3 (145 kDa), and γ2 (155 and 80 kDa) chains (lane 1). However, upon incubation of Ln-332 with 0.13 or 1.3 μm hepsin (lanes 2 and 3, respectively), an additional ∼100 kDa band was seen (indicated by a solid arrow), indicating a cleavage event. Those lanes including hepsin treatment (lanes 2–4) also produced an ∼30 kDa band, which represents the protease domain of hepsin (indicated by an open arrow). B, after incubation of Ln-332 alone at various time points (0, 2, and 6 h), the same uncleaved Ln-332 chains are visible. However, co-incubation of Ln-332 (0.2 μm) with hepsin (1.3 μm) resulted in the generation of a new band (∼100 kDa, indicated by a closed arrow), again indicating cleavage. Again, the ∼30 kDa band corresponds to the hepsin protease domain (indicated by an open arrow; lanes 2, 4, and 6).
In a time course experiment, we incubated Ln-332 and hepsin in reaction buffer for 0, 2, or 6 h (Fig. 1B). As expected, no cleavage product of Ln-332 was observed at 0 h (lane 2), whereas the intensity of the ∼100 kDa band increased from 2 h (lane 4) to 6 h(lane 6) (closed arrow). As shown in Fig. 1B, those treatments including hepsin (lanes 2, 4, and 6) also showed the ∼30 kDa hepsin band (open arrow). These results confirmed that untreated Ln-332 resolved as bands corresponding to α3 (190 kDa), β3 (145 kDa), and γ2 chains (155 and 80 kDa) and that no cleavage band was detectable in the absence of hepsin (lanes 1, 3, and 5). These results suggest that the ∼100 kDa band is a unique product of hepsin cleavage.
Ln-332 Is Specifically Cleaved by Hepsin—In order to confirm that Ln-332 was cleaved by hepsin specifically and not by another contamination protease, we performed an enzyme inhibition assay. We added a specific inhibitor of hepsin, KD1, in the cleavage reaction with Ln-332 and hepsin during incubation and analyzed by SDS-PAGE (Fig. 2A). Hepsin-treated Ln-332, as expected, contained a ∼100 kDa band (solid arrow; lane 2) not present in untreated Ln-332 (lane 1). However, the addition of the hepsin inhibitor KD1 nearly abolished this band (lanes 3 and 4). Those lanes with hepsin treatment (lanes 2–4) again revealed ∼30 kDa protease domain bands (open arrow). Additionally, those treatments with KD1 inhibitor (lanes 3 and 4) produced a band at ∼10 kDa (double arrow). This experiment indicates that the ∼100 kDa band is a product of cleavage of Ln-332 by hepsin.
FIGURE 2.
Specificity of hepsin cleavage of laminin-332. A, purified rat Ln-332 (0.2 μm) was incubated alone, with recombinant hepsin (1.3 μm), or with hepsin and KD1 inhibitor (5.6 or 11.2 μm) for 1.5 h at 37 °C. All treatments were electrophoresed on 4–12% gradient gel under reducing conditions and stained with Coomassie Blue. Ln-332 incubation alone displayed three strong bands, one for each chain (lane 1). Ln-332 coincubated with hepsin displayed an additional band of cleavage product (indicated by a solid arrow; ∼100 kDa; lane 2). However, Ln-332 incubated with both hepsin and KD1 inhibitor displayed greatly diminished bands of cleavage product compared with the band in the absence of KD1 (lanes 3 and 4, respectively). Those treatments with hepsin (lanes 2–4) again produced an ∼30 kDa band (indicated by an open arrow). Lanes with KD1 also showed an ∼11 kDa band, representing the presence of this inhibitor (indicated by a double arrow). B, to further confirm that Ln-332 was cleaved by hepsin specifically, we performed another SDS-PAGE assay. In this setup, Ln-332 was incubated either alone (lane 1) or in the presence of hepsin (lane 2), inactive EGR-hepsin (lane 3), or Ctrl-hepsin (lane 4). Ln-332 incubated with catalytically active hepsin or Ctrl-hepsin resulted in an ∼100-kDa band (shown with an arrow; lanes 2 and 4, respectively). In contrast, Ln-332 alone or Ln-332 incubated with inactive EGR-hepsin did not produce this band (lanes 1 and 3, respectively). Those lanes with only hepsin (lane 7), Ctrl-hepsin (lane 6), or inactive EGR-hepsin (lane 5) only produced a single band at ∼30 kDa. These results further strengthen the idea that Ln-332 is cleaved specifically by active hepsin molecule and no other contaminating protease.
Inactive Hepsin Does Not Cleave Ln-332—To further examine the specificity of Ln-332 cleavage, we performed experiments with catalytically inactive EGR-hepsin. In enzymatic assays with synthetic S2366 substrate, EGR-hepsin displayed less than 1% activity of uninhibited Ctrl-hepsin. In the experiments (Fig. 2B), we incubated Ln-332 either alone (lane 1) or in the presence of hepsin (lane 2), inactive EGR-hepsin (lane 3), or Ctrl-hepsin (lane 4). As shown in Fig. 2B, Ln-332 incubated with active hepsin or Ctrl-hepsin resulted in an ∼100 kDa band (solid arrow; lanes 2 and 4, respectively). In contrast, Ln-332 alone or Ln-332 incubated with inactive EGR-hepsin did not produce this band (lanes 1 and 3, respectively). Those lanes with only hepsin (lane 7), Ctrl-hepsin (lane 6), or inactive EGR-hepsin (lane 5) only produced a single band at ∼30 kDa. These results further support the conclusion that Ln-332 is cleaved specifically by catalytically active hepsin molecule and no other contaminating protease.
Characterization of Cleaved Ln-332 Band—To determine the identity of the unique band that appeared after hepsin treatment of Ln-332, we resorted to using an antibody specific for the Ln-332 chains. A pAb raised against the C-terminal sequence of Ln-332 β3 chain (46) reacted in Western blotting (Fig. 3A), both with the full-length β3 chain in Ln-332 alone (lane 1) and the 100-kDa hepsin-cleaved fragment (lane 2). This result suggested that hepsin cleaves the β3 chain of Ln-332, possibly removing an N-terminal sequence. To confirm this possibility, we further established the identity of the cleaved fragment by mass spectrometry (performed by the Mass Spectrometry Research Center at Vanderbilt University). Briefly, trypsin digestion of the ∼100 kDa cleavage band produced 17 distinct peptides, all identical to peptides located in the ∼100 kDa C-terminal region of rat Ln-332 β3 chain (Fig. 3B). This result indicates that hepsin cleaves the Ln-332 β3 chain in the vicinity of the N terminus. To confirm this possibility and to positively identify the cleavage site, we performed N-terminal sequencing of the cleaved band. The strongest signal was obtained for the following sequence: NH2-LQGSCFC (note that cysteine residues (underlined) are not detected by Edman sequencing and were deduced from cDNA), which corresponds exactly to a sequence in rat Ln-332 starting at Leu246. Therefore, we conclude that the hepsin cleavage site on the Ln-332 β3 chain is located between Arg245 and Leu246 (Fig. 3C).
FIGURE 3.
Identification of hepsin-cleaved product as laminin-332 β3 chain. A, Western blot was performed on Ln-332 alone (0.1 μm) and in combination with hepsin (0.7 μm) and probed with a polyclonal antibody specific for the β3 chain. Application of β3 antibody identified the ∼100-kDa cleaved product of Ln-332 β3 chain and the uncleaved β3 chain. B, trypsin digestion of the ∼100 kDa band was performed for 2 h at 37°C before subjection to matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. The mass spectral data were used to examine protein data bases to generate statistically significant candidate identifications using GPS Explorer software running the Mascot data base search algorithm. Seventeen individual peptides (highlighted in gray) were identified from the digestion, which directly aligned with the sequence of rat Ln-332 β3 chain. N-terminal sequencing was also applied to the digested band, which generated the sequence LQGSCF (underlined). C, schematic representation of Ln-332 including the specific hepsin cleavage site determined to be between the Arg245-Leu246 residues of the rat Ln-332 β3 chain. Note that cysteine residues (underlined) are not detected by Edman sequencing and were deduced from cDNA. The three chains (α3, β3, and γ2), including domains I–VI and LG domains of Ln-332, are also indicated. One-letter amino acid codes are shown.
Migration of DU145 Cells on Ln-332 Is Enhanced by Hepsin Cleavage—To determine the potential biological significance of the cleavage of Ln-332 β3 chain, we examined the migratory behavior of DU145 prostate cancer cells on hepsin-cleaved Ln-332 versus untreated Ln-332 substrate (Fig. 4). Using a modified Boyden chamber assay, we applied various substrates to transwells, as described under “Experimental Procedures.” Hepsin-cleaved Ln-332 promoted a significant increase in migration (1.7-fold) compared with untreated Ln-332 (n = 4, in duplicate; p < 0.05). To confirm that hepsin cleavage of Ln-332 caused increased migration of cells, we added hepsin inhibitor KD1 to test its ability to abolish increased activity. As expected, in the presence of KD1, cells migrated similarly to cells on Ln-332 (n = 4, in duplicate). As a control, almost no cells were seen on PBS-, KD1-, or hepsin alone-treated inserts, (i.e. without ECM substrate). These results suggest that cleavage of Ln-332 by hepsin may physiologically enhance migration.
FIGURE 4.
DU145 prostate cancer cells exhibit enhanced migration on hepsin-cleaved laminin-332. DU145 cells (2 × 104) in serum-free Dulbecco's modified Eagle's medium were added to pretreated upper chambers of transwell inserts and allowed to migrate for 5 h at 37 °C. After incubation, nonmigratory cells and media were washed from transwells, and those cells that migrated to the bottom of the filters were stained with the Hema® kit, fixed, and imaged using a Zeiss LSM-510. A, representative images (5 fields) were taken of pretreated (Ln-332, Ln-332 + Hepsin, Ln-332 + Hepsin + KD1, PBS, hepsin, or KD1) filters with fixed cells. Scale bar, 20 μm. B, cells plated on hepsin-cleaved Ln-332-treated inserts had a significant (n = 4; p < 0.001) increase in migration, compared with cells on either Ln-332 alone or Ln-332 with hepsin and KD1 inhibitor. Cells on PBS-, hepsin-, or KD1-treated inserts migrated significantly (n = 4, in duplicate; p < 0.01 in all cases) less that all other treatments.
Migration of Hepsin-overexpressing Cells Is Enhanced on Ln-332 Substrate—To further determine the potential biological significance of hepsin cleavage of Ln-332 β3 chain, we also examined the migratory behavior of hepsin-overexpressing LNCaP-34 prostate cancer cells on Ln-332 versus low hepsin-expressing LNCaP-17 cells on the same substrate (Fig. 5). We verified LNCaP cell expression by real time PCR and Western blot (results not shown) and obtained data consistent with published findings that LNCaP-34 cells express ∼5-fold higher levels of hepsin than LNCaP-17 cells (40). Using a modified Boyden chamber assay, we applied either Ln-332 (10 μg/ml) or PBS substrate to transwells and allowed cells to migrate at 37 °C for 24 h, as described under “Experimental Procedures” (Fig. 5A). As shown in Fig. 5A, LNCaP-34 cells exhibited a significant increase in migration on Ln-332 (∼2.1-fold) compared with LNCaP-17 cells on Ln-332 (n = 3, in duplicate; p < 0.01). To confirm that degradation of Ln-332 by cells influenced migration, we also measured both cell lines on PBS-treated inserts. As expected, both cell clones migrated slowly on PBS-treated transwells (n = 3, in duplicate; p > 0.05). Additionally, a pAb directed against the β3 chain of Ln-332 in Western blot analysis (Fig. 5B) revealed an additional ∼100-kDa band unique to hepsin-overexpressing LNCaP-34 cells incubated with Ln-332 (lane 2), which was not exhibited by LNCaP-17 cells (lanes 1 and 3) or by LNCaP-34 cells in the absence of Ln-332 (lane 4). The bands in lanes 3 and 4 are background bands, possibly due to endogenous expression of β3 by these LNCaP cells. Taken together, these results suggest that cleavage of Ln-332 by hepsin may physiologically enhance migration.
FIGURE 5.
Migration of LNCaP hepsin-overexpressing prostate cancer cells is enhanced on Ln-332. A, to determine the potential biological significance of hepsin cleavage of the Ln-332 β3 chain, we examined the migratory behavior of hepsin-overexpressing LNCaP-34 prostate cancer cells on Ln-332 versus low hepsin-expressing LNCaP-17 cells. Using a modified Boyden chamber assay, we applied either Ln-332 (10 μg/ml) or PBS to transwells, and allowed cells (5 × 104) to migrate at 37 °C for 24 h, as described under “Experimental Procedures.” LNCaP-34 cells exhibited a significant increase in migration on Ln-332 (∼2.1-fold) compared with LNCaP-17 cells on Ln-332 (n = 3, in duplicate; p < 0.01). To confirm that degradation of Ln-332 by cells influenced migration, we also measured both cell lines on PBS-treated inserts. As expected, both clones weakly migrated on PBS-treated transwells (n = 3, in duplicate; p > 0.05). These results suggest that cleavage of Ln-332 by hepsin may physiologically enhance migration. B, Western blot analysis of LNCaP-17 and LNCaP-34 cells, each in the presence or absence of Ln-332, revealed that hepsin-overexpressing cells (LNCaP-34) created an additional band at ∼100 kDa. The bands in lanes 3 and 4 (cells alone) are background, possibly due to endogenous expression of β3 by these LNCaP cells.
DISCUSSION
In this study, we establish that Ln-332, an important basement membrane component that is apparently down-regulated in human prostate cancer (13–16), is cleaved by hepsin, a serine protease that is overexpressed in more than 90% of human prostate cancer cases (36). In this report, the experimental evidence that hepsin proteolytically cleaves the Ln-332 β3 chain is as follows: 1) treatment of purified Ln-332 with catalytically active hepsin produces an ∼100-kDa fragment both in a time- and dose-dependent fashion; 2) production of this fragment is abolished by KD1, a specific inhibitor of hepsin; 3) catalytically inactive EGR-hepsin does not promote cleavage; 4) the ∼100-kDa fragment reacts by Western blot with antibodies against the C terminus of the β3 chain; and 5) the sequences of peptides from the ∼100 kDa band, determined by mass spectrometry, are identical to peptides from the C-terminal region of the rat Ln-332 β3 chain.
Since all of our cleavage experiments were carried out with Ln-332 purified from a rat cell line, an important question is whether Ln-332 cleavage by hepsin applies to other species as well, particularly humans. Although this is likely, due to the general functional interchangeability of ECM macromolecules across mammalian species (47), we sought additional evidence by locating the precise hepsin cleavage site on rat Ln-332 and then determining whether this cleavage site is present on human Ln-332. N-terminal sequencing of the hepsin-generated Ln-332 β3 fragment identified the hepsin cleavage site at Arg245-Leu246 of the β3 chain of rat Ln-332. The sequence around this cleavage site, SQLR↓LQGSCFC, agrees well with target sequences for hepsin, identified by a high-throughput combinatorial approach (48). In this recent study, P4-P1 substrate specificity of hepsin was determined by using a tetrapeptide positional scanning-synthetic combinatorial library screening approach. On the basis of peptide profiling and amidolytic activity measurements, the authors identified optimal P4–P1 cleavage motifs for hepsin (P1 represents the residue immediately N-terminal to the cleavage site). It was reported that hepsin exhibited a strong preference for arginine at the P1 position and moderately favored threonine, leucine, or asparagine at P2, glutamine or lysine at P3, and proline or lysine at the P4 position. Accordingly, we found that hepsin cleaves Ln-332 after a QLR sequence (P3–P1). It should be noted that the target sequence for pro-hepatocyte growth factor, which turned up at the top of the list of hepsin substrates identified by Herter et al. (48), is KQLR, almost identical to the Ln-332 hepsin target sequence, SQLR. The only difference is at the P4 position (Lys for pro-hepatocyte growth factor and Ser for Ln-332), which was found to be more promiscuous and less critical than P3–P1 (48). The same author also identified serine as a possible residue in position P4 (Fig. 1 in Ref. 48). Visual inspection for homology showed that the hepsin substrate sequence SQLR↓LQGSCFC is completely conserved between rat, mouse, and human Ln-332 β3 sequences (NCBI data base rat LAMB3 accession number XM_001069930, mouse LAMB3 accession number NM_008484, and human LAMB3 accession number NM_000228). Sequence conservation strongly supports a functional significance in terms of hepsin activity. Nonetheless, cleavage of human Ln-332 could not be verified directly, because purified human Ln-332 is not available at this time and remains to be determined.
Interestingly, the SQLR245 is repeated downstream in the rat Ln-332 β3 sequence, SQLR743. However, this is an unlikely cleavage site of hepsin for three reasons: 1) we found no evidence of cleavage products matching a corresponding size; 2) the sequence SQLR743 is not conserved in human Ln-332 β3 chain; and 3) residue 743 falls within the predicted coiled-coil region of Ln-332, presumably inaccessible to proteases. Therefore, it is unlikely that SQLR743 is a site first used by hepsin for cleavage. However, once the residue 245 site is cleaved, it is theoretically possible that the coiled-coil might open up and allow for a second cut at SQLR743. This, of course, could only occur in the rat protein (absent in the human protein). To this end, we performed a time course experiment to determine if cleavage occurred at this second site; however, we did not see additional cleavage product, even at longer incubation times (Fig. 1B). Future studies will address whether or not hepsin further cleaves Ln-332. In summary, the SQLR245 cleavage sequence, directly identified by N-terminal sequencing, appears to be a convincing argument that ties hepsin activity with Ln-332.
Among the laminin chains expressed in prostate, α1 (Ln-111) is expressed in fetal and newborn infants and is replaced in adults by α3 (Ln-332) and α5 (Ln-511/Ln-521) (14). Ln-511/Ln-521 and Ln-211 are present in normal gland and in prostate cancer. In contrast, Ln-332 is present in normal gland and lost in prostate cancer (14). Therefore, Ln-332 seems especially interesting as a substrate for hepsin during prostate cancer progression. It is to be noted that Ln-332 may be unique among laminins as a substrate for hepsin, since it is the only heterotrimer featuring the β3 chain. It is unlikely that the other laminin β chains, β1 and β2, are substrates for hepsin since, whereas homologous to β3, they do not contain sequences resembling the hepsin substrate sequence (SQLR↓LQGSCFC). However, in future studies, it will be interesting to determine experimentally whether other ECM macromolecules, including other laminins, may be substrates for hepsin.
It is intriguing to note that the hepsin cleavage site is located almost exactly at the predicted boundary between domains V and VI of the Ln-332 β3 chain (9). It is therefore possible that hepsin cleavage may release domain VI of β3. It has been reported that Ln-332 interacts with collagen VII via domain VI of the β3 chain (49, 50). Therefore, release of domain VI by hepsin could disrupt the interaction of Ln-332 with collagen VII. The consequences of this disruption may be at least 2-fold. First, β3 domain VI has a key role in the assembly of hemidesmosomes (51), integrin-based adhesion complexes that anchor epithelial cells to underlying tissue via Ln-332 and collagen VII (52, 53). It is therefore possible that hepsin may prevent or down-regulate hemidesmosome formation. Second, FNC1, the specific region of collagen VII that physically interacts with the Ln-332 β3 chain (50), was reported to promote tumor invasion in an Ln-332-dependent manner (54). These two previous observations suggest possible mechanisms whereby hepsin cleavage may play a physiological role in epithelial organization or a pathological role in tumor development. In this respect, it is worth noting that loss of hemidesmosomal complexes in prostate cancer was reported (55). These considerations warrant further studies to determine whether hepsin cleavage of Ln-332 β3 chain has an effect on the interaction of collagen VII with Ln-332.
We have also shown that hepsin cleavage produces a clear enhancement of prostate cancer cell migration on Ln-332 in vitro. DU145 cells exhibited significantly enhanced motility on hepsin-cleaved Ln-332 in transwell inserts, compared with untreated substrate. Further, hepsin-overexpressing LNCaP-34 cells also displayed significantly increased migration on Ln-332, compared with low hepsin-expressing LNCaP-17 cells. The basis for this enhancement remains to be elucidated. A possibility is that domain VI of Ln-332 may support “nonspecific” interactions with filters onto which migration assays were performed, thus effectively producing a cell-anchoring effect that may be released by hepsin. In any case, enhancement of migration fits well with the protumorigenic and proinvasive effects of hepsin in tumor animal models (37). These results may, in part, explain the findings of an orthotopic prostate cancer model, whereby LNCaP-34 tumors grew larger than LNCaP-17 tumors. Also, LNCaP-34 tumors showed 100% contralateral prostate invasion, which is not observed in animals with LnCaP-17 tumors (56).
Although hepsin overexpression has been linked to prostate cancer in many reports, the mechanism(s) by which it affects tumor progression have remained elusive (57). In a recent report by Klezovitch et al. (37), it was shown that specific overexpression of hepsin in mouse prostate epithelium did not cause changes in cell proliferation or differentiation but rather resulted in disorganized BM and weak or absent staining of Ln-332. Interestingly, these areas of disorganized BM coincided with areas of epithelial cells with the highest expression of hepsin, reinforcing a link between hepsin and Ln-332, a major component of BM. In the same report, hepsin transgenic mice, when crossed with the LPB-Tag 12T-7f mouse model of prostate cancer, caused faster progression of tumorigenesis, including bone metastasis, making it one of the very few mouse models of prostate cancer that develop bone metastasis. This report indicates that hepsin overexpression in itself is incapable of initiating tumorigenesis but can influence tumor progression. Our findings raise the possibility that one mechanism whereby hepsin may play this role is via its proteolytic activity on Ln-332. Experimental verification of this hypothesis could further elucidate the significance of BM integrity in tumor progression.
Further, many studies have independently shown that hepsin is consistently up-regulated in human prostate cancer (36, 58–65). For example, Stephan et al. (36) reported hepsin overexpression, as much as 10 times higher, in human prostate cancer tissue, compared with noncancerous tissue. Conversely, a number of studies have reported that expression of Ln-332 is down-regulated in human prostate cancer tissue (13–16). Taken together, these reports constitute circumstantial evidence that cleavage of Ln-332 by hepsin may occur in vivo. Defining exact ratios of hepsin/Ln-332 in tumor tissue quantitatively remains an important goal, albeit difficult, but based on these reports, we tentatively conclude that in human prostate cancer, the enzyme/substrate ratio is increased. In our in vitro experiments, Ln-332 cleavage by hepsin was detectable at an enzyme/substrate ratio of 1:1.5, although in order to maximize the yield of cleavage product, we generally used an enzyme/substrate ratio of 1:0.15. The need for this high ratio might be in part due to various reasons, including the multichain nature and high molecular size of Ln-332 (490 kDa), which may hinder the accessibility of cleavage sites in vitro. Other reports of cleavage of laminin by various MMPs have also used relatively high ratios in vitro (20, 21, 25, 27). In a similar study, Bair et al. (66) reported cleavage of Ln-511 by MT1-MMP at an enzyme/substrate ratio of 1:2. MT1-MMP, like hepsin, is a transmembrane cell surface protease. These authors speculated that, due to MT1-MMP spatial localization to the invading tumor front, the local ratio of Ln-511/MT1-MMP might approach 1:1 in vivo (66). Further, in vivo proteolysis may be favored by additional factors like temperature, pH, cations, and the relative topology of enzyme and substrate. In this respect, it is worth stressing the transmembrane location of the hepsin serine protease, since this topology places it in close vicinity to the BM (67). According to the structure of hepsin, its catalytic domain is extracellular and should lie flat on the plasma membrane (67) (i.e. in an ideal position to access BM substrates, such as Ln-332). In summary, our report should stimulate additional studies aimed at molecular mechanisms of interaction between epithelial cells and their immediate microenvironment, the BM.
Supplementary Material
Acknowledgments
We thank Dr. Myron Crawford, director of W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University, for protein sequencing and amino acid analyses. We thank Dr. David Friedman (Vanderbilt University Mass Spectrometry Research Center) for mass spectral analysis. Also, we thank Dr. Manju Bala (Department of Pharmacology, Vanderbilt University) for help with peptide analysis. We thank Drs. Lourdes Estrada, Kam Yoonseok, Jérôme Jourquin, Mohamed Hassanein, and Brandy Weidow (Department of Cancer Biology, Vanderbilt University) for helpful discussion and assistance with manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grants CA47858-17A2 and GM067221-03 (to V. Q.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: BM, basement membrane; ECM, extracellular matrix; EGR-cmk, Glu-Gly-Arg chloromethyl ketone; KD1, Kunitz domain-1; Ln, laminin; MALDI, matrix-assisted laser desorption/ionization; TOF, time-of-flight; MS, mass spectrometry; MMP, matrix metalloproteinase(s); pAb, polyclonal antibody; PBS, phosphate-buffered saline; EGR-hepsin, EGR-cmk-inactivated hepsin; Ctrl-hepsin, control hepsin for EGR-hepsin; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
References
- 1.Jemal, A., Siegel, R., Ward, E., Hao, Y., Xu, J., Murray, T., and Thun, M. J. (2008) CA-Cancer J. Clin. 58 71-96 [DOI] [PubMed] [Google Scholar]
- 2.Mundy, G. R. (2002) Nat. Rev. Cancer 2 584-593 [DOI] [PubMed] [Google Scholar]
- 3.Gupta, G. P., and Massague, J. (2006) Cell 127 679-695 [DOI] [PubMed] [Google Scholar]
- 4.Birkedal-Hansen, H. (1995) Curr. Opin. Cell Biol. 7 728-735 [DOI] [PubMed] [Google Scholar]
- 5.Woodward, J. K., Holen, I., Coleman, R. E., and Buttle, D. J. (2007) Bone 41 912-927 [DOI] [PubMed] [Google Scholar]
- 6.Noel, A., Gilles, C., Bajou, K., Devy, L., Kebers, F., Lewalle, J. M., Maquoi, E., Munaut, C., Remacle, A., and Foidart, J. M. (1997) Invasion Metastasis 17 221-239 [PubMed] [Google Scholar]
- 7.Quaranta, V., and Giannelli, G. (2003) Tumori. 89 343-348 [DOI] [PubMed] [Google Scholar]
- 8.Rousselle, P., Lunstrum, G. P., Keene, D. R., and Burgeson, R. E. (1991) J. Cell Biol. 114 567-576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aumailley, M., Bruckner-Tuderman, L., Carter, W. G., Deutzmann, R., Edgar, D., Ekblom, P., Engel, J., Engvall, E., Hohenester, E., Jones, J. C., Kleinman, H. K., Marinkovich, M. P., Martin, G. R., Mayer, U., Meneguzzi, G., Miner, J. H., Miyazaki, K., Patarroyo, M., Paulsson, M., Quaranta, V., Sanes, J. R., Sasaki, T., Sekiguchi, K., Sorokin, L. M., Talts, J. F., Tryggvason, K., Uitto, J., Virtanen, I., von der Mark, K., Wewer, U. M., Yamada, Y., and Yurchenco, P. D. (2005) Matrix Biol. 24 326-332 [DOI] [PubMed] [Google Scholar]
- 10.Meneguzzi, G., Marinkovich, M. P., Aberdam, D., Pisani, A., Burgeson, R., and Ortonne, J. P. (1992) Exp. Dermatol. 1 221-229 [DOI] [PubMed] [Google Scholar]
- 11.Ryan, M. C., Christiano, A. M., Engvall, E., Wewer, U. M., Miner, J. H., Sanes, J. R., and Burgeson, R. E. (1996) Matrix Biol. 15 369-381 [DOI] [PubMed] [Google Scholar]
- 12.Marinkovich, M. P. (2007) Nat. Rev. Cancer 7 370-380 [DOI] [PubMed] [Google Scholar]
- 13.Hao, J., Jackson, L., Calaluce, R., McDaniel, K., Dalkin, B. L., and Nagle, R. B. (2001) Am. J. Pathol. 158 1129-1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagle, R. B. (2004) J. Cell. Biochem. 91 36-40 [DOI] [PubMed] [Google Scholar]
- 15.Calaluce, R., Beck, S. K., Bair, E. L., Pandey, R., Greer, K. A., Hoying, A. M., Hoying, J. B., Mount, D. W., and Nagle, R. B. (2006) Prostate 66 1381-1390 [DOI] [PubMed] [Google Scholar]
- 16.Davis, T. L., Cress, A. E., Dalkin, B. L., and Nagle, R. B. (2001) Prostate 46 240-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schenk, S., and Quaranta, V. (2003) Trends Cell Biol. 13 366-375 [DOI] [PubMed] [Google Scholar]
- 18.Hintermann, E., and Quaranta, V. (2004) Matrix. Biol. 23 75-85 [DOI] [PubMed] [Google Scholar]
- 19.Koshikawa, N., Schenk, S., Moeckel, G., Sharabi, A., Miyazaki, K., Gardner, H., Zent, R., and Quaranta, V. (2004) FASEB J. 18 364-366 [DOI] [PubMed] [Google Scholar]
- 20.Giannelli, G., Falk-Marzillier, J., Schiraldi, O., Stetler-Stevenson, W. G., and Quaranta, V. (1997) Science 277 225-228 [DOI] [PubMed] [Google Scholar]
- 21.Pirila, E., Sharabi, A., Salo, T., Quaranta, V., Tu, H., Heljasvaara, R., Koshikawa, N., Sorsa, T., and Maisi, P. (2003) Biochem. Biophys. Res. Commun. 303 1012-1017 [DOI] [PubMed] [Google Scholar]
- 22.Wang, B., Sun, J., Kitamoto, S., Yang, M., Grubb, A., Chapman, H. A., Kalluri, R., and Shi, G. P. (2006) J. Biol. Chem. 281 6020-6029 [DOI] [PubMed] [Google Scholar]
- 23.Veitch, D. P., Nokelainen, P., McGowan, K. A., Nguyen, T. T., Nguyen, N. E., Stephenson, R., Pappano, W. N., Keene, D. R., Spong, S. M., Greenspan, D. S., Findell, P. R., and Marinkovich, M. P. (2003) J. Biol. Chem. 278 15661-15668 [DOI] [PubMed] [Google Scholar]
- 24.Amano, S., Scott, I. C., Takahara, K., Koch, M., Champliaud, M. F., Gerecke, D. R., Keene, D. R., Hudson, D. L., Nishiyama, T., Lee, S., Greenspan, D. S., and Burgeson, R. E. (2000) J. Biol. Chem. 275 22728-22735 [DOI] [PubMed] [Google Scholar]
- 25.Mydel, P., Shipley, J. M., Adair-Kirk, T. L., Kelley, D. G., Broekelmann, T. J., Mecham, R. P., and Senior, R. M. (2008) J. Biol. Chem. 283 9513-9522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Udayakumar, T. S., Chen, M. L., Bair, E. L., Von Bredow, D. C., Cress, A. E., Nagle, R. B., and Bowden, G. T. (2003) Cancer Res. 63 2292-2299 [PubMed] [Google Scholar]
- 27.Remy, L., Trespeuch, C., Bachy, S., Scoazec, J. Y., and Rousselle, P. (2006) Cancer Res. 66 11228-11237 [DOI] [PubMed] [Google Scholar]
- 28.Nakashima, Y., Kariya, Y., Yasuda, C., and Miyazaki, K. (2005) J. Biochem. (Tokyo) 138 539-552 [DOI] [PubMed] [Google Scholar]
- 29.Morrissey, C., True, L. D., Roudier, M. P., Coleman, I. M., Hawley, S., Nelson, P. S., Coleman, R., Wang, Y. C., Corey, E., Lange, P. H., Higano, C. S., and Vessella, R. L. (2008) Clin. Exp. Metastasis 25 377-388 [DOI] [PubMed] [Google Scholar]
- 30.Narla, G., Heath, K. E., Reeves, H. L., Li, D., Giono, L. E., Kimmelman, A. C., Glucksman, M. J., Narla, J., Eng, F. J., Chan, A. M., Ferrari, A. C., Martignetti, J. A., and Friedman, S. L. (2001) Science 294 2563-2566 [DOI] [PubMed] [Google Scholar]
- 31.Wang, S. I., Parsons, R., and Ittmann, M. (1998) Clin. Cancer Res. 4 811-815 [PubMed] [Google Scholar]
- 32.Jenkins, R. B., Qian, J., Lieber, M. M., and Bostwick, D. G. (1997) Cancer Res. 57 524-531 [PubMed] [Google Scholar]
- 33.MacGrogan, D., and Bookstein, R. (1997) Semin. Cancer Biol. 8 11-19 [DOI] [PubMed] [Google Scholar]
- 34.Turner, D. P., and Watson, D. K. (2008) Expert Rev. Anticancer Ther. 8 33-42 [DOI] [PubMed] [Google Scholar]
- 35.Landers, K. A., Burger, M. J., Tebay, M. A., Purdie, D. M., Scells, B., Samaratunga, H., Lavin, M. F., and Gardiner, R. A. (2005) Int. J. Cancer 114 950-956 [DOI] [PubMed] [Google Scholar]
- 36.Stephan, C., Yousef, G. M., Scorilas, A., Jung, K., Jung, M., Kristiansen, G., Hauptmann, S., Kishi, T., Nakamura, T., Loening, S. A., and Diamandis, E. P. (2004) J. Urol. 171 187-191 [DOI] [PubMed] [Google Scholar]
- 37.Klezovitch, O., Chevillet, J., Mirosevich, J., Roberts, R. L., Matusik, R. J., and Vasioukhin, V. (2004) Cancer Cell 6 185-195 [DOI] [PubMed] [Google Scholar]
- 38.Wu, Q., and Parry, G. (2007) Front. Biosci. 12 5052-5059 [DOI] [PubMed] [Google Scholar]
- 39.Kazama, Y., Hamamoto, T., Foster, D. C., and Kisiel, W. (1995) J. Biol. Chem. 270 66-72 [DOI] [PubMed] [Google Scholar]
- 40.Moran, P., Li, W., Fan, B., Vij, R., Eigenbrot, C., and Kirchhofer, D. (2006) J. Biol. Chem. 281 30439-30446 [DOI] [PubMed] [Google Scholar]
- 41.Kirchhofer, D., Peek, M., Lipari, M. T., Billeci, K., Fan, B., and Moran, P. (2005) FEBS Lett. 579 1945-1950 [DOI] [PubMed] [Google Scholar]
- 42.Xuan, J. A., Schneider, D., Toy, P., Lin, R., Newton, A., Zhu, Y., Finster, S., Vogel, D., Mintzer, B., Dinter, H., Light, D., Parry, R., Polokoff, M., Whitlow, M., Wu, Q., and Parry, G. (2006) Cancer Res. 66 3611-3619 [DOI] [PubMed] [Google Scholar]
- 43.Falk-Marzillier, J., Domanico, S. Z., Pelletier, A., Mullen, L., and Quaranta, V. (1998) Biochem. Biophys. Res. Commun. 251 49-55 [DOI] [PubMed] [Google Scholar]
- 44.Plopper, G., Falk-Marzillier, J., Glaser, S., Fitchmun, M., Giannelli, G., Romano, T., Jones, J. C., and Quaranta, V. (1996) J. Cell Sci. 109, 1965-1973 [DOI] [PubMed] [Google Scholar]
- 45.Shia, S., Stamos, J., Kirchhofer, D., Fan, B., Wu, J., Corpuz, R. T., Santell, L., Lazarus, R. A., and Eigenbrot, C. (2005) J. Mol. Biol. 346 1335-1349 [DOI] [PubMed] [Google Scholar]
- 46.Zapatka, M., Zboralski, D., Radacz, Y., Bockmann, M., Arnold, C., Schoneck, A., Hoppe, S., Tannapfel, A., Schmiegel, W., Simon-Assmann, P., and Schwarte-Waldhoff, I. (2007) Oncogene 26 1417-1427 [DOI] [PubMed] [Google Scholar]
- 47.Yurchenco, P. D., and Wadsworth, W. G. (2004) Curr. Opin. Cell Biol. 16 572-579 [DOI] [PubMed] [Google Scholar]
- 48.Herter, S., Piper, D. E., Aaron, W., Gabriele, T., Cutler, G., Cao, P., Bhatt, A. S., Choe, Y., Craik, C. S., Walker, N., Meininger, D., Hoey, T., and Austin, R. J. (2005) Biochem. J. 390 125-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brittingham, R., Uitto, J., and Fertala, A. (2006) Biochem. Biophys. Res. Commun. 343 692-699 [DOI] [PubMed] [Google Scholar]
- 50.Chen, M., Marinkovich, M. P., Jones, J. C., O'Toole, E. A., Li, Y. Y., and Woodley, D. T. (1999) J. Invest. Dermatol. 112 177-183 [DOI] [PubMed] [Google Scholar]
- 51.Waterman, E. A., Sakai, N., Nguyen, N. T., Horst, B. A., Veitch, D. P., Dey, C. N., Ortiz-Urda, S., Khavari, P. A., and Marinkovich, M. P. (2007) Cancer Res. 67 4264-4270 [DOI] [PubMed] [Google Scholar]
- 52.Jones, J. C., Hopkinson, S. B., and Goldfinger, L. E. (1998) BioEssays 20 488-494 [DOI] [PubMed] [Google Scholar]
- 53.Litjens, S. H., de Pereda, J. M., and Sonnenberg, A. (2006) Trends Cell Biol. 16 376-383 [DOI] [PubMed] [Google Scholar]
- 54.Ortiz-Urda, S., Garcia, J., Green, C. L., Chen, L., Lin, Q., Veitch, D. P., Sakai, L. Y., Lee, H., Marinkovich, M. P., and Khavari, P. A. (2005) Science 307 1773-1776 [DOI] [PubMed] [Google Scholar]
- 55.Nagle, R. B., Hao, J., Knox, J. D., Dalkin, B. L., Clark, V., and Cress, A. E. (1995) Am. J. Pathol. 146 1498-1507 [PMC free article] [PubMed] [Google Scholar]
- 56.Li, W., Wang, B.-E., Gao, W.-Q., Gogineni, A., Cole, M., DeGuzman, L., Bunting, S., and Kirchhofer, D. (2007) The American Association for Cancer Research Annual Meeting, April 14–18, 2007, AACR, Los Angeles, CA, Abstr. 3096
- 57.Vasoiukhin, V. (2004) Cell Cycle 3 1394-1397 [DOI] [PubMed] [Google Scholar]
- 58.Luo, J., Duggan, D. J., Chen, Y., Sauvageot, J., Ewing, C. M., Bittner, M. L., Trent, J. M., and Isaacs, W. B. (2001) Cancer Res. 61 4683-4688 [PubMed] [Google Scholar]
- 59.Halvorsen, O. J., Oyan, A. M., Bo, T. H., Olsen, S., Rostad, K., Haukaas, S. A., Bakke, A. M., Marzolf, B., Dimitrov, K., Stordrange, L., Lin, B., Jonassen, I., Hood, L., Akslen, L. A., and Kalland, K. H. (2005) Int. J. Oncol. 26 329-336 [PubMed] [Google Scholar]
- 60.Dhanasekaran, S. M., Barrette, T. R., Ghosh, D., Shah, R., Varambally, S., Kurachi, K., Pienta, K. J., Rubin, M. A., and Chinnaiyan, A. M. (2001) Nature 412 822-826 [DOI] [PubMed] [Google Scholar]
- 61.Magee, J. A., Araki, T., Patil, S., Ehrig, T., True, L., Humphrey, P. A., Catalona, W. J., Watson, M. A., and Milbrandt, J. (2001) Cancer Res. 61 5692-5696 [PubMed] [Google Scholar]
- 62.Chen, Z., Fan, Z., McNeal, J. E., Nolley, R., Caldwell, M. C., Mahadevappa, M., Zhang, Z., Warrington, J. A., and Stamey, T. A. (2003) J. Urol. 169 1316-1319 [DOI] [PubMed] [Google Scholar]
- 63.Ernst, T., Hergenhahn, M., Kenzelmann, M., Cohen, C. D., Bonrouhi, M., Weninger, A., Klaren, R., Grone, E. F., Wiesel, M., Gudemann, C., Kuster, J., Schott, W., Staehler, G., Kretzler, M., Hollstein, M., and Grone, H. J. (2002) Am. J. Pathol. 160 2169-2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lai, Y., Wu, B., Chen, L., and Zhao, H. (2004) Bioinformatics 20 3146-3155 [DOI] [PubMed] [Google Scholar]
- 65.Singh, D., Febbo, P. G., Ross, K., Jackson, D. G., Manola, J., Ladd, C., Tamayo, P., Renshaw, A. A., D'Amico, A. V., Richie, J. P., Lander, E. S., Loda, M., Kantoff, P. W., Golub, T. R., and Sellers, W. R. (2002) Cancer Cell 1 203-209 [DOI] [PubMed] [Google Scholar]
- 66.Bair, E. L., Chen, M. L., McDaniel, K., Sekiguchi, K., Cress, A. E., Nagle, R. B., and Bowden, G. T. (2005) Neoplasia 7 380-389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Somoza, J. R., Ho, J. D., Luong, C., Ghate, M., Sprengeler, P. A., Mortara, K., Shrader, W. D., Sperandio, D., Chan, H., McGrath, M. E., and Katz, B. A. (2003) Structure 11 1123-1131 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.