Abstract
The G-protein-coupled receptor (GPCR) GPR54 is essential for the development and maintenance of reproductive function in mammals. A point mutation (L148S) in the second intracellular loop (IL2) of GPR54 causes idiopathic hypogonadotropic hypogonadism, a disorder characterized by delayed puberty and infertility. Here, we characterize the molecular mechanism by which the L148S mutation causes disease and address the role of IL2 in Class A GPCR function. Biochemical, immunocytochemical, and pharmacological analysis demonstrates that the mutation does not affect the expression, ligand binding properties, or protein interaction network of GPR54. In contrast, diverse GPR54 functional responses are markedly inhibited by the L148S mutation. Importantly, the leucine residue at this position is highly conserved among class A GPCRs. Indeed, mutating the corresponding leucine of the α1A-AR recapitulates the effects observed with L148S GPR54, suggesting the critical importance of this hydrophobic IL2 residue for Class A GPCR functional coupling. Interestingly, co-immunoprecipitation studies indicate that L148S does not hinder the association of Gα subunits with GPR54. However, fluorescence resonance energy transfer analysis strongly suggests that L148S impairs the ligand-induced catalytic activation of Gα. Combining our data with a predictive Class A GPCR/Gα model suggests that IL2 domains contain a conserved hydrophobic motif that, upon agonist stimulation, might stabilize the switch II region of Gα. Such an interaction could promote opening of switch II of Gα to facilitate GDP-GTP exchange and coupling to downstream signaling responses. Importantly, mutations that disrupt this key hydrophobic interface can manifest as human disease.
A diverse network of signaling pathways have evolved within the hypothalamic-pituitary-gonadal axis to ensure precise neuroendocrine regulation of reproductive function in mammals (1). An essential feature of this physiological system is the pulsatile release of gonadotropin-releasing hormone from hypothalamic neurons, which subsequently initiates follicle-stimulating hormone and luteinizing hormone release from the pituitary and ultimately impinges on the gonads to elicit sex steroid secretion (2). Together, the components of the hypothalamic-pituitary-gonadal axis function with precise temporal and spatial accuracy to regulate the development and maintenance of proper reproductive function, including puberty onset and the estrous cycle (3). Thus, functional mutations in key elements of this critical physiological system can result in the development of various reproductive disorders. For example, idiopathic hypogonadotropic hypogonadism (IHH),2 which is characterized by delayed or absent puberty, immature reproductive organs, low levels of sex steroids and infertility, is commonly associated with loss-of-function mutations in the gonadotropin-releasing hormone receptor (4, 5). More recently, IHH-causing mutations were identified in a relatively uncharacterized orphan G-protein-coupled receptor (GPCR), GPR54 (6-8). GPR54 subsequently emerged as a novel gatekeeper of the reproductive cascade that initiates puberty. Myriad animal studies have demonstrated that engagement of GPR54 by endogenous peptide ligands, termed kisspeptins, potently stimulates gonadotropin-releasing hormone release from hypothalamic neurons to activate the hypothalamic-pituitary-gonadal axis (7, 9-12). Furthermore, the characterization of GPR54 KO mice, which phenocopy the human condition of IHH, confirmed the essential role of GPR54 for reproductive function (7, 13).
In an elegant study, Seminara et al. (7) demonstrated that a single leucine to serine (L148S) mutation in the second intracellular loop (IL2) of GPR54 causes autosomal recessive IHH in humans. At the physiochemical level, the L148S mutation results in the substitution of a hydrophobic with a hydrophilic amino acid in the central portion of the IL2 of GPR54. Although the L148S GPR54 mutant displayed diminished functional responses in vitro, whether these deficits were due to improper expression, trafficking to the plasma membrane, or ligand-binding capacity was not examined (7) and remains an important unexplored question.
A number of previous reports utilizing site-directed or alanine-scanning mutagenesis approaches have suggested that the IL2 is an important determinant for the functional coupling of Class A GPCRs (14-19), yet in vivo evidence to substantiate this hypothesis is limited (20). Numerous studies have also suggested that the third intracellular loop (IL3) of GPCRs is paramount for G-protein activation (21-25), establishing the likelihood that a multidomain GPCR-G-protein interface is required for G-protein activation. Unfortunately, however, in the absence of a crystal structure of a GPCR in complex with a G-protein, the specific sites that comprise the GPCR-G-protein interface remain relatively undefined. Thus, enhancing our understanding of the exact mechanism by which agonist binding to a GPCR engages G-proteins and activates intracellular signaling cascades remains one of the most elusive and intriguing areas of research in the GPCR field.
Herein, we utilize biochemical and pharmacological techniques to elucidate the molecular mechanism by which the clinically relevant L148S mutation of GPR54 causes disease and assess the importance of this IL2 residue for proper Class A GPCR-G-protein coupling. Specifically, we have developed an in vitro model to ascertain whether the L148S mutation causes defects in the expression, trafficking, or signaling and/or alters the protein interaction network of GPR54. Importantly, characterization of L148S hGPR54 revealed that conserved residues in the IL2 of Class A GPCRs are essential for functional interactions between the GPCR and G-protein, specifically Gα. Docking analysis of Gα to the recently solved β2-adrenergic receptor (β2-AR) crystal structure (26) predicts a molecular model whereby hydrophobic interactions between the IL2 of Class A GPCRs and conserved residues of Gα subunits stabilize the switch II region of activated Gα in a conformation that facilitates GDP-GTP exchange and thus maximizes downstream effector signaling. Thus, the IL2 of Class A GPCRs could act as a guanine nucleotide exchange factor (GEF) during agonist-mediated activation of Gα.
EXPERIMENTAL PROCEDURES
Constructs—cDNA for human GPR54 (hGPR54) in pEFIN3 was kindly provided by Dr. Marc Parmentier (University of Brussels). hGPR54 was amplified by PCR to add 5′ EcoRI and 3′ BamHI sites for subcloning into pcDNA3.1 containing an N-terminal FLAG or hemagglutinin (HA) epitope tag. 5′ BamHI and 3′ XhoI sites were added using PCR to subclone hGPR54 into pEGFP-N3 containing a C-terminal GFP epitope tag. QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA) was used to introduce the L148S mutation into hGPR54 and L132S mutation into the human α1A-AR and to create various Leu148 amino acid substitutions. Gα15/16 and Gαq in pcDNA3.1 were obtained from the UMR cDNA Resource Center (available on the World Wide Web), and an N-terminal HA tag was added to Gα15/16 to facilitate immunodetection. cDNA for mouse Gαq-eCFP, bovine eYFP-Gβ1, and bovine GRK2 were kindly provided by Bertil Hille (University of Washington).
Cell Culture and Transfection—Human embryonic kidney (HEK) 293 cells were propagated in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 100 μg/ml streptomycin, and 100 units/ml penicillin at 37 °C in 5% CO2. Plates were grown to ∼70% confluence for transfection. cDNA constructs were transfected using Lipofectamine 2000 (Invitrogen), and cells were used for experimentation 24-48 h post-transfection. Cells were treated with 400 μg/ml Geneticin (Invitrogen) to generate polyclonal stable cell lines.
Tandem Affinity Purification and Mass Spectrometry—Tandem affinity purification (TAP) was conducted as previously described with minor modifications (27, 28). The expression vector was designed to position tandem streptavidin-binding protein, HA, and calmodulin-binding protein affinity tags at the N terminus of GPR54. Stable HEK293 cell lines expressing wild type (WT) or L148S hGPR54 were lysed overnight in buffer containing 1% digitonin. Solubilized protein was incubated with streptavidin-Sepharose, washed, and eluted with 50 mm d-biotin. Eluate was incubated with calmodulin-Sepharose, washed, and eluted with 50 mm ammonium bicarbonate (pH 8.0) plus 25 mm EDTA. Final eluate was analyzed via mass spectrometry (LTQ-FT or LTQ Orbitrap). We used a PeptideProphet™ score of greater than 0.65 to identify positive hits from the TAP/MS results. This cut-off provided a sensitivity of 0.744 and 0.737 for WT hGPR54 and L148S hGPR54, respectively, and an error rate of 0.074 and 0.064 for WT hGPR54 and L148S hGPR54, respectively.
Confocal Microscopy—HEK293 cells stably expressing GFP-tagged hGPR54 constructs were split onto 35-mm dishes containing 25-mm glass coverslips and allowed to adhere overnight. Cells were washed with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde in 0.1 m phosphate buffer for 30 min at room temperature. Cells were washed three times with PBS containing 4% horse serum, stained for 2 min with 300 nm 4′,6-diamidino-2-phenylindole/PBS (Invitrogen), washed again with PBS, and mounted onto slides. Cells were imaged with a Zeiss LSM 510 laser-scanning confocal microscope. GFP was excited using an argon laser (λ = 488 nm) and detected at 510-520 nm.
GFP Immunoprecipitation/Immunoblotting—Confluent 150-mm plates of HEK293 cells were washed with PBS, scraped, and centrifuged at 5,000 × g at 4 °C for 5 min. Pellets were resuspended in 0.5 ml of lysis buffer (50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1% Triton-X) and passed through a 20-gauge needle 10 times at 4 °C. Samples were rotated at 4 °C for 15 min and then centrifuged at 10,000 × g for 15 min. Proteins concentrations were determined using a BCA assay (Pierce). Cell lysates were then incubated with mouse anti-GFP antibody (1 mg of cell lysate per 2 μg of antibody) (ab1218; Abcam, Cambridge, MA) in cell lysis buffer without detergent overnight at 4 °C. Protein G-Sepharose beads were then added to the mixture and incubated for 4 h at 4 °C. To pellet beads, samples were centrifuged at 500 × g, and supernatants were collected, resuspended in 50 μl laurel dodecyl sulfate sample buffer, and heated to 95 °C for 5 min. Samples were then run on 10% BisTris gels, transferred to nitrocellulose, and probed with a rabbit anti-GFP antibody (ab6556; Abcam) in 3% milk/TBST. Blots were washed with TBST and probed with IRDye® 800CW goat α-rabbit IgG (LI-COR, Lincoln, NB). Bands were detected by fluorescence at 680 and 800 nm using the Odyssey Detection System.
Radioligand Binding—Confluent 150-mm plates of HEK293 cells were scraped into PBS at 4 °C, subjected to Polytron homogenization, and centrifuged at 15,000 × g for 20 min. Pellets were resuspended in PBS, rehomogenized, and recentrifuged at 15,000 × g. Protein concentrations were determined using a BCA assay (Pierce). For hGPR54 radioligand binding assays, samples were incubated with increasing concentrations of 125I-labeled KP-10 (PerkinElmer Life Sciences), a synthetic full agonist for GPR54, in the absence (total bound) or presence (nonspecific bound) of 0.5 μm unlabeled KP-10. For α1A-AR radioligand binding assays, samples were incubated with increasing concentrations of [3H]prazosin (PerkinElmer Life Sciences) in the absence (total bound) or presence (nonspecific bound) of 10 μm phentolamine. Samples were incubated at 37 °C with gentle shaking. Bound drug was determined by passing samples through a Brandel Cell Harvester (Brandel, Gaithersburg, MD) containing glass fiber filter paper. For [3H]prazosin, 5 ml of liquid scintillation fluid was added per sample. Samples were counted, and specific bound drug was determined by subtracting nonspecific from total bound. Data were then analyzed using GraphPad Prism software (GraphPad, San Diego, CA) to calculate KD and Bmax values using nonlinear regression.
[3H]Phosphoinositol Hydrolysis—HEK293 cells were split into 24-well plates (1 million cells/well) and incubated with 1 μCi of myo-[3H]inositol. 24 h later, the medium was aspirated and replaced with 1 ml of Krebs buffer (129 mm NaCl, 5.5 mm KCl, 2.5 mm CaCl2, 1.2 mm NaH2PO4, 1.2 mm MgCl2, 20 mm NaHCO3, 11 mm glucose, 0.029 mm Na2EDTA) containing 10 mm LiCl. Cells were incubated with or without agonist for 1 h, and reactions were stopped by aspirating Krebs buffer and adding 750 μl of ice-cold methanol. 100 μm Kisspeptin (Phoenix Pharmaceuticals, Burlingame, CA) stocks were prepared in sterile PBS. Samples were scraped into 500 μl of chloroform, ultrasonicated, and separated using anion exchange chromatography. Total [3H]inositol phosphates produced were quantified using a Beckman liquid scintillation counter. Data were analyzed using GraphPad Prism software (GraphPad) to calculate agonist EC50 values and intrinsic activities.
ERK1/2 Activation—The protocol previously described in Ref. 29 was used with minor modifications. HEK293 cells were seeded in 96-well plates at ∼150,000 cells/well. Cells were serum-starved for 4 h to reduce background ERK1/2 activation, and agonists were then added to wells and incubated at 37 °C for the indicated times. To terminate the reaction, plates were put on ice, medium was aspirated, and cells were fixed with 4% paraformaldeyde at room temperature for 20 min. Cells were then washed with PBS containing 0.1% Triton-X and probed with anti-phospho-ERK1/2 and anti-total ERK1/2 antibodies (Cell Signaling, Danvers, MA) overnight at 4 °C. Cells were then washed with PBS plus 0.1% Tween 20 buffer and incubated for 1 h with IRDye®800CW- and IRDye®680-conjugated second antibodies (LiCor Biotechnology, Lincoln, NE). Cells were then washed three times with PBS, and the PBS was removed by gently tapping the plate upside down onto a paper towel. Plates were imaged using the Odyssey Detection System, and GraphPad Prism software was used to quantify data, which are expressed as percentage of the epidermal growth factor maximal response.
Fluorescence-activated Cell Sorting Analysis—Approximately 200,000 cells were washed and pelleted in PBS plus 2% BSA (PBS+) in 5-ml polystyrene round bottom test tubes (BD Falcon, San Jose, CA). 100 μl of PBS+ with primary antibody (5 μg/ml; catalog number 12CA5; Roche Applied Science) was added to each tube, and cells were dislodged by gentle flicking. Cells were incubated for 1 h at 4 °C, pelleted, washed three times in PBS+, resuspended in secondary antibody at 1 μg/ml (Invitrogen GAM Alexa Fluor®), and incubated for 30 min at 4 °C. Cells were then pelleted and washed three times in PBS+. Fluorescence intensity was measured using an LSRII flow cytometer (BD Biosciences). As a negative control, primary antibody was omitted.
Structural Modeling—Limited structural information on the β2-AR IL3 led us to use the TASSER-generated structural model of GPR54 (30) for initial docking of the crystal structure of the activated Gαq subunit (Protein Data Bank code 2BCJ). We used the RosettaDock protein-protein docking server for docking procedures (31). The GPR54 computational model was refined and replaced using the crystal structure of the β2-AR (Protein Data Bank code 2RH1).
Co-immunoprecipitation of GPR54 and Gq Family Members—150-mm plates of HEK293 cells were transiently transfected with TAP-WT or L148S hGPR54 and Gαq or Gα15/16. After 24 h, cells were harvested, lysed with TAP buffer, cleared, and incubated overnight with streptavidin-Sepharose. Beads were washed with TAP buffer minus digitonin and boiled in 50 μl of laurel dodecyl sulfate sample buffer. Immunoblotting was performed as previously described with anti-Gq (1:1000; catalog number G612704; BD Biosciences) and anti-HA (1:1000; Cell Signaling) antibodies.
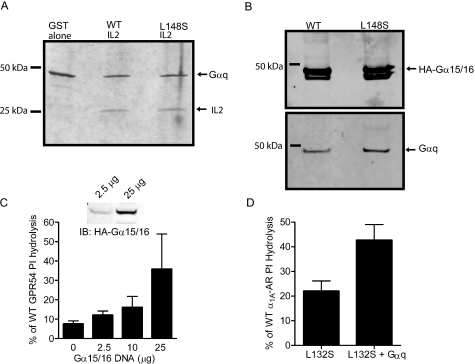
Co-purification of GPR54 IL2 and Gαq—An overlapping primer strategy was used to clone the WT and L148S hGPR54 IL2 into a pGEX-2T-based, N-terminal GST expression vector. Gαq was subcloned into the pAL vector for the expression of N-terminal His6-Gαq fusion protein. The IL2 and Gαq fusions were co-transformed into Escherichia coli. A colony was picked and grown in 1 liter of Luria broth at 37 °C. isopropyl 1-thio-β-d-galactopyranoside (0.02 m) was added to the cell culture when the A600 reached 1.0, and culture was continued at 18 °C overnight. Cells were harvested and suspended in His lysis buffer (20 mm Tris, pH 8.0, 300 mm NaCl, 2 mm imidazole, 3 mm β-mercaptoethanol, and protease inhibitors (Roche Applied Science)). After sonication and centrifugation, the supernatant was loaded on a 2-ml Ni2+-nitrilotriacetic acid column (Qiagen, Valencia, CA). Bound His-Gαq was eluted using 200 mm imidazole. The eluted proteins were run on a 12% SDS-PAGE and stained with Coomassie Blue. To determine the strength of the interaction, the column was washed with increasing amounts of NaCl (300-900 mm).
Fluorescence Resonance Energy Transfer (FRET)—To measure ratiometric FRET between CFP and YFP, we performed photometric measurements on single cells excited by a 75-watt xenon arc lamp and measured on an inverted Nikon diaphot microscope. Excitation light was filtered by a 0.3 neutral density filter and a cube containing a 436 ± 10-nm bandpass filter and a 460-nm dichroic mirror. Emitted light was separated by two cubes in series; a 505-nm dichroic mirror with a 480 ± 15-nm bandpass filter directed light to one photomultiplier tube (“short wavelength channel”), and a 570-nm dichroic mirror with a 535 ± 12.5-nm bandpass filter directed light to the other photomultiplier tube (“long wavelength channel”). The shutter was opened for 24 ms every 500 ms. Solution was exchanged using a θ tube moved by a step-driven motor and was complete within 100 ms. The FRET ratio was taken as the ratio of YFP to CFP emission after single-fluorophore corrections for background fluorescence, bleed-through, and direct excitation. FRET ratio = FYFP/FCFP = 0.98 (LW - 0.17®SW)/SW, where SW and LW are counts in the short and long wavelength channels. Changes in FRET ratio provide a good measure of interaction kinetics. To check membrane expression, cells were imaged using a Zeiss SP1 confocal microscope with a 100× oil immersion objective.
RESULTS
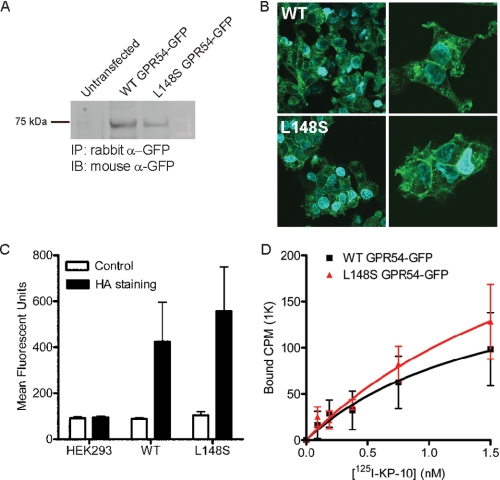
L148S hGPR54 Expression, Localization, and Ligand Binding Are Equivalent to Those of WT hGPR54—To address the mechanism by which a single L148S amino acid conversion results in GPR54 loss of function, we compared the expression, localization, and pharmacological characteristics of L148S and WT GPR54. We thus generated polyclonal HEK293 cell lines stably expressing WT or L148S mutant human GPR54 (L148S hGPR54) with either N-terminal (HA or FLAG) or C-terminal (GFP) epitope tags. Immunoprecipitation of WT and L148S hGPR54-GFP showed that both WT and mutant receptors were expressed at the expected molecular mass of ∼70 kDa, demonstrating that the mutation does not interfere with protein expression (Fig. 1A). Confocal analysis revealed that WT and L148S hGPR54-GFP cellular localization patterns were indistinguishable, suggesting that the L148S mutation does not cause trafficking or expression deficits of GPR54 (Fig. 1B). To obtain a quantitative measurement of plasma membrane expression, we employed fluorescence-activated cell sorting analysis to detect the N-terminal HA tag of WT and L148S hGPR54. As shown in Fig. 1C, the cell surface expression of L148S HA-hGPR54 was statistically equivalent to WT HA-hGPR54 (Fig. 1C), suggesting that this mutation does not prevent GPR54 from reaching the plasma membrane in vitro.
FIGURE 1.
The L148S mutation does not alter the expression, localization, or ligand-binding properties of hGPR54. A, immunoprecipitation of WT and L148S hGPR54-GFP. B, confocal microscopy showing the localization of WT and L148S hGPR54-GFP. C, cell surface expression levels of WT and L148S HA-hGPR54 measured using fluorescence-activated cell sorting analysis. D, saturation binding of 125I-labeled KP-10 to membranes from WT and L148S hGPR54 expressing HEK293 cells. The results presented in A and B are representative of three experiments performed in duplicate, and C and D show the means ± S.E. of three or four experiments performed in triplicate. IP, immunoprecipitation; IB, immunoblot.
To address the potential effects of this mutation on ligand binding, we performed 125I-labeled KP-10 saturation radioligand binding assays on HEK293 cells stably expressing WT and L148S hGPR54. KP-10 is a synthetic 10-amino acid peptide that displays full agonist properties for GPR54. Saturation analysis demonstrated that L148S hGPR54 bound 125I-labeled KP-10 in a manner indistinguishable from the WT receptor (Fig. 1D). A single, high affinity binding site with comparable equilibrium dissociation constants was identified in cells expressing both WT (KD = 1.6 nm) and L148S hGPR54 (KD = 2.5 nm). Collectively, data obtained with multiple, independent techniques demonstrate that L148S GPR54 loss-of-function cannot be accounted for by defects in receptor expression, trafficking, or ligand-binding properties.
The L148S Mutation Does Not Alter the Protein Interaction Network of GPR54—GPCRs are now recognized to exist as multiprotein complexes composed of GPCR-interacting proteins (GIPs) that impart precise spatial and temporal regulation of expression, trafficking, ligand binding, and signaling (32). We hypothesized that alterations in the protein interaction network of GPR54 caused by the L148S mutation might result in the clinical manifestation of IHH. To address this question, we used a TAP strategy to isolate GPR54 protein complexes from mammalian cells in combination with mass spectrometry (TAP/MS). We confirmed the functional activity of TAP-WT hGPR54 in stable HEK293 cell lines by measuring phosphoinositol (PI) hydrolysis in response to a saturating dose of kisspeptin (data not shown). TAP/MS data of isolated WT and L148S hGPR54 complexes identified both previously suspected and novel interacting proteins involved in signal transduction, protein quality control, trafficking, and transcriptional regulation (selected GIPs presented in Table 1). Importantly, direct comparison of WT and L148S hits suggested that the mutation did not alter the protein interactome of GPR54. To perform a functional comparison, we created a study set of all identified WT and L148S GPR54 GIPs using Gene Ontology annotations to proteins in the UniProt Knowledgebase and performed an automated classification to assign WT and L148S hGPR54 GIPs into categories for cellular localization, involvement in a biological process, and molecular function. Functional comparison of WT and L148S hGPR54 clearly demonstrated equivalent GIP profiles (Fig. S1), suggesting that the L148S mutation does not significantly alter the proteomic network of GPR54.
TABLE 1.
Proteins associated with WT and L148S hGPR54
TAP-WT hGPR54 and TAP-L148S hGPR54 were purified from HEK293 cells and analyzed by mass spectrometry. Hits shown displayed a peptide prophet score of greater than 0.65 for both WT and L148S hGPR54.
|
Swiss-Prot accession number
|
Protein identity
|
WT GPR54
|
L148S GPR54
|
||
|---|---|---|---|---|---|
| Hits | Coverage | Hits | Coverage | ||
| % | % | ||||
| Bait protein | |||||
| Q969F8 | G-protein-coupled receptor 54 (GPR54) | 2 | 7 | 2 | 5 |
| Signal Transduction | |||||
| Q9NQ66 | Phospholipase cβ1 (PLCβ1) | 1 | 2 | 1 | 1 |
| P68104 | Elongation factor 1α1 (EEF1A1) | ||||
| Protein quality control | |||||
| O75477 | ER lipid raft-associated protein 1 (ERLIN1, SPFH1) | 9 | 27 | 21 | 34 |
| O95816 | BAG family molecular chaperone regulator 2 (BAG2) | 2 | 5 | 2 | 5 |
| P07339 | Cathepsin D precursor (CSTD) | 1 | 4 | 2 | 4 |
| P11142 | Heat shock cognate 71-kDa protein (HSPA8, HSC70) | 6 | 11 | 1 | 2 |
| Q9Y385 | Ubiquitin-conjugating enzyme 1 (UB2J1) | 2 | 5 | 5 | 9 |
| Trafficking | |||||
| Q9P0L0 | Vesicle-associated membrane protein-associated protein A (VAPA) | 1 | 6 | 3 | 12 |
| Transcriptional Regulation | |||||
| Q6AW86 | Zinc finger protein 324B (ZNF324B) | 2 | 1 | 1 | 1 |
| Q53AQ2 | Hepatocyte odd protein shuttling protein (HOPS) | 4 | 17 | 5 | 22 |
| Q9NWT6 | Hypoxia-inducible factor 1α inhibitor (HIF1AN) | 1 | 2 | 1 | 2 |
| Other | |||||
| P29508 | SERPINB3 | 1 | 3 | 4 | 8 |
| P31025 | Lipocalin-1 (LCN1) | 1 | 4 | 2 | 4 |
| Q02413 | Desmoglein-1 precursor (DSG1) | 1 | 2 | 1 | 2 |
| Q61B90 | Cystatin A (CSTA) | 1 | 23 | 2 | 28 |
| Unknown | |||||
| Q71RG4 | Transmembrane and ubiquitin-like domain-containing protein 2 (TMUB2) | 1 | 7 | 2 | 7 |
| Q9BVC6 | Transmembrane protein 109 (TMEM109) | 1 | 5 | 2 | 10 |
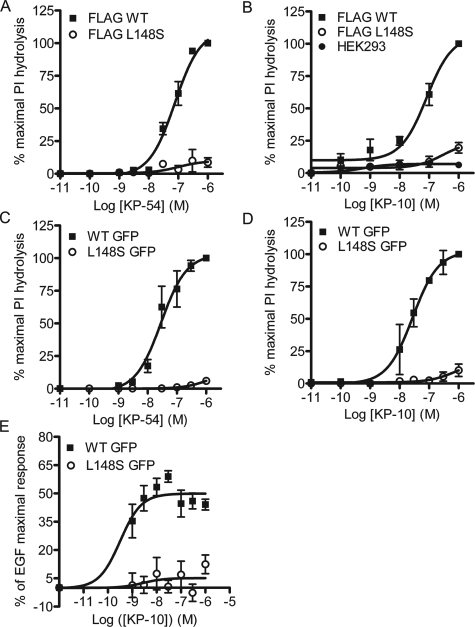
Leucine 148 of GPR54 Is a Critical Regulator of G-protein Coupling—To determine the impact of the L148S mutation on functional coupling of GPR54, we next utilized the PI hydrolysis assay to generate concentration-response curves for the full agonists, KP-54 and KP-10. Although KP-10 is a synthetic agonist, KP-54 is presumably an endogenous GPR54 ligand. Both KP-54 (EC50 = 28 nm) (Fig. 2A) and KP-10 (EC50 = 28 nm) (Fig. 2B) stimulated N-terminal FLAG-tagged WT hGPR54 PI hydrolysis with equal potency. This effect was reproduced with C-terminal GFP-tagged WT hGPR54 (KP-54 EC50 = 82 nm; KP-10 EC50 = 77 nm), indicating that epitope tags are noninterfering (Fig. 2, C and D). In contrast, L148S hGPR54 demonstrated >90% decreases in KP-10- and KP-54-stimulated maximal responses (Fig. 2, A-D). Although the low coupling efficiency of L148S hGPR54 precluded determination of EC50 values, we observed that KP-10 displayed a slightly higher intrinsic activity (IA = 0.1) than KP-54 (IA = 0.06) for the mutant receptor (Fig. 2, A-D). To determine whether the L148S hGPR54 functional deficits could be reproduced with a different functional readout, the ability of KP-10 to stimulate ERK1/2 activation was compared between WT and L148S hGPR54 using a high throughput 96-well in-cell Western assay. Time course experiments showed that maximal stimulation occurred at 10 min (data not shown); thus, this time point was used for subsequent experiments. Similar to the PI hydrolysis experimental results, L148S hGPR54 displayed >90% reductions in KP-10 intrinsic activity for ERK1/2 activation (Fig. 2E). Taken together, these data demonstrate that leucine 148 of human GPR54 is critical for GPR54/G-protein coupling to diverse functional responses.
FIGURE 2.
Leucine 148 plays an important role in GPR54 functional coupling. A and B, the L148S mutation decreased the intrinsic activity as measured by PI hydrolysis in response to KP-54 (A) or KP-10 (B) of FLAG-hGPR54 cells. C and D, the L148S mutation also decreased the intrinsic activity, measured by PI hydrolysis in response to KP-54 (C) or KP-10 (D) of hGPR54-GFP cells. E, KP-10-mediated ERK activation was suppressed in L148S hGPR54-GFP cells. All results are expressed as the means ± S.E. of 3-5 experiments performed in triplicate. For A-D, results are normalized to the response of WT hGPR54 to 1 μm KP (maximal response). For the ERK assay (E), results are normalized to ERK activation in response to 1 nm epidermal growth factor.
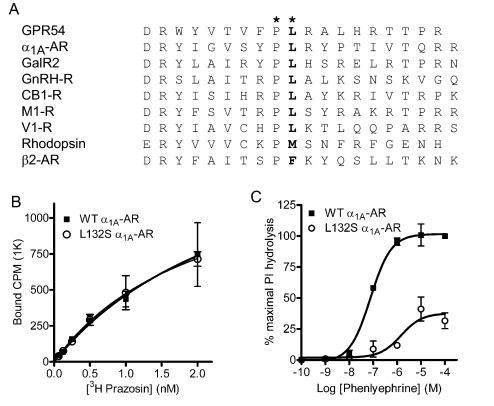
A Conserved, Hydrophobic IL2 Residue Regulates Class A GPCR Function—Class A GPCR IL2 domains contain both single conserved amino acid residues and conserved amino acid motifs (14, 33). In fact, the overwhelming majority of Class A GPCRs contain a hydrophobic residue at the position corresponding to leucine 148 of GPR54, which we designate as IL2-10 (Fig. 3A). The importance of this conserved IL2-10 leucine residue for Class A GPCR functional coupling was further examined by mutating the analogous leucine to serine (L132S) in the α1A-AR. Similar to our results with GPR54, L132S α1A-AR-GFP showed no alterations in cellular localization (data not shown) nor any detectable changes in total receptor density (WT = 1794 cpm; L132S = 1579 cpm) or radioligand equilibrium dissociation constants (KD for WT = 2.9 nm; L132S = 2.2 nm) for the α1-AR-selective antagonist [3H]prazosin (Fig. 3B). Next, concentration-response curves for stimulation of PI hydrolysis were generated for the α1-AR-selective agonist phenylephrine. Equivalent to the results of the L148S hGPR54 functional analysis, the L132S α1A-AR mutation markedly decreased the potency (WT EC50 = 76 nm; L132S = 1.5 μm) and intrinsic activity (L132S IA = 0.4) of phenylephrine (Fig. 3C). Taken together, these findings demonstrate the importance of the IL2-10 leucine for Class A GPCR functional coupling. Moreover, these results suggest that the IL2-10 residue is a crucial component of the ligand-induced Class A GPCR structural rearrangement cascade that ultimately results in G-protein activation.
FIGURE 3.
The highly conserved Leu148 of GPR54 is a critical determinant of effective G-protein coupling for diverse Class A GPCRs. A, alignment of IL2 of a number of Class A GPCRs highlights the conservation of proline and leucine residues (*) downstream of the DRY motif that initiates IL2. B, [3H]prazosin saturation radioligand binding shows that mutation of leucine 132 to serine does not alter the ligand-binding properties of the α1A-AR. Results in B show the means ± S.E. of three experiments performed in triplicate. C, the L132S mutation markedly inhibits α1A-AR functional coupling, as measured by phenylephrine-stimulated PI hydrolysis. Results in C are normalized to the response of WT α1A-AR to 100 μm phenylephrine (maximal response) and presented as the means ± S.E. of four experiments performed in triplicate.
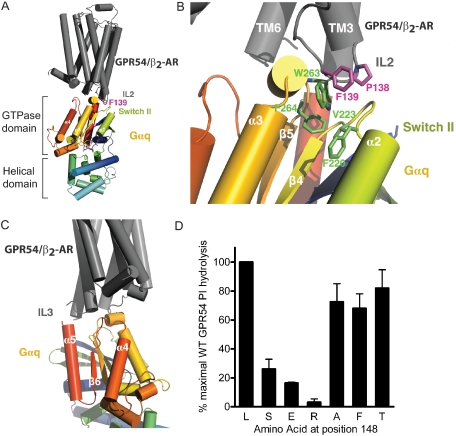
Structural Modeling of Class A GPCR/G-protein Coupling—The observed functional deficits of the L148S GPR54 and L132S α1A-AR mutants led us to examine the potential structural role of the conserved hydrophobic IL2-10 residue in G-protein activation. Thus, we devised a structural model to investigate the molecular mechanism by which IL2-10 could participate in Class A GPCR/G-protein engagement. First, we docked a previously generated TASSER structural prediction of GPR54 (30) to the solved crystal structure of activated Gαq (34). This computational GPCR model was then examined and replaced using the recently described β2-AR 2.4 Å crystal structure (26) to generate a hybrid GPR54/β2-AR structural model (Fig. 4A). Interestingly, our modeling analysis indicates that the β2-AR IL2-10 hydrophobic residue (Phe139), which corresponds to Leu148 in GPR54, docks in close proximity to the GTPase domain of the activated Gαq (Fig. 4A). It is well established that the activation of heterotrimeric G-proteins is accompanied by the liberation of Gβγ subunits and a dramatic structural rearrangement of the Gα subunit that facilitates GDP release, specifically in the flexible switch I, II, and III regions of the Gα GTPase domain (35). Closer examination of our structural model reveals that Pro138 of the β2-AR (which is also highly conserved among Class A GPCRs) initiates a change in the orientation of IL2 that enables Phe139 to fit into a hydrophobic groove that is created by the α2/β4 loop and the α3/β5 loop of Gαq (Fig. 4B). Specifically, our model predicts that the highly conserved Pro138 and Phe139 residues of the β2-AR undergo hydrophobic interactions with highly conserved Gα residues Phe220 and Val223 that immediately follow switch II, as well as Phe264 and Trp263. Interestingly, our model also suggests that the β2-AR IL3 lies in close proximity to the C-terminal α4 helix and β6 loop of Gαq (Fig. 4C). Together, these observations highlight the potential importance of both IL2 and IL3 in establishing multisite structural interactions between GPCRs and G-proteins to promote efficient functional coupling.
FIGURE 4.
A model to suggest the involvement of IL2 in hydrophobichydrophobic interactions between GPCR and G-protein. A, docking analysis reveals that the IL2 of GPR54/β2-AR comes into close proximity to the GTPase domain of the activated Gαq subunit. B, Pro138 of the β2-AR positions Phe139 so that a productive hydrophobic interaction face is formed with highly conserved Phe220, Val223, Trp263, and Phe264 residues of Gαq. C, the third intracellular loop of GPR54/β2-AR also comes into close contact with the α4/β6 loop of the Gαq subunit. D, mutational analysis of GPR54 demonstrates that the presence of a highly hydrophilic, acidic, or basic amino acid at position 148 inhibits functional coupling, as measured by PI hydrolysis. Data are normalized to the WT response to 1 μm KP-10, which is set as 100%. Results are expressed as the means ± S.E. of four experiments performed in triplicate.
Mutational Analysis of GPR54 Amino Acid 148—Cumulatively, our data suggest that the hydrophobic milieu encompassing IL2-10 is imperative for effective coupling between GPCR and G-protein. To understand how the precise physiochemical characteristics of the IL2-10 residue 148 might dictate the strength of GPR54 functional coupling, site-directed mutagenesis was performed to convert leucine 148 to glutamic acid (L148E hGPR54), arginine (L148R hGPR54), alanine (L148A hGPR54), phenylalanine (L148F hGPR54), or threonine (L148T hGPR54) in hGPR54-GFP. Confocal microscopy imaging verified that the expression pattern of each GPR54 variant was indistinguishable from WT hGPR54-GFP in HEK293 cells (data not shown). Next, transiently transfected HEK293 cells expressing individual hGPR54-GFP variants were stimulated with 1 μm KP-10 and assayed for PI hydrolysis (Fig. 4D). Replacement of leucine 148 with small (L148A) or aromatic (L148F) amino acids caused a minor decrease in GPR54 functional responses. In contrast, substitution of leucine 148 with basic (L148R) or acidic residues (L148E) impaired GPR54 function even more severely than that of the disease-causing L148S mutant. Although the structural similarity between threonine and serine led us to expect that L148T hGPR54 would be devoid of functional activity, surprisingly, L148T caused only a small decrease in WT hGPR54 functional coupling. Therefore, these data demonstrate that precise amino acid properties at position 148 in IL2 are required for proper GPR54/G-protein coupling. More specifically, only a major disruption of hydrophobicity at position 148 via the introduction of a highly exposed hydroxyl (OH) group or a charged amino acid will significantly impair GPR54 functional responses.
Is Gαq Association with GPR54 Affected by the L148S Mutation?—It has been proposed that upon agonist binding, GPCRs interact with G-proteins through a cascade of sequential protein-protein interactions, yet the precise molecular mechanism by which GPCRs activate G-proteins remains largely enigmatic (35). Thus far, our experimental evidence examining the mechanism by which L148S uncouples GPR54 suggests that the IL2 is a critical determinant in G-protein coupling. However, we are unsure if L148S could interfere with either the binding of Gαq to GPR54 or the catalytic activation of Gαq by GPR54. To address the possibility that L148S is affecting Gαq/GPR54 binding, we first determined whether Gαq directly interacts with purified hGPR54-IL2. Thus, GST fusions of WT and L148S hGPR54 IL2 were generated and co-expressed in E. coli with His-Gαq. Cell lysates were then applied to nickel columns to bind Gαq, and eluate was analyzed by Coomassie staining. We found that WT hGPR54 IL2 co-purified with Gαq (Fig. 5A), suggesting that these two proteins form direct protein-protein interactions. Importantly, GST alone did not co-purify with Gαq (Fig. 5A), demonstrating that the interaction was mediated by the IL2. Indeed, the interaction affinity between Gαq and IL2 was remarkably strong, since stringent washes with up to 900 mm NaCl did not disrupt binding (data not shown). Strikingly, the interaction between GPR54 IL2 and Gαq was completely unaffected by the L148S mutation (Fig. 5A), suggesting that the mechanism by which this mutation uncouples functional responses is not due to abrogating GPCR/Gαq interactions. We confirmed these results with full-length receptors expressed in HEK293 cells by using co-immunoprecipitation of TAP-hGPR54 and blotting for overexpressed HA-Gα15/16, the promiscuous Gq family member, or Gαq. Similar to our results using purified GST fusion constructs, we found that the ability of GPR54 to bind Gαq or Gα15/16 was unaltered by L148S (Fig. 5B), again suggesting that mutation does not alter the Gα binding affinity to GPR54.
FIGURE 5.
The L148S mutation does not abolish G-protein binding, and overexpression of G-proteins partially rescues the functional deficits of IL2-10 mutants. A, in E. coli, both WT and L148S mutant GST-hGPR54 IL2 fusion proteins co-purify with Gαq. GST alone does not co-purify with Gαq. B, co-immunoprecipitation experiments reveal that both TAP-WT hGPR54 and TAP-L148S hGPR54 proteins pull down Gαq and Gα15/16. C, overexpression of Gα15/16 DNA in L148S hGPR54-GFP cells leads to a concentration-dependent rescue of functional coupling, as measured by PI hydrolysis. L148S hGPR54 results (C) are normalized to the WT response to 1 μm KP-10, which was set at 100%. Inset, Western blot shows that increasing the amount of Gα15/16 DNA transfected increases the relative levels of Gα15/16 protein in L148S hGPR54-GFP cells. D, co-overexpression of L132S α1A-AR and Gαq modestly rescues the deficit in PI hydrolysis. The results presented in A and B are representative of three experiments, and C and D show the means ± S.E. of three experiments performed in triplicate.
Overexpression of Gα Subunits Can Partially Rescue the Functional Deficits of IL-10 Mutants—We previously observed minimal activity of L148S GPR54 and L132S α1A-AR at saturating concentrations of agonist. Combined with our results demonstrating that the L148S mutation did not disrupt G-protein docking, we speculated that flooding the cells with high levels of G-proteins might compensate for the apparent catalytic defect of the IL2-10 GPCR mutants. Indeed, overexpression of increasing concentrations of Gα15/16 partially rescued the functional deficits of L148S hGPR54, measured by KP-10-stimulated PI hydrolysis (Fig. 5C). Immunoblotting confirmed the increase in relative levels of Gα15/16 protein. Importantly, overexpression of Gα15/16 did not affect KP-10-stimulated PI hydrolysis of WT hGPR54 (data not shown), demonstrating that this effect was specific for the mutant receptor. We also attempted to rescue L132S α1A-AR functional responses by overexpressing the cognate G-protein, Gαq. Reminiscent of our results with L148S hGPR54, L132S α1A-AR coupling to PI hydrolysis increased by ∼20% in the presence of overexpressed Gαq, and again, no effect of Gαq overexpression was observed with WT α1A-AR. For both mutant receptors, we observed at best a ∼20% increase in functional activity relative to WT with 25 μg of Gα DNA, suggesting that the IL2-10 mutation dramatically affects the ability of Class A GPCRs to catalytically engage their cognate Gα proteins.
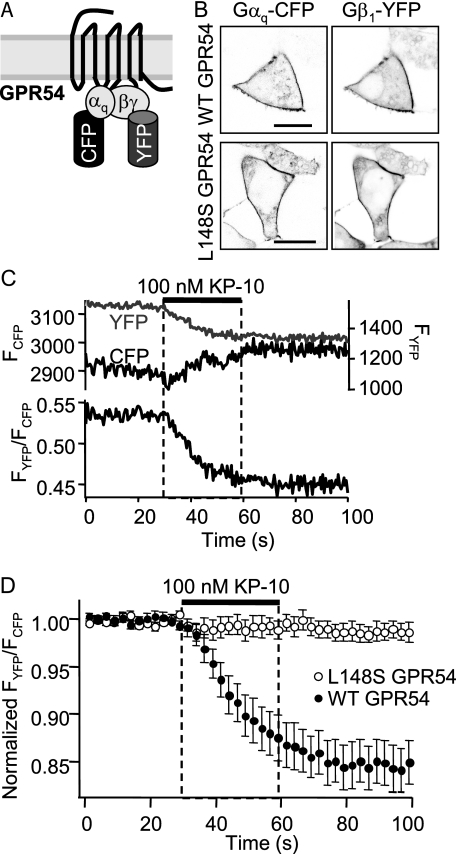
The L148S Mutation of GPR54 Inhibits the Catalytic Activation of Gαq—Next, we utilized a novel FRET assay to investigate whether the L148S mutation inhibits catalytic activation of the Gαq subunit by GPR54. This assay allows one to monitor the kinetics of G-protein activation in real time for WT and L148S hGPR54, as determined by the structural rearrangement of the G-protein heterotrimer, specifically the liberation of Gβγ subunits from the activated Gα subunit, that occurs in response to GPR54 activation by kisspeptin. We transiently transfected HEK293 cells with WT or L148S hGPR54, Gαq-CFP, Gβ1-YFP, Gγ2, and GRK2 and examined the resting FRET ratio between Gαq-CFP and Gβ1-YFP (Fig. 6A). Although initial experiments were performed in the absence of GRK2 (data not shown), we found that GRK2 enhanced the amplitude of G-protein FRET changes and improved the signal-to-noise ratio (47) and thus included GRK2 in all subsequent experiments. Confocal microscopy verified that Gαq-CFP and Gβ1-YFP were expressed at the plasma membrane (Fig. 6B). Next, we measured the average resting FRET ratios for WT (0.37 ± 0.03) and L148S (0.32 ± 0.02) hGPR54 (Fig. 6, C and D). When cells were treated with 100 nm KP-10, WT hGPR54 elicited a robust 16 ± 2% reduction in the FRET ratio with a mean single exponential time constant of 19.6 ± 0.8 s. In sharp contrast, KP-10 treatment of L148S hGPR54 cells caused no change in the FRET ratio (Fig. 6, C and D). These results demonstrate that the L148S disease-causing mutation in the IL2 of GPR54 directly impairs G-protein activation by inhibiting the dissociation of Gα and Gβγ subunits and suggest that the IL2-10 residue is a critical component of GPCR-mediated GEF activity.
FIGURE 6.
The L148S mutation abolishes the agonist-induced catalytic activation of Gαq by GPR54. A, schematic demonstrating that under basal conditions, FRET occurs between the Gαq-CFP and Gβ1-YFP proteins that are docked to GPR54. B, representative confocal microscopy images showing the membrane localization of Gαq-CFP and Gβ1-YFP in both WT hGPR54 (top) and L148S hGPR54 (bottom) cells. C, data from a representative WT hGPR54 cell demonstrating that stimulation with 100 nm KP-10 causes a decrease in FRET between YFP and CFP. D, averaged FRET data from 11-13 cells from three distinct experiments expressing either WT or L148S hGPR54. WT hGPR54 shows a robust decrease in FRET in response to KP-10, whereas L148S hGPR54 fails to respond. For clarity in display, data are pooled in 2.5-5 bins.
DISCUSSION
Myriad studies analyzing the molecular mechanisms by which GPCR mutations cause disease have revealed key fundamental concepts in GPCR biology (36, 37). For example, inactivating GPCR mutations are now classified as causing defects in either receptor biosynthesis, trafficking to the cell surface, ligand binding, or receptor activation (36). Due to the inherent complexities involved in the folding of a seven-transmembrane protein, GPCRs must pass stringent quality control mechanisms in order to be trafficked and properly expressed at the cell surface (38). Thus, it is not surprising that intracellular retention is the most prevalent mechanism by which inactivating GPCR mutations cause disease (36, 39). Notably, of the 21 mutations in the gonadotropin-releasing hormone receptor that have been found to cause IHH, extensive in vitro analysis has shown that ∼17 are caused by deficits in trafficking and/or plasma membrane expression (37). Due to the overwhelming percentage of disease-causing GPCR mutations that result in inefficient plasma membrane expression, we suspected that the L148S mutation of GPR54 might cause intracellular retention and thus explain the etiology of IHH. However, in the present study, we determined that the L148S GPR54 mutant retains normal expression, localization, and kisspeptin binding properties. Moreover, the protein interaction network of GPR54 is unaltered by the L148S mutation, demonstrating that the mutant protein is not directed toward the cellular protein degradation machinery. Instead, we find that L148S GPR54 is essentially uncoupled from all functional responses, since the mutant receptor neither initiates G-protein dissociation nor activates phospholipase C or ERK1/2. The ability of this single hydrophobic to hydrophilic amino acid substitution to cause disease implicates the central portion of IL2 as an important determinant of GPCR/G-protein coupling in vivo.
Although the overall sequence similarity among GPCRs is low, several stretches of highly conserved amino acids stand out as likely candidates for regulating GPCR function. For example, the highly conserved (D/E)R(Y/W) motif, recognized for its importance in modulating the activation state of GPCRs, initiates the IL2 in most Class A GPCRs (40). Moreover, in ∼75% of Class A GPCRs (41), the 10th residue of IL2 (IL2-10) is a hydrophobic amino acid. This IL2-10 residue corresponds to Leu148 of GPR54. Interestingly, site-directed mutagenesis studies have suggested that IL2-10 is important for Class A GPCR/G-protein coupling (14, 16, 18, 19). However, in contrast to previous findings where mutation of IL2-10 to alanine abolished GPCR functional coupling (14, 16, 19), we demonstrated that the L148A GPR54 mutant displayed only moderately decreased functional responses to kisspeptin. In fact, markedly decreased functional responses were only observed after mutating leucine 148 to amino acids with strongly divergent R group characteristics. Examination of hydropathy values for serine (-1.1), glutamic acid (-2.6), and arginine (-7.5) revealed a correlation between score and degree of functional inhibition. Interestingly, in contrast to the L148S mutant, L148T GPR54 (threonine hydropathy score = -0.75) displayed nearly a WT functional response, indicating that the methyl group may sufficiently buffer the detrimental effect of the hydroxyl group on the hydrophobic environment of IL2. We propose that the exposed hydroxyl of the disease-associated serine 148 significantly disrupts the hydrophobic milieu of IL2, which compromises a structural interface required for efficient GPCR/G-protein coupling. Collectively, our findings suggest that in vivo, conservative mutations of the IL2 leucine may be tolerated, whereas significant changes in the size, charge, or hydrophobicity of IL2-10 are sufficient to cause disease.
Recent crystallographic studies have suggested that G-protein activation requires conformational changes in the flexible switch I, II, and III regions of Gα to relieve a GDP clamp formed between the GTPase and α-helical domains (42). Although activated GPCRs are known to act as GEFs and promote G-protein activation, the structural basis for GPCR-catalyzed nucleotide exchange remains unclear (35). We hypothesize that the IL2-10 residue characterized in this study directly interacts with Gα and is critical for Class A GPCR GEF activity. Our structural model of an activated Gαq subunit docked to the hybrid GPR54/β2-AR suggests that IL2 might stabilize switch II of Gα in a favorable orientation for GDP release. Although the switch II region is sequestered by Gβγ in the inactive Gαβγ heterotrimer, one theory proposes that GPCR activation enables the receptor to use Gβγ as a lever against the β3/α2 loop of Gα, which encompasses switch II, to relieve the block on GDP release (43). It is plausible that IL2 could first lever Gβγ away from switch II and subsequently catch and stabilize switch II in a more open state. Thus, we suggest that in response to GPCR activation the switch II region of Gα is structurally stabilized by hydrophobic interactions between the IL2-10 residue and conserved Gα residues to establish a productive egress route for GDP. We acknowledge, however, that the process by which a GPCR activates a G-protein is complex and probably operates through a series of sequential and/or synergistic steps that involve numerous points of contact between GPCR and G-protein. In fact, our model supports recent findings from the solved crystal structure of Gαi1 in complex with a peptide derived from the N-terminal region of the D2 dopamine receptor IL3 (D2N) and a Gβγ peptide mimetic (KB-752) (25). In this study, Johnston and Siderovski (25) showed that D2N binds to the α4/β6 loop of Gα in the crystal structure and, furthermore, the D2N peptide demonstrated significant GEF activity toward Gαi1. Accordingly, our structural modeling data demonstrate that the β2-AR IL3 could contact the α4/β6 loop of Gα. Thus, we suggest that IL2, acting on switch II, and IL3, acting on the C-terminal region of Gα to disrupt critical contacts between the β6/α5 loop and GDP, may synergistically stabilize the GDP-GTP transition state to facilitate maximal nucleotide exchange. Interestingly, the Gα residues proposed to interact with the β2-AR in our model were highly conserved among all classes of G-proteins, suggesting that other, nonconserved regions probably determine specificity of G-protein coupling and participate in the multisite process of agonist-mediated G-protein activation. Thus, our structural modeling data support the emerging paradigm that a multisite interaction motif between GPCRs and G-proteins is required for efficient activation and GDP-GTP exchange (44).
Interestingly, we observed that saturating agonist concentrations elicited minor increases in functional activity for both the L148S GPR54 and L132S α1A-AR mutants and that overexpressing Gα partially rescued L148S GPR54 and L132S α1A-AR functional responses. Although increasing the relative concentration of Gα subunits caused minor increases in functional coupling, we were unable to obtain even a modest rescue of L148S GPR54 or L132S α1A-AR function, indicating that increasing the concentration of reactants >25-fold is unable to compensate for the deficits in G-protein engagement. Our immunoprecipitation data show that the L148S mutation does not affect the ability of GPR54 to bind Gα; thus, it is more likely that the structural disruption caused by the introduction of a hydrophilic residue into a hydrophobic environment alters the catalytic activation of Gα. Accordingly, our FRET data demonstrate a clear defect of the L148S GPR54 in catalytically engaging the Gα subunit to promote G-protein activation and functional coupling. This finding leads us to the surprising suggestion that the IL2 of Class A GPCRs may act as a GEF to promote the dissociation of Gβγ subunits from Gα, thus promoting efficient release of GDP, which is the rate-limiting step in G-protein activation, and exchange for GTP. We suspect that such a stabilizing interaction between the IL2 and Gα would be transient, since the switch II region of Gα has previously been shown to form a hydrophobic interaction domain that can interface with multiple downstream signaling proteins such as adenylyl cyclase and RGS4 (45, 46).
In conclusion, our characterization of the IHH-causing L148S GPR54 mutant has revealed fundamental concepts concerning GPCR/G-protein signaling. Importantly, the IL2-10 residue may be a key player in the ligand-induced structural rearrangement of Class A GPCRs that ultimately results in G-protein activation subsequent to ligand-induced receptor activation. This information should prove useful for continued exploration of GPCR structure-function predictions and drug discovery for ligands targeting GPCRs or GPCR-interacting proteins.
Supplementary Material
Acknowledgments
We thank Dr. Bertil Hille for assistance with manuscript preparation and FRET experiments, Dr. Marc Parmentier (Institute of Multidisciplinary Research in Human and Molecular Biology, Brussels, Belgium) for the hGPR54 pEFIN3 construct, Dr. Robert Steiner (Department of Physiology and Biophysics, University of Washington) for reagents and helpful discussion, Dr. Nephi Stella for technical expertise (Department of Pharmacology, University of Washington), and Dr. Ning Zheng (Department of Pharmacology, University of Washington) for help with structural modeling and manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grant T32 HD07453 (to J. L. W.); NIGMS National Research Service Award T32 GM07270 (to J. S. L.); T32 GM07750 (to M. C. D.); and T32 GM07108, RO1 NS08174 (BH), and U54HD12629 (to J. B. J.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: IHH, idiopathic hypogonadotropic hypogonadism; AR, adrenergic receptor; FRET, fluorescence resonance energy transfer; GEF, guanine nucleotide exchange factor; GPCR, G-protein-coupled receptor; HEK, human embryonic kidney; IL2 and IL3, second and third intracellular loop, respectively; KP, kisspeptin; PI, phosphatidylinositol; TAP, tandem affinity purification; MS, mass spectrometry; hGPR54, human GPR54; HA, hemagglutinin; GFP, green fluorescent protein; WT, wild type; PBS, phosphate-buffered saline; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; CFP, cyan fluorescent protein; YFP, yellow fluorescent protein; GIP, GPCR-interacting protein; GST, glutathione S-transferase; ERK, extracellular signal-regulated kinase.
References
- 1.Tena-Sempere, M., and Huhtaniemi, I. T. (2003) in Reproductive Medicine: Molecular, Cellular and Genetic Fundamentals (Fauser, B., ed) pp. 225-244, Parthenon Publishing, New York
- 2.Belchetz, P. E., Plant, T. M., Nakai, Y., Keogh, E. J., and Knobil, E. (1978) Science 202 631-633 [DOI] [PubMed] [Google Scholar]
- 3.Seminara, S. B. (2006) Nat. Clin. Pract. Endocrinol. Metab. 2 328-334 [DOI] [PubMed] [Google Scholar]
- 4.Iovane, A., Aumas, C., and de Roux, N. (2004) Eur. J. Endocrinol. 151 Suppl. 3, 83-88 [DOI] [PubMed] [Google Scholar]
- 5.Bhagavath, B. M. D., and Layman, L. M. D. (2007) Semin. Reprod. Med. 25 272-286 [DOI] [PubMed] [Google Scholar]
- 6.de Roux, N., Genin, E., Carel, J.-C., Matsuda, F., Chaussain, J.-L., and Milgrom, E. (2003) Proc. Natl. Acad. Sci. U. S. A 100 10972-10976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seminara, S. B., Messager, S., Chatzidaki, E. E., Thresher, R. R., Acierno, J. S., Jr., Shagoury, J. K., Bo-Abbas, Y., Kuohung, W., Schwinof, K. M., Hendrick, A. G., Zahn, D., Dixon, J., Kaiser, U. B., Slaugenhaupt, S. A., Gusella, J. F., O'Rahilly, S., Carlton, M. B. L., Crowley, W. F., Jr., Aparicio, S. A. J. R., and Colledge, W. H. (2003) N. Engl. J. Med. 349 1614-1627 [DOI] [PubMed] [Google Scholar]
- 8.Semple, R. K., Achermann, J. C., Ellery, J., Farooqi, I. S., Karet, F. E., Stanhope, R. G., O'Rahilly, S., and Aparicio, S. A. (2005) J. Clin. Endocrinol. Metab. 90 1849-1855 [DOI] [PubMed] [Google Scholar]
- 9.Gottsch, M. L., Cunningham, M. J., Smith, J. T., Popa, S. M., Acohido, B. V., Crowley, W. F., Seminara, S., Clifton, D. K., and Steiner, R. A. (2004) Endocrinology 145 4073-4077 [DOI] [PubMed] [Google Scholar]
- 10.Irwig, M. S., Fraley, G. S., Smith, J. T., Acohido, B. V., Popa, S. M., Cunningham, M. J., Gottsch, M. L., Clifton, D. K., and Steiner, R. A. (2004) Neuroendocrinology 80 264-272 [DOI] [PubMed] [Google Scholar]
- 11.Navarro, V. M., Castellano, J. M., Fernandez-Fernandez, R., Tovar, S., Roa, J., Mayen, A., Nogueiras, R., Vazquez, M. J., Barreiro, M. L., Magni, P., Aguilar, E., Dieguez, C., Pinilla, L., and Tena-Sempere, M. (2005) Endocrinology 146 156-163 [DOI] [PubMed] [Google Scholar]
- 12.Dhillo, W. S., Chaudhri, O. B., Patterson, M., Thompson, E. L., Murphy, K. G., Badman, M. K., McGowan, B. M., Amber, V., Patel, S., Ghatei, M. A., and Bloom, S. R. (2005) J. Clin. Endocrinol. Metab. 90 6609-6615 [DOI] [PubMed] [Google Scholar]
- 13.Funes, S., Hedrick, J. A., Vassileva, G., Markowitz, L., Abbondanzo, S., Golovko, A., Yang, S., Monsma, F. J., and Gustafson, E. L. (2003) Biochem. Biophys. Res. Commun. 312 1357-1363 [DOI] [PubMed] [Google Scholar]
- 14.Moro, O., Lameh, J., Hogger, P., and Sadee, W. (1993) J. Biol. Chem. 268 22273-22276 [PubMed] [Google Scholar]
- 15.Blin, N., Yun, J., and Wess, J. (1995) J. Biol. Chem. 270 17741-17748 [DOI] [PubMed] [Google Scholar]
- 16.Arora, K. K., Sakai, A., and Catt, K. J. (1995) J. Biol. Chem. 270 22820-22826 [DOI] [PubMed] [Google Scholar]
- 17.Burstein, E. S., Spalding, T. A., and Brann, M. R. (1998) J. Biol. Chem. 273 24322-24327 [DOI] [PubMed] [Google Scholar]
- 18.Zhou, H., Yan, F., Yamamoto, S., and Tai, H.-H. (1999) Biochem. Biophys. Res. Commun. 264 171-175 [DOI] [PubMed] [Google Scholar]
- 19.Gaborik, Z., Jagadeesh, G., Zhang, M., Spat, A., Catt, K. J., and Hunyady, L. (2003) Endocrinology 144 2220-2228 [DOI] [PubMed] [Google Scholar]
- 20.Costa, E. M. F., Bedecarrats, G. Y., Mendonca, B. B., Arnhold, I. J. P., Kaiser, U. B., and Latronico, A. C. (2001) J. Clin. Endocrinol. Metab. 86 2680-2686 [DOI] [PubMed] [Google Scholar]
- 21.Cai, K., Itoh, Y., and Khorana, H. G. (2001) Proc. Natl. Acad. Sci. 98 4877-4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang, H., Lee, W. K., Choi, Y. H., Vukoti, K. M., Bang, W. G., and Yu, Y. G. (2005) Biochem. Biophys. Res. Commun. 329 684-692 [DOI] [PubMed] [Google Scholar]
- 23.Geiser, A. H., Sievert, M. K., Guo, L.-W., Grant, J. E., Krebs, M. P., Fotiadis, D., Engel, A., and Ruoho, A. E. (2006) Protein Sci. 15 1679-1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nanoff, C., Koppensteiner, R., Yang, Q., Fuerst, E., Ahorn, H., and Freissmuth, M. (2006) Mol. Pharmacol. 69 397-405 [DOI] [PubMed] [Google Scholar]
- 25.Johnston, C. A., and Siderovski, D. P. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 2001-2006 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Cherezov, V., Rosenbaum, D. M., Hanson, M. A., Rasmussen, S. G. F., Thian, F. S., Kobilka, T. S., Choi, H.-J., Kuhn, P., Weis, W. I., Kobilka, B. K., and Stevens, R. C. (2007) Science 318 1258-1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daulat, A. M., Maurice, P., Froment, C., Guillaume, J.-L., Broussard, C., Monsarrat, B., Delagrange, P., and Jockers, R. (2007) Mol. Cell Proteomics 6 835-844 [DOI] [PubMed] [Google Scholar]
- 28.Lyssand, J. S., DeFino, M. C., Tang, X. B., Hertz, A. L., Feller, D. B., Wacker, J. L., Adams, M. E., and Hague, C. (2008) J. Biol. Chem. 283 18792-18800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen, H., Kovar, J., Sissons, S., Cox, K., Matter, W., Chadwell, F., Luan, P., Vlahos, C. J., Schutz-Geschwender, A., and Olive, D. M. (2005) Anal. Biochem. 338 136-142 [DOI] [PubMed] [Google Scholar]
- 30.Zhang, Y., Devries, M. E., and Skolnick, J. (2006) PLoS Comput. Biol. 2 88-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray, J. J., Moughon, S., Wang, C., Schueler-Furman, O., Kuhlman, B., Rohl, C. A., and Baker, D. (2003) J. Mol. Biol. 331 281-299 [DOI] [PubMed] [Google Scholar]
- 32.Hall, R. A., and Lefkowitz, R. J. (2002) Circ. Res. 91 672-680 [DOI] [PubMed] [Google Scholar]
- 33.Dohlman, H. G., Caron, M. G., and Lefkowitz, R. J. (1987) Biochemistry 26 2657-2664 [DOI] [PubMed] [Google Scholar]
- 34.Tesmer, V. M., Kawano, T., Shankaranarayanan, A., Kozasa, T., and Tesmer, J. J. G. (2005) Science 310 1686-1690 [DOI] [PubMed] [Google Scholar]
- 35.Oldham, W. M., and Hamm, H. E. (2008) Nat. Rev. Mol. Cell. Biol. 9 60-71 [DOI] [PubMed] [Google Scholar]
- 36.Tao, Y.-X. (2006) Pharmacol. Ther. 111 949-973 [DOI] [PubMed] [Google Scholar]
- 37.Conn, P. M., Ulloa-Aguirre, A., Ito, J., and Janovick, J. A. (2007) Pharmacol. Rev. 59 225-250 [DOI] [PubMed] [Google Scholar]
- 38.Conn, P. M., Janovick, J. A., Brothers, S. P., and Knollman, P. E. (2006) J. Endocrinol. 190 13-16 [DOI] [PubMed] [Google Scholar]
- 39.Tan, C. M., Brady, A. E., Nickols, H. H., Wang, Q., and Limbird, L. E. (2004) Annu. Rev. Pharmacol. Toxicol. 44 559-609 [DOI] [PubMed] [Google Scholar]
- 40.Rovati, G. E., Capra, V., and Neubig, R. R. (2007) Mol. Pharmacol. 71 959-964 [DOI] [PubMed] [Google Scholar]
- 41.Marion, S., Oakley, R. H., Kim, K. M., Caron, M. G., and Barak, L. S. (2006) J. Biol. Chem. 281 2932-2938 [DOI] [PubMed] [Google Scholar]
- 42.Oldham, W. M., and Hamm, H. E. (2006) Q. Rev. Biophys. 39 117-166 [DOI] [PubMed] [Google Scholar]
- 43.Iiri, T., Farfel, Z., and Bourne, H. R. (1998) Nature 394 35-38 [DOI] [PubMed] [Google Scholar]
- 44.Johnston, C. A., and Siderovski, D. P. (2007) Mol. Pharmacol. 72 219-230 [DOI] [PubMed] [Google Scholar]
- 45.Tesmer, J. J. G., Sunahara, R. K., Gilman, A. G., and Sprang, S. R. (1997) Science 278 1907-1916 [DOI] [PubMed] [Google Scholar]
- 46.Tesmer, J. J. G., Berman, D. M., Gilman, A. G., and Sprang, S. R. (1997) Cell 89 251-261 [DOI] [PubMed] [Google Scholar]
- 47.Schliefenbaum, J., Kreile, A. K., Lohse, M. J., and Bunemann, M. (2008) Biophysical Society Meeting, Long Beach, CA, February 2-6, Abstr. 1977
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.