Abstract
The 5′ ends of all picornaviral RNAs are linked covalently to the genome-encoded peptide, VPg (or 3B). VPg linkage is thought to occur in two steps. First, VPg serves as a primer for production of diuridylylated VPg (VPg-pUpU) in a reaction catalyzed by the viral polymerase that is templated by an RNA element (oriI). It is currently thought that the viral 3AB protein is the source of VPg in vivo. Second, VPg-pUpU is transferred to the 3′ end of plus- and/or minus-strand RNA and serves as primer for production of full-length RNA. Nothing is known about the mechanism of transfer. We present biochemical and biological evidence refuting the use of 3AB as the donor for VPg uridylylation. Our data are consistent with precursors 3BC and/or 3BCD being employed for uridylylation. This conclusion is supported by in vitro uridylylation of these proteins, the ability of a mutant replicon incapable of producing processed VPg to replicate in HeLa cells and cell-free extracts and corresponding precursor processing profiles, and the demonstration of 3BC-linked RNA in mutant replicon-transfected cells. These data permit elaboration of our model for VPg uridylylation to include the use of precursor proteins and invoke a possible mechanism for location of the diuridylylated, VPg-containing precursor at the 3′ end of plus- or minus-strand RNA for production of full-length RNA. Finally, determinants of VPg uridylylation efficiency suggest formation and/or collapse or release of the uridylylated product as the rate-limiting step in vitro depending upon the VPg donor employed.
The picornavirus family of viruses causes a wide variety of diseases in humans and animals (1). Poliovirus (PV),3 the causative agent of poliomyelitis, is the most extensively studied member of this family and has proven to be a useful model system for understanding picornavirus molecular biology, including genome replication (1). PV is a nonenveloped virus with a single-stranded RNA genome of positive polarity that is ∼7500 nt in length. As shown in Fig. 1A, the genome encodes a single open reading frame flanked by 5′- and 3′-nontranslated regions (NTRs). The genome contains a 22-amino acid peptide (referred to as either VPg, 3B or primer for RNA synthesis) covalently linked to its 5′ end and is polyadenylated at its 3′ end. Translation of the genome is initiated from an internal ribosome entry site located in the 5′-NTR, producing a 247-kDa polyprotein that is co- and post-translationally processed by the virus-encoded 2A, 3C, and 3CD proteases (1). Although polyprotein processing ultimately yields a set of processed end products, processing intermediates will form during the course of an infection, greatly increasing the functional proteome of the virus. Processing intermediates located in the P2 (e.g. 2ABC, 2BC, etc.) and P3 regions of the polyprotein are likely important for genome replication.
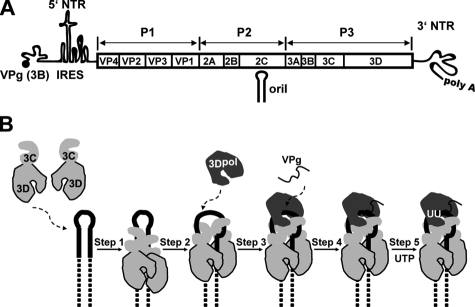
FIGURE 1.
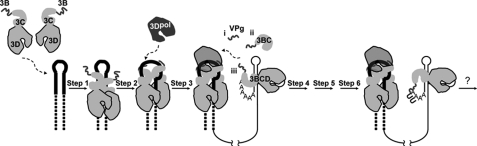
Poliovirus genome organization and model for VPg uridylylation. A, schematic of the poliovirus genome. The 5′ end of the genome is covalently linked to a peptide (VPg) encoded by the 3B region of the genome. The 3′ end contains a poly(rA) tail. Three cis-acting replication elements are known. oriL is located in 5′-NTR. oriR is located in the 3′-NTR. oriI is located in 2C-coding sequence for poliovirus; the position of this element is virus-dependent. oriI is the template for VPg uridylylation. Translation initiation employs an internal ribosome entry site (IRES). The single open reading frame encodes a polyprotein. P1 encodes virion structural proteins as indicated. P2 encodes proteins thought to participate in virus-host interactions required for genome replication. P3 encodes proteins thought to participate directly in genome replication. Polyprotein processing is mediated by protease activity residing in 2A, 3C, and/or 3CD proteins. B, model for VPg uridylylation. Step 1, two 3C(D) molecules bind to oriI with the 3C domains contacting the upper stem (solid lines) and the 3D domains contacting the lower stem (dashed lines). Step 2, 3C dimer opens the RNA stem by forming a more stable interaction with single strands forming the stem. Step 3, 3Dpol is recruited to and retained in this complex by a physical interaction between the back of the thumb sub-domain of 3Dpol and a surface formed by the 3C dimer. Step 4, VPg is recruited to the complex. Step 5, two successive cycles of UMP incorporation yields VPg-pUpU by using a slide-back mechanism. Adapted from Ref. 3.
Our laboratory has been quite interested in defining the molecular details of VPg attachment to the 5′ end of picornaviral RNAs (2–5). This reaction is thought to occur in two independent half-reactions catalyzed by the viral RNA-dependent RNA polymerase, 3Dpol. First, VPg is uridylylated to produce VPg-pUpU; second, VPg-pUpU serves as a primer for full-length RNA synthesis (1). VPg uridylylation requires a template. To date, two templates have been described. The first is the poly(rA) tail at the 3′ end of the genome (6). The second is an RNA stem-loop structure found at different positions in the genomes of different picornaviruses but most often occurring in protein-coding sequence (7–11). We refer to this latter template as oriI (origin of replication internal). PV oriI is located in 2C-coding sequence (Fig. 1A) (7).
Our current model for oriI-templated VPg uridylylation is shown in Fig. 1B. This model is consistent with much of what is known about this reaction (2–5, 7, 8, 11–19), but the details have emerged from studies performed in vitro on minimal templates and minimal protein domains (2–5). Briefly, two molecules of 3C(D) bind to oriI (step 1) (3). This complex isomerizes, unwinding the stem and extending the loop (step 2) (3, 4). 3Dpol associates with the complex, directed and stabilized by an interaction between the back of the “thumb” subdomain of 3Dpol and a convex surface formed by the top of both subunits of the 3C dimer (step 3) (2–5). VPg joins the complex, perhaps by binding to the RNA-primer-binding site of 3Dpol in an extended conformation (step 4) (19). In the presence of UTP, Tyr-3 hydroxyl of VPg is used as a nucleophile to form VPg-pU, the 3′-OH of which, in turn, serves as the nucleophile to form VPg-pUpU (step 5) (13). Both uridylate residues are templated by a single adenylate residue in the oriI loop by using a slide-back mechanism (13). VPg-pUpU must occur processively as VPg-pU appears to be catalytically incompetent (20).
This model provides a clear mechanistic framework for the study of oriI-templated VPg uridylylation in vitro. Importantly, this model explains and predicts biological phenotypes (2, 4, 5, 20). However, VPg uridylylation occurs 10–50-fold slower in vitro than necessary to support the rate constant (∼0.1/s) for initiation calculated from biological data (see Ref. 21). In addition, this model assumes that the processed VPg peptide is the primer employed in vivo, which may not be the case. Finally, this model does not provide any hints of how a VPg-pUpU molecule produced in the middle of the genome could be transferred to the 3′ end of plus- and/or minus-strand RNA for production of the complementary RNA strand. Because the end products of polyprotein processing may not be the forms of the protein employed for assembly and/or function of the genome-replication complexes in vivo, it is possible that the use of polyprotein processing intermediates may be essential to recapitulate VPg uridylylation in vitro that occurs on a biologically relevant time scale, providing additional insight into the mechanism of this reaction that is absolutely essential for picornavirus genome replication.
In this study, we show that P3 precursor proteins containing VPg at the amino terminus (3BC and 3BCD) can be uridylylated in vitro. 3BC-containing precursors bind to oriI more efficiently than processed proteins and serve as VPg donors that can be recruited to and/or retained in the uridylylation complex better than processed VPg. We demonstrate an optimal affinity of precursor binding to oriI for maximal accumulation of uridylylated product in the steady state, suggesting formation and/or collapse as a rate-limiting step for uridylylation in vitro. The use of precursor proteins during replication in cells was queried by preventing the production of processed VPg. Preventing VPg formation in cells is not lethal and leads to production of 3BC-linked RNA. Therefore, processed VPg is not essential for uridylylation and/or full-length plus- and/or minus-strand RNA synthesis in vivo. Analysis of the polyprotein processing profile of the VPg-processing-defective mutant in cell-free extracts uncovered two pathways of P3 precursor processing, major and minor, and only the major pathway was perturbed. We propose that the minor pathway is responsible for production of proteins (processing intermediates) required for uridylylation and RNA synthesis. These studies provide new insight into the mechanism of VPg uridylylation and suggest mechanisms for transfer of the diuridylylated protein primer from the middle of the genome to the 3′ end of plus- and/or minus-strand RNA.
EXPERIMENTAL PROCEDURES
Materials—Deep Vent DNA polymerase and restriction enzymes were from New England Biolabs; shrimp alkaline phosphatase was from U. S. Biochemical Corp.; T4 DNA ligase was from Invitrogen; Difco-NZCYM was from BD Biosciences; QIAEX beads were from Qiagen; RNases A and T1 were from Sigma; Ultrapure UTP solution was from GE Healthcare; [α-32P]UTP (6000 Ci/mmol) was from PerkinElmer Life Sciences; synthetic VPg peptide was from Alpha Diagnostic International (San Antonio, Texas); all other reagents and apparatuses were available through Fisher, VWR, or as indicated.
Construction of Expression Plasmids for 3BC, 3BCD, 3AB, and 3Cpro—Standard PCR and cloning procedures were used to generate expression plasmids for 3BC, 3BCD, 3AB, and 3Cpro. 3Cpro refers to 3C protein with an active protease. Oligonucleotides used in PCRs for this study were purchased from Invitrogen or Integrated DNA Technologies, Inc; sequences are provided in supplemental Table S1. Clones were verified by sequencing at the Pennsylvania State Nucleic Acid Facility. A detailed description of the cloning is provided in the supplemental material.
Bacterial Expression and Purification of 3BC, 3BC-Y3F, 3BCD, 3Cpro, 3AB, 3Dpol, and 3C—3Dpol and 3C were purified as described previously (2). Purification procedures for wild-type 3BC, the 3BC Y3F mutant, 3BCD, 3Cpro, and 3AB represented modifications of our published protocol. A detailed description of the purification protocols is provided in the supplemental material.
Transcription and Purification of 61-nt oriI— oriI for the VPg uridylylation reactions and for the filter-binding assays was transcribed from the pUC18–61-nt oriI plasmid (2) linearized using the BstZ17I site. A complete protocol is provided in the supplemental material.
VPg Uridylylation Assays—Reactions were performed essentially as described previously (2), with slight modifications. Reaction mixtures contained 1 μm 3Dpol, 1 μm 61-nt oriI, in reaction buffer (50 mm HEPES, pH 7.5, 10% glycerol, 5 mm magnesium acetate, 10 mm β-mercaptoethanol, 10 μm UTP, and 0.04 μm [α-32P]UTP (6000 Ci/mmol)). The concentrations of 3BC, VPg, and 3C were 1 μm unless these were being titrated. All reactions were adjusted to a final NaCl concentration of 20 mm. All components were diluted to working concentrations immediately prior to use. Reactions were assembled on ice with oriI, 3C, and VPg and/or 3BC, or 3BCD in reaction buffer. The reactions were then transferred to 30 °C for 5 min and initiated by addition of 3Dpol. Reactions were incubated at 30 °C for 20 min or the indicated amount of time during a time course and quenched with an equal volume of 100 mm EDTA in 75% formamide containing 0.05% bromphenol blue dye. Quenched reactions were analyzed by using Tris-Tricine SDS-PAGE as described previously (2).
3BC Cleavage by Using 3Cpro—Processing of 3BC by 3Cpro was performed as follows. PV 3Cpro was freshly diluted to 6 μm in dilution buffer (50 mm HEPES, pH 7.5, 10% glycerol, 5 mm magnesium acetate, 10 mm β-mercaptoethanol). Following uridylylation of 3BC for 20 min as described above, 3Cpro (60 pmol, 10 μl of 6 μm) was added to a 10-μl aliquot of the uridylylation reaction. This was then incubated at 30 °C for 60 min. The reaction was quenched with an equal volume of quench dye (100 mm EDTA in 75% formamide containing 0.05% bromphenol blue), and the sample was analyzed by using Tris-Tricine SDS-PAGE as described previously (2).
RNA Filter-binding Assays—Reaction mixtures (20 μl) contained 10 nm oriI and varying concentrations of 3BC, 3C, or 3C and VPg in reaction buffer (50 mm HEPES, pH 7.5, 10% glycerol, 5 mm magnesium acetate, 10 mm β-mercaptoethanol). Binding reactions were initiated by the addition of freshly diluted 3BC (3C or 3C and VPg) to the RNA in reaction buffer. Reactions were incubated at 30 °C for 15–20 min. Membranes and Whatman 3MM paper were presoaked in equilibration buffer (50 mm HEPES, pH 7.5, and 5 mm magnesium acetate, and 10% glycerol) for 2 min and assembled, in order from top to bottom, nitrocellulose, nylon, and Whatman paper, in a slot blotter (GE Healthcare). After assembly, the binding reactions (20 μl) were loaded into the slot blotter, and vacuum was applied for 2 min at 200 mm Hg. Membranes were air-dried and visualized by using a Typhoon 8600 scanner in the storage phosphor mode and quantified by using ImageQuant software.
Construction of Mutated Replicons—Standard PCR and cloning procedures were used to generate poliovirus subgenomic replicons containing the mutation of the Gln-Gly cleavage site between 3B and 3C to Gly-Gly and for construction of the Y3F mutant with a Gly-Gly mutation between 3B and 3C. A complete description of the cloning is provided in the supplemental material.
Transcription of Subgenomic Replicons and Luciferase Assays—RNA transcripts for performing luciferase assays were generated from the pRLucRA plasmids after linearization with ApaI. Luciferase assays were performed as described previously (2) with slight modifications. A complete description of the transcription reactions and the luciferase assays is provided in the supplemental material.
Western Blot Analysis of Replicon Proteins—Cells were transfected with replicon RNA and incubated at 34 °C for 20 h. Cells were harvested and lysed, and Western blot analysis was performed as described in the supplemental material.
Northern Blot Analysis—RNA isolation for Northern blot analysis was performed as follows. HeLa cells (6 × 106) were transfected by using electroporation as described above with 25 μg of the indicated subgenomic RNAs (5 μg/1.2 × 106 cells). Cells were suspended in 30 ml of normal growth media and incubated at 34 °C. Total RNA was then isolated from the cells at the indicated times post-transfection using TRIzol reagent (Invitrogen). Briefly, cells were pelleted by centrifugation at 1000 × g for 4 min at 4 °C, washed with phosphate-buffered saline (10 ml), and lysed in TRIzol reagent (1 ml). Lysed cells were incubated at room temperature for 5 min, and chloroform (0.2 ml) was added. The samples were vortexed vigorously (15 s) and incubated at room temperature for 3 min. Samples were centrifuged at 12,000 × g for 15 min at 4 °C. Isopropyl alcohol (0.5 ml) was added to the aqueous phase for each sample and incubated at room temperature for 10 min. Following centrifugation at 12,000 × g for 10 min at 4 °C, the RNA pellet was washed with 75% ethyl alcohol (1 ml) and then dissolved in water (50 μl). Concentration was determined by measuring the absorbance at 260 nm. The quality of the RNA was assayed by agarose gel electrophoresis, and the concentration of all of the RNAs was normalized to the 18 S rRNA band by using the Typhoon 8600 scanner in the fluorescence mode.
For Northern blot analysis, total RNA (5 μg) was separated on a 0.6% agarose gel containing 0.8 m formaldehyde. The gel was then washed twice in water for 30 min each and then in 20× SSC (3 m sodium chloride, 0.3 m sodium citrate, pH 7.2) for 30 min. RNA was transferred to nylon membrane (Hybond XL, GE Healthcare) by using overnight capillary blotting using 10× SSC as the transfer buffer. RNA was cross-linked to the membrane by using a Stratalinker 2400 UV cross-linker (Stratagene). The membrane was dried and washed twice in wash buffer (1× SSC, 0.1% SDS) at 65 °C for 30 min each. Prehybridization was performed in modified Church buffer (0.5 m sodium phosphate, pH 7.2, 7% SDS, 1 mm EDTA) for 4 h at 65°C. Hybridization probes were denatured at 95 °C for 5 min and chilled on ice for 1 min prior to addition to the membrane (1 × 107 cpm). Hybridization was performed in the modified Church buffer for 16 h at 65 °C. The membrane was washed twice in wash buffer for 20 min at 65 °C and one time at room temperature. The membrane was dried, wrapped in plastic wrap, and exposed to a phosphor screen and scanned on a Typhoon 8600 scanner in the storage phosphor mode and quantified by using ImageQuant software.
Hybridization probes were made by PCR using oligonucleotides 16 and 18 (supplemental Table S1) and pRLuc as template. [α-32P]dATP (1 mCi/ml and 3000 Ci/mmol; GE Healthcare) was included in the PCR; the concentration of cold dNTPs was 3 mm for dCTP, dGTP, and dTTP and 500 μm for dATP. The quality of the PCR product was assayed by agarose gel electrophoresis. Scintillation counting was performed to determine the counts/min for the probe.
Immunoprecipitation of RNA—Total RNA was prepared, and concentration was measured as described above from HeLa cells transfected with the wild-type, Gly-Gly mutant, or Y3F/Gly-Gly mutant RNA. Total RNA was aliquoted equally into three microcentrifuge tubes (30 μg per aliquot), and IP buffer (50 mm Tris, pH 7.4, 0.5 m NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) was added to a final volume of 500 μl. Purified polyclonal anti-VPg, anti-3C, or anti-NS5A sera were added to each tube. The mixtures were rotated at room temperature for 45 min. Protein A magnetic beads (New England Biolabs, 50 μl) were conditioned by washing twice in IP buffer and then suspended in IP buffer (50 μl). These conditioned beads were added to total RNA and anti-sera mixture and incubated for 1.5 h at room temperature. The beads were collected by a magnetic stand (Dynal, Oslo, Norway) and washed four times with IP buffer followed by two times with Tris buffer (50 mm Tris, pH 7.4, 150 mm NaCl). For Northern blotting, 5 μl of water and 15 μl of formaldehyde loading dye (20 mm MOPS, 63.3% formamide, 23.3% formaldehyde, 5 mm sodium acetate, 4.3 mm EDTA, 0.066% bromphenol blue, 0.066% xylene cyanol) were added to the beads and incubated for 15 min at 65 °C. The beads were then pelleted and the entire volume loaded on a denaturing gel for Northern blot analysis as described above.
Generation and Purification of Polyclonal PV VPg, PV 3C, PV 3D, and HCV NS5A Antisera—Polyclonal antibodies raised in rabbits against viral proteins PV VPg, PV 3C-His, PV 3D, and HCV NS5A-His were purified using ammonium sulfate precipitation and DEAE-Affi-Gel blue (Bio-Rad) column purification as described in detail in the supplemental material.
HeLa/S10 Translation/Replication Reactions—Reactions were performed as described previously (22) (method 3) with the following modifications. Nuclease-treated rabbit reticulocyte lysate (10% v/v) was used rather than initiation factors for both the translation and replication reactions. [α-32P]UTP rather than [α-32P]CTP was used for the replication reactions.
Radioimmunoprecipitations were performed as follows. A 100-μl in vitro translation reaction set up in the presence of 1 mm guanidine HCl containing 50 μCi of [35S]methionine was allowed to proceed at 30 °C for 3 h after which 8 μl were removed into 80 μl of 1× SDS-PAGE sample buffer to serve as the translation control. 1 ml of RIPA buffer (50 mm Tris, pH 8, 250 mm NaCl, 1% Triton X-100, 0.1% SDS) was then added to the remainder of the reaction, and 100 μl of a 50% slurry of protein-G-agarose in RIPA buffer was added to preclear the lysate. This was rotated at room temperature for 15–30 min. The agarose was centrifuged at low speed (1000 rpm) for 1 min and the supernatant removed into a fresh tube. An aliquot of this pre-cleared lysate (150 μl) was then transferred into a fresh tube containing 3 μl of rabbit specific antisera (3AB, 3B, 3C, 3D, or preimmune antisera)4 and the samples incubated at room temperature for 15 min. 70 μl of a 50% slurry of protein G-agarose was then added, and the tubes were rotated overnight at 4 °C. The agarose was pelleted by centrifugation at low speed (1000 rpm) for 1 min, washed four times in RIPA buffer, and suspended in 50 μl of 1× SDS-PAGE sample buffer. The samples were heated to 80 °C and briefly spun prior to loading 15 μl on a gel. The gels were fixed and then treated with amplify solution before being exposed to autorad film.
RESULTS
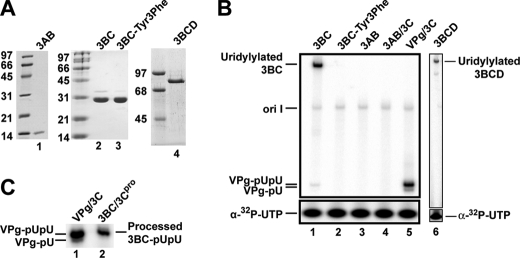
Uridylylation of VPg Precursor Proteins—Two observations in vitro point to the use of a VPg donor other than the processed VPg peptide. First, the kinetics of VPg uridylylation are slow, and the concentration of VPg required for maximal incorporation is high (2). It is generally believed that the 3AB precursor protein serves as the donor for VPg in vivo given the high concentration of this protein in virus-infected cells (1, 23). However, other precursor proteins, for example 3BC and/or 3BCD, are known to be present in picornavirus-infected cells (24). Therefore, we expressed and purified poliovirus (PV) 3AB, 3BC, and 3BCD proteins (Fig. 2A) to determine whether or not any of these proteins could serve as VPg donors in 3Dpol-catalyzed, oriI-dependent uridylylation in vitro. 3AB did not display any propensity to serve as a VPg donor (Fig. 2B, lane 3). 3C protein has been shown to function as a stimulatory factor for 3Dpol in the VPg uridylylation reaction (2). Addition of 3C to the 3AB uridylylation reaction did not alter the ability of 3AB to serve as a VPg donor (Fig. 2B, lane 4). The 3BC protein, on the other hand, was uridylylated efficiently by 3Dpol (Fig. 2B, lane 1). 3BC not only functioned as a VPg donor but also functioned as the recruitment/retention factor for 3Dpol, as this reaction did not require addition of 3C. Like 3BC, 3BCD was uridylylated in the absence of 3C (Fig. 2B, lane 6).
FIGURE 2.
3BC and 3BCD are substrates for oriI-templated uridylylation in vitro but 3AB is not. A, SDS-PAGE analysis of 2 μg of each bacterially expressed, purified protein employed in this study. 3AB, 3BC, 3BC-Y3F, and 3BCD are in lanes 1–4, respectively. B, uridylylation of 3B-containing precursors. oriI-templated uridylylation reactions were performed with VPg or the indicated 3B-containing precursors (1 μm). Reactions contained 61-nt oriI (1 μm) and proceeded for 20 min. 3C (1 μm) was present only when indicated explicitly. The positions of uridylylated 3BCD, 3BC, and VPg are indicated. C, proteolytic processing of uridylylated 3BC. Following the 20-min 3BC uridylylation reaction, 3C protease was added for 1 h and resolved on a Tris-Tricine gel to determine the extent of uridylylation (lane 2) relative to control reactions performed with VPg peptide (lane 1).
To demonstrate that the 3BC(D) uridylylation product occurred at the biologically relevant position, Tyr-3, we constructed, expressed, and purified a 3BC derivative in which Tyr-3 was changed to Phe (3BC-Y3F). The 3BC-Y3F derivative was not uridylylated by 3Dpol (Fig. 2B, lane 2).
Uridylylation of PV VPg occurs processively to form VPg-pUpU (20). Processive uridylylation is likely essential as VPg-pU does not chase into VPg-pUpU in vitro (20). In addition, a highly active 3Dpol derivative that produces much more VPg-pU than VPg-pUpU under physiological conditions is incapable of producing any RNA in cells (20). To determine the stoichiometry of 3BC uridylylation, uridylylated 3BC was cleaved in vitro by PV 3Cpro, and the uridylylated VPg product was resolved by gel electrophoresis and compared with VPg-pUpU produced in vitro from VPg peptide (Fig. 2C). Only VPg-pUpU was observed (compare lane 2 with lane 1 in Fig. 2C), suggesting that 3BC uridylylation occurs processively.
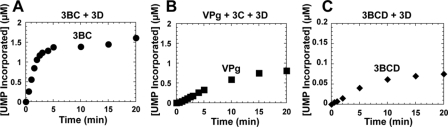
3BC Is the Optimal VPg Donor in Vitro—To compare the efficiency of 3BC(D) uridylylation to that of VPg, we evaluated the kinetics of the reactions under conditions in which 3BC(D) or VPg was present at a concentration of 1 μm, stoichiometric with 3Dpol and oriI (Fig. 3). These experiments showed that 3BC uridylylation was ∼10-fold faster than VPg uridylylation (compare Fig. 3, A to B). At least 75% of the input 3BC was uridylylated (Fig. 3A); only 25% of the VPg was uridylylated, even after an incubation time of 20 min (Fig. 3B). In contrast, the efficiency of 3BCD uridylylation was ∼10-fold lower than VPg uridylylation (compare Fig. 3, C with B). The efficiency of 3BCD uridylylation could not be improved by increasing the concentration of 3BCD employed (data not shown).
FIGURE 3.
3BC is the most efficient substrate for ori-templated uridylylation in vitro. Kinetics of 3BC (A), VPg (B), and 3BCD (C) uridylylation were performed in reactions containing 1 μm 3Dpol, 61-nt oriI template, and either 1 μm 3BC, VPg, or 3BCD. The VPg reaction contained 3C (1 μm). Rates of uridylylation were obtained from the linear portion of each curve. The rates were as follows: 0.54 ± 0.02 μm/min for 3BC (•), 0.062 ± 0.002 μm/min for VPg (▪), and 0.007 ± 0.002 μm/min for 3BCD (♦).
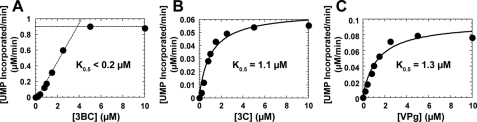
These data suggested that 3BC is the preferred VPg donor in vitro. To probe the mechanistic basis for this observation, we evaluated the 3BC concentration dependence of the rate of uridylylation (Fig. 4A). This experiment revealed a cooperative titration of 3BC under conditions in which oriI and 3Dpol were present at a concentration of 1 μm. Therefore, the K0.5 value for 3BC must be less than 0.2 μm. Importantly, the rate of 3BC was maximal at a stoichiometry of three 3BC molecules per oriI and 3Dpol. These data are consistent with two 3BC molecules binding to oriI cooperatively and with a higher affinity than 3C (Fig. 4B). A third molecule of 3BC then serves as the VPg donor. Given a maximal rate of 3BC uridylylation at a stoichiometry of three or four, the K0.5 value of 3BC for 3Dpol is likely lower than observed for VPg (Fig. 4C). Therefore, the efficiency (kcat/K0.5) of 3BC uridylylation was at least 75-fold higher than VPg uridylylation because the (3BC)2-oriI complex forms more efficiently, is more stable, and/or the VPg-donating 3BC associates with this complex more readily than VPg.
FIGURE 4.
3BC enhances uridylylation complex assembly and serves as a better VPg donor. A, 3BC concentration dependence of the rate of 3BC uridylylation. The rate of 3BC uridylylation was determined from reactions containing 1 μm 3Dpol, 61-nt oriI template, and 3BC (0–10 μm) and then plotted as a function of 3BC concentration. The K0.5 value for 3BC is estimated to be less than 0.2 μm based on the stoichiometric binding under the conditions employed (1 μm oriI and 3Dpol). The dotted lines define the two phases of the titration as determined by linear regression. The point of intersection was obtained algebraically. The intersection was 3.7 ± 0.5 μm. B, 3C concentration dependence of the rate of VPg uridylylation. The rate of VPg uridylylation was determined from reactions containing 1 μm 3Dpol, 61-nt oriI template, VPg, and 3C (0–10 μm). Rates were plotted as a function of 3C concentration and fit to a hyperbola, yielding a K0.5 value of 1.1 ± 0.04 μm for 3C. C, VPg concentration dependence of the rate of VPg uridylylation. The rate of VPg uridylylation was determined from reactions containing 1 μm 3Dpol, 61-nt oriI template, 3C, and VPg (0–10 μm). Rates were plotted as a function of VPg concentration and fit to a hyperbola, yielding a K0.5 value of 1.3 ± 0.04 μm for VPg.
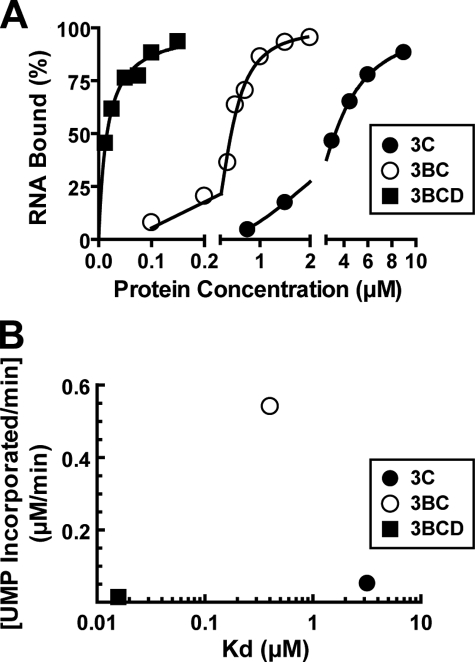
Formation and Collapse of (3C)2-oriI-VPg Complex as Rate-limiting Steps for Uridylylation—To follow up on the observation that uridylylation efficiency appeared to correlate with the stability of the 3C-containing protein-oriI complex, we used a filter-binding assay to measure the dissociation constants (Kd) for the complexes of 3C, 3BC, or 3BCD with oriI (Fig. 5A). Labeled oriI was incubated with increasing concentrations of the indicated 3C-containing protein. The binding reactions were given 15–20 min to reach equilibrium and then pulled sequentially through nitrocellulose (binds protein-RNA complex) and nylon (binds free RNA) membranes by attaching a vacuum to a slot-blot apparatus (25). The filters were visualized and quantified by using a PhosphorImager. Percentage of RNA bound was plotted as a function of 3C-containing protein concentration and fit to the appropriate model as indicated in the legend. In all cases, an end point of 80–90% was observed (Fig. 5A). The Kd values for 3C, 3BC, and 3BCD were as follows: 3 ± 0.1, 0.4 ± 0.03, and 0.02 ± 0.002 μm, respectively.
FIGURE 5.
Maximal uridylylation requires an optimal affinity for oriI implicating formation and/or collapse of the uridylylation complex as a rate-limiting step for uridylylation in vitro. A, binding of 3C, 3BC, or 3BCD to oriI. RNA binding was monitored by using a filter-binding assay. Radiolabeled RNA (1 nm) was incubated with various concentrations of 3C, 3BC, or 3BCD for 15–20 min prior to separation of protein bound and free RNA on nitrocellulose and nylon membranes, respectively, by using a slot-blot apparatus. The percentage of RNA bound was plotted as a function of protein concentration. The data for 3C (•) and 3BC (○) fit best to a Hill equation (B = BmaxPn/(Pn + K0.5n), where B is total RNA bound; P is the concentration of protein, and n is the Hill coefficient). The K0.5 values for 3C and 3BC were 3.2 ± 0.5 and 0.42 ± 0.05 μm, respectively, where n = 2. The data for 3BCD (▪) fit best to a hyperbola yielding a K0.5 value of 0.016 ± 0.002 μm. B, changes in uridylylation efficiency as a function of oriI-binding affinity. The rate of VPg/3C (•), 3BC (○), and 3BCD (▪) uridylylation plotted as a function of the K0.5 values for the interaction with oriI as determined in A for 3C, 3BC, and 3BCD.
Although previous studies demonstrated the ability of the 3D domain to contribute to RNA binding by 3C (3), the ability for the 3B domain to contribute to RNA binding by 3C is novel. An optimal Kd value exists for maximal uridylylation (Fig. 5B). The actual uridylylation reaction is bracketed by two steps, each of which can be extensively subdivided. The macro step prior to uridylylation is formation of the uridylylation complex, which includes association of the uridylylation substrate (VPg, 3BC, or 3BCD). The macro step after uridylylation is collapse of the complex, which, at a minimum, requires release of the uridylylated product. The observation of an optimal K0.5 value for maximal uridylylation efficiency suggests that formation and collapse of the 3C-containing protein-oriI complex can contribute to the rate-limiting step for uridylylation in vitro. When complex dissociation is fast (e.g. (3C)2-oriI) or slow (e.g. (3BCD)2-oriI) relative to uridylylation, then the observed steady-state rate of uridylylation will be diminished. The existence of multiple steps other than uridylylation that can alter the observed steady-state rate of uridylylation mandates caution when comparing and contrasting uridylylation in vitro and in vivo, when comparing and contrasting uridylylation in different picornaviruses, and when interrogating the biochemical mechanism by altering solution conditions and reaction components.
Use of 3B-linked Precursors for Genome Replication in Vivo—As alluded to above, the current models for picornavirus genome replication indicate that 3AB is the donor for VPg and that processed VPg is used in trans for production of VPg-pUpU, which, in turn, is employed for minus- and/or plus-strand RNA synthesis (15, 16, 26, 27). In contrast, our data are consistent with a model in which 3BC(D) is employed as the VPg donor, and formation of 3BC-pUpU would be essential for turnover. If processed VPg peptide must be produced and employed in trans for genome replication, then a genome incapable of producing VPg should be incapable of replication.
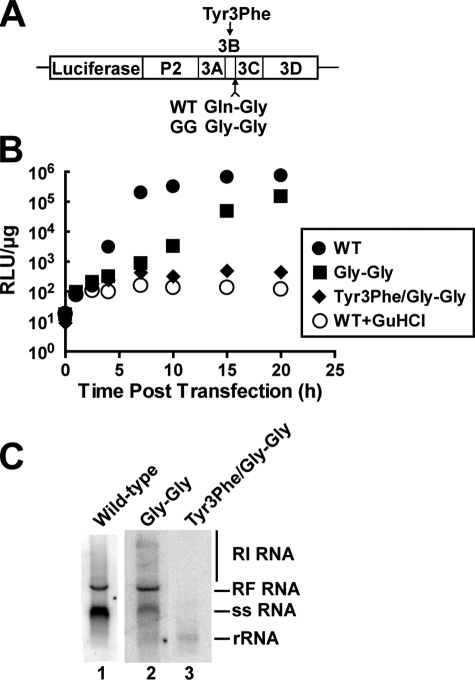
We mutated the cleavage site between 3B and 3C from Gln-Gly to Gly-Gly to preclude formation of VPg (Fig. 6A). The subgenomic replicon, pRLuc, was employed for this study as it permits RNA synthesis to be monitored indirectly by monitoring luciferase production (Fig. 6A). Here the mutated replicon will be referred to as the Gly-Gly mutant. In addition, this Gly-Gly mutation was introduced into a replicon encoding a substitution of the tyrosine nucleophile of VPg to phenylalanine to ensure that any RNA produced is initiated by using VPg instead of some alternative, irrelevant mechanism (Fig. 6A). This replicon will be referred to as the Y3F/Gly-Gly mutant. These replicons were electroporated into HeLa cells and maintained at 37 or 34 °C; luciferase activity was monitored for 20 h post-transfection (Fig. 6B). At both temperatures, the Gly-Gly replicon was capable of replication (Fig. 6B and data not shown). At 37 °C, the Gly-Gly mutant replicated to within 15-fold of wild-type replicon (data not shown); at 34 °C, the Gly-Gly mutant replicated to within 5-fold of wild-type replicon (Fig. 6B). In all cases, Gly-Gly mutant replication employed VPg priming as the Y3F/Gly-Gly mutant did not display any significant increase in luciferase activity over that attributable to translation of the input RNA. The kinetics of luciferase production for this mutant was on par with that observed for wild-type replicon in the presence of 3 mm guanidine hydrochloride, a replication inhibitor for poliovirus (Fig. 6B). These data suggest that processed VPg is not essential for genome replication.
FIGURE 6.
Release of 3B(VPg) from the polyprotein is not essential for genome replication in HeLa cells or cell-free extracts, providing evidence for the use of precursor forms of 3B in vivo. A, schematic of PV subgenomic replicon. The Gln-Gly (or QG) processing site between 3B and 3C in wild-type (WT) was changed to Gly-Gly (or GG) to prevent release of 3B peptide. The Gly-Gly junction was also introduced into a replicon encoding a 3B with a substitution of Tyr-3 to Phe. Tyr-3 is the nucleophile employed to form VPg-pU. B, replication of in vitro transcribed replicon RNA in HeLa cells: wild type (•), Gly-Gly mutant (▪), and Y3F/Gly-Gly mutant (♦). As a control, wild-type replicon RNA was evaluated in the presence of guanidine hydrochloride, an inhibitor of replication (○). HeLa cells were transfected with in vitro transcribed replicon RNA, placed at 34 °C, and luciferase activity was monitored for 20 h post-transfection. C, replication of in vitro transcribed replicon RNA in HeLa cell-free extracts. HeLa S10 extracts containing [α-32P]UTP were primed with wild-type, Gly-Gly, or Y3F/Gly-Gly subgenomic replicon RNA and then analyzed by 1% native agarose gel electrophoresis. Shown is a phosphorimage after a 2-day exposure for wild-type (lane 1) and after a 3-day exposure for Gly-Gly mutant (lane 2) and Y3F/Gly-Gly mutant (lanes 3). The three forms of newly synthesized RNA products (replicative form (RF), replicative intermediate (RI), and single-stranded, genomic RNA (ss)) are indicated. Ribosomal RNA (rRNA) was used as a loading control. RLU, relative light units.
Given the reduced kinetics and temperature sensitivity of Gly-Gly mutant replication, it was possible that a revertant was selectively amplified in the cell-based experiments. It is important to note that this possibility has never been documented. Nevertheless, we evaluated RNA synthesis of mutant replicons by using a cell-free system (22) that would preclude this possibility. Briefly, HeLa S10 extracts containing [α-32P]UTP were programmed with the mutated replicon RNAs; reaction products were resolved by agarose gel electrophoresis and visualized by autoradiography. Reaction products include the following: single-stranded, genomic RNA (ssRNA in Fig. 6C); minus-strand RNA product hybridized to plus-strand template, the so-called replicative form RNA (RF RNA in Fig. 6C); and minus-strand RNA template harboring multiple copies of nascent plus-strand RNA of various lengths, the so-called replicative intermediate (RI RNA in Fig. 6C). The Gly-Gly mutant was capable of replicating as all expected reaction products were observed (lane 2 of Fig. 6C). The level of Gly-Gly mutant replication was reduced relative to wild-type replicon (compare lane 2 to lane 1 in Fig. 6C). Notably, wild-type RNA could be observed after a 2-day exposure; however, detection of mutant RNA required a 3-day exposure. Therefore, the Gly-Gly mutant may be more debilitated in RNA synthesis than evident from monitoring luciferase activity. Interestingly, the ratio of plus-strand RNA:minus-strand RNA (ssRNA:RF RNA) was reduced ∼2-fold. The significance of this observation is unclear. RNA synthesis by the Gly-Gly mutant required VPg priming as the Y3F/Gly-Gly mutant failed to produce any RNA (lane 3 of Fig. 6C). These data provide further support for the notion that processed VPg is not essential for genome replication.
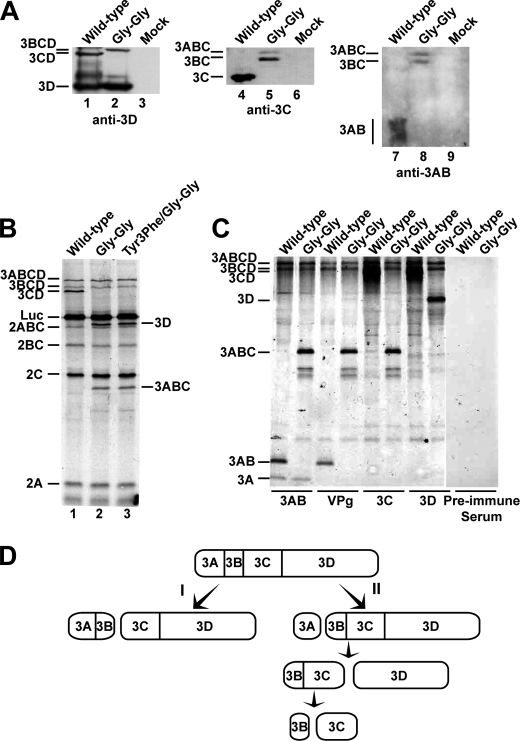
To evaluate processing of 3B-containing precursors directly, cells were transfected with wild-type or Gly-Gly mutant replicons and harvested at 20 h post-transfection. Lysates were analyzed by Western blotting using polyclonal antibodies raised against poliovirus 3D, 3C, and 3AB proteins (Fig. 7A). Wild-type replicon produced 3CD and 3D; however, the Gly-Gly mutant replicon failed to produce 3CD (anti-3D in Fig. 7A). Wild-type replicon produced 3C; however, the Gly-Gly mutant replicon failed to produce 3C but accumulated 3ABC and 3BC (anti-3C in Fig. 7A). Finally, wild-type replicon produced 3AB, but the Gly-Gly mutant replicon failed to produce 3AB but produced 3ABC and 3BC instead (anti-3AB in Fig. 7A). We conclude that cryptic processing events leading to production of processed VPg that could account for RNA synthesis are unlikely.
FIGURE 7.
Analysis of polyprotein processing by the Gly-Gly mutant reveals the existence of two independent pathways for P3 precursor processing. A, processing evaluated by Western blotting. HeLa cells were transfected with wild-type or Gly-Gly mutant RNA, placed at 34 °C, harvested 20 h post-transfection, and extracts prepared and processed for Western blotting as described under “Experimental Procedures.” Antisera against 3Dpol, 3C, and 3AB were employed. The bands corresponding to the different precursor and processed proteins are indicated. B, processing evaluated by cell-free translation. HeLa cell-free translation extracts containing [35S]methionine and [35S]cysteine were programmed with wild-type, Gly-Gly mutant, or Y3F/Gly-Gly mutant RNA. Radiolabeled proteins were separated by 15% SDS-PAGE and detected by phosphorimaging. The bands corresponding to the different precursor and processed proteins expected for wild-type replicon are indicated on the left. Luc denotes the luciferase reporter. The identity of bands unique to the mutant are indicated on the right. C, assignment of bands unique to the Gly-Gly mutants by immunoprecipitation. Products from cell-free translation reactions of wild-type and Gly-Gly mutant replicon RNAs were immunoprecipitated using antiserum raised against 3AB, VPg, 3C, or 3D protein; preimmune serum was employed as a negative control. The precipitated proteins were separated by 15% SDS-PAGE and detected by phosphorimaging. The location of the various precursor and processed proteins are indicated on the left. D, processing of the P3 precursor occurs by two independent pathways. There are major (I) and minor (II) pathways. In pathway I, processing between 3B and 3C yields 3AB and 3CD. In pathway II, processing between 3A and 3B yields 3A and 3BCD. 3BCD processing yields 3BC and 3D; 3BC processing yields 3B and 3C. Pathway II is proposed to function in genome replication and is not perturbed in the Gly-Gly mutant.
The observation that the Gly-Gly mutant replicon failed to produce 3C and 3CD was expected (Fig. 7A). However, this observation highlighted the possibility that the reduced kinetics and temperature sensitivity of Gly-Gly mutant replication could be attributable to incomplete polyprotein processing as dogma states that 3C and 3CD are the proteases that function in the infected cell (1, 23). To address this issue, it was important to use an approach that would be as unbiased as possible. Therefore, we again turned to the cell-free system. HeLa S10 extracts containing 35S-Met and 35S-Cys were programmed with wild-type or mutant replicon RNA. Translated products were resolved by denaturing PAGE and visualized by autoradiography. Surprisingly, the processing profile of the mutant polyproteins (lanes 2 and 3 of Fig. 7B) was very similar to the wild-type polyprotein (lane 1 of Fig. 7B). The key difference was that two new products appeared that were consistent with processed 3D and the 3ABC precursor (lanes 2 and 3 of Fig. 7B). We conclude that precursor forms of 3C other than 3CD are capable of processing the P2-P3 polyprotein.
To identify the bands that were unique to the Gly-Gly mutant polyprotein, products present in translation reactions were immunoprecipitated with antiserum raised against 3AB, VPg, 3C, or 3D; preimmune serum was employed as a negative control (Fig. 7C). The pulldown experiments were consistent with the unique bands being 3D (precipitated only by anti-3D serum) and 3ABC (precipitated by all sera except the anti-3D serum) (Fig. 7C).
Unexpectedly, this experiment revealed the existence of two independent processing pathways for the P3 (3ABCD) precursor, and only one of these was altered by impairing cleavage at the 3B-3C junction. Processing of P3 via pathway I was the major pathway and produces 3AB and 3CD (Fig. 7D). 3AB and 3CD were the primary products of P3 cleavage observed for wild-type replicon (Fig. 7, B and C). Pathway II is the minor pathway and produces 3A and 3BCD (Fig. 7D). 3A and 3BCD are the second-most abundant products of P3 cleavage observed for wild-type replicon (Fig. 7, B and C). This minor pathway may also give rise to 3C and 3D (the active form of the polymerase) (Fig. 7D) as high levels of these proteins could not be detected for wild-type replicon by using metabolic labeling or immunoprecipitation (Fig. 7, B and C). The observation that PV multiplication becomes resistant to cycloheximide 3 h post-infection suggests that very little 3Dpol is required for replication (28).
In this model, pathway II would yield VPg, originating from 3BC rather than 3AB (Fig. 7D). Pathway II was unchanged for the Gly-Gly mutant replicon as the levels of 3A and 3BCD were unchanged relative to wild-type replicon (Fig. 7C). In contrast, inactivation of the 3B-3C cleavage site led to activation of the 3C-3D cleavage site, yielding 3ABC and 3D (Fig. 7, B and C). Although it is possible that 3ABC performs all of the function of 3AB, the literature would suggest 3D and 3CD have distinct, nonoverlapping functions. The reduced kinetics and temperature sensitivity of genome replication may be caused by the absence of 3CD functions.
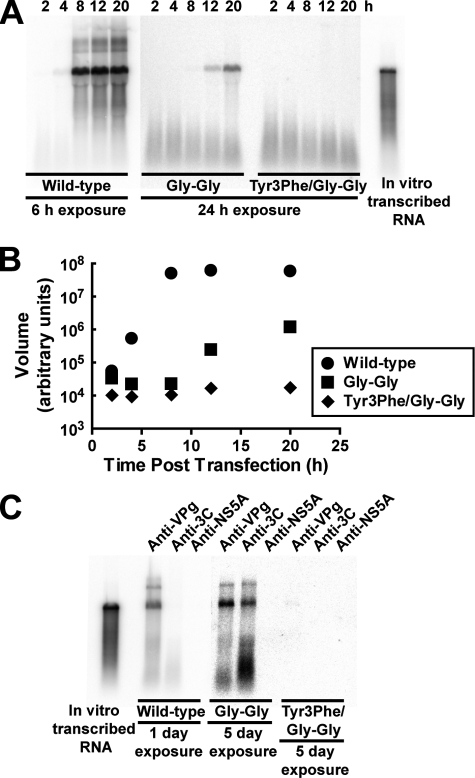
Gly-Gly Mutant Produces Full-length, 3BC-linked RNA—The use of luciferase activity as a measure of RNA synthesis by a mutant replicon does not establish unambiguously that the RNA produced is full length or that the kinetics of RNA synthesis are truly equivalent to wild-type replicon. Therefore, we isolated total RNA from cells transfected with wild-type, Gly-Gly mutant, or Y3F/Gly-Gly mutant replicon as a function of time post-transfection. RNA was run on a denaturing agarose gel, transferred to a nylon membrane, and probed with a 32P-labeled PCR product representing a portion of the 3D gene. Hybridized probe was visualized and quantified by phosphorimaging. The Gly-Gly mutant produced full-length RNA that was dependent on VPg priming (compare 3B-3C-Gly-Gly with 3BC-Gly-Gly-Y3F in Fig. 8A). Surprisingly, the kinetics of RNA synthesis appeared slower, and the final yield of RNA appeared lower than anticipated based on the luciferase activity for the Gly-Gly mutant relative to wild type. Quantitation of the Northern blot showed that the Gly-Gly mutant exhibited a longer lag phase than observed by monitoring luciferase activity (compare Fig. 8B to Fig. 6B). In addition, the final yield of Gly-Gly mutant RNA was 30-fold lower than observed for wild-type RNA (Fig. 8B). This value is 6-fold lower than observed by monitoring luciferase activity (compare Fig. 8B to Fig. 6B). These data suggest that each plus-strand, Gly-Gly-mutant RNA molecule synthesized is translated for a longer time than each wild-type RNA molecule. We conclude that the Gly-Gly mutant exhibits a defect in the switch from translation to replication.
FIGURE 8.
RNA produced by the Gly-Gly mutant is full length and covalently linked to 3BC. A, kinetics of RNA synthesis by wild-type, Gly-Gly mutant, and Y3F/Gly-Gly mutant monitored by Northern blotting. HeLa cells were transfected with in vitro transcribed replicon RNA and placed at 34 °C. At the indicated times, total RNA was isolated from transfected HeLa cells, separated on a 0.6% agarose gel containing 0.8 m formaldehyde, transferred to nylon membrane, and hybridized with a 32P-labeled DNA probe. The hybridized DNA probe was visualized by phosphorimaging. Shown is a phosphorimage after a 6-h exposure for wild-type and after a 24-h exposure for Gly-Gly and Y3F/Gly-Gly mutants. In vitro transcribed RNA is shown as a reference. B, quantification of data shown in A: wild-type (•), Gly-Gly mutant (GG) (▪), and Y3F/Gly-Gly mutant (Y3F/GG) (♦). C, interrogation of protein linkage to wild-type and Gly-Gly mutant RNA by RNA immunoprecipitation and Northern blotting. HeLa cells were transfected with wild-type, Gly-Gly mutant, or Y3F/Gly-Gly mutant RNA and placed at 34 °C. Total RNA was isolated from transfected HeLa cells and immunoprecipitated using antibodies against VPg, 3C, or HCV NS5A (as a negative control). The immunoprecipitated RNA was detected by Northern blotting as described above. Shown is a phosphorimage after a 1-day exposure for wild-type and after a 5-day exposure for Gly-Gly mutant and Y3F/Gly-Gly mutant.
All of the data accumulated to this point were consistent with a 3B(VPg)-containing precursor being employed for Gly-Gly mutant RNA synthesis. To identify the precursors associated with Gly-Gly mutant RNA, total RNA was immunoprecipitated by using antisera raised against PV VPg(3B), 3C, 3Dpol, or hepatitis C virus NS5A (as a negative control). RNA was processed for Northern blotting as described above. The Gly-Gly mutant RNA was immunoprecipitated by anti-VPg and anti-3C sera (Fig. 8C) but not by anti-3D (data not shown) or anti-NS5A serum (Fig. 8C). Importantly, only anti-VPg serum was capable of immunoprecipitating wild-type RNA (Fig. 8C). A signal could not be detected with any antiserum when Y3F/Gly-Gly mutant-transfected cells were the source of the total RNA employed for the experiment. Therefore, this experiment is detecting replicated RNA not transfected RNA. We conclude that Gly-Gly mutant RNA is covalently linked to the 3BC precursor protein.
DISCUSSION
The 5′ ends of picornaviral plus- and minus-strand RNAs are linked covalently to a short peptide termed VPg (virion protein genome-linked) encoded by the 3B region of the genome. It has been thought for a long time that VPg-RNA is produced in two steps as follows: 1) production of VPg-pUpU from VPg; and 2) production of VPg-RNA from VPg-pUpU (1, 23). The discovery that VPg uridylylation is templated by a cis-acting replication element (CRE) located at an internal position (oriI) rather than the 3′-poly(rA) tail further complicated the mechanism of VPg-RNA synthesis as VPg-pUpU needed to be transferred from the internal site of uridylylation to the 3′-terminal site for elongation (7). Whether VPg or some precursor thereof is uridylylated is not clear. Most models suggest that the viral 3AB protein is either uridylylated or the source of VPg that goes on to be uridylylated (Fig. 9) (15, 16, 26, 27). However, it is possible that precursors other than 3AB serve as substrates for uridylylation, for example 3BC or 3BCD (Fig. 9). The use of larger precursors like 3BCD provides a mechanism for transfer. For example, 3BCD bound to the CRE at the 3′ end of the genome (oriR) could be used for uridylylation at oriI, and release from the uridylylation complex would return uridylylated 3BCD to the 3′ end for extension and production of full-length RNA (Fig. 9). oriR has been shown to influence the start site for minus-strand synthesis in Coxsackievirus B3 (29). The proposed organization of the uridylylation complex predicts that use of this complex will always be in trans (Fig. 9) (5). However, the origin of 3BCD-CRE employed for uridylylation could be intra- (Fig. 9) or intermolecular. An intermolecular origin would permit 3BCD bound to a CRE located in the minus-strand to use this complex for plus-strand synthesis, consistent with the known requirement of oriI for plus-strand synthesis (11, 15, 16). Therefore, knowledge of the precursor employed for uridylylation has significant implications for mechanism(s) employed for production of picornaviral VPg-linked RNA.
FIGURE 9.
Hypothetical model for use of precursors bound to 3′ end of plus- or minus-strand RNA for uridylylation and production of full-length RNA. Assembly and organization of the uridylylation complex are essentially as indicated in Fig. 1B. This study demonstrates that 3CD can be replaced by 3BCD in vitro and in vivo, forming a more stable interaction with oriI. Two 3BCD molecules bind to oriI to form the (3BCD)2-oriI complex (step 1) that isomerizes (step 2) to form the complex competent for recruitment of and utilization by 3Dpol (step 3). The VPg donor associates with the complex. Three possible VPg donors have been shown in this study as follows: VPg, 3BC, and 3BCD. Use of 3BCD is the preferred hypothesis because this protein can bind to the 3′ end of plus- and minus-strand RNA and join the uridylylation complex via an intra- or intermolecular interaction (step 4; data not shown). 3BCD uridylylation would occur (step 5; data not shown) and the uridylylated 3BCD would be released and returned to the 3′ end for extension (step 6). The mechanism for elongation, timing of 3BCD proteolysis relative to RNA elongation, etc. remain unexplored.
As indicated above, most models for picornavirus genome replication show 3AB as the VPg-containing precursor protein contributing to uridylylation (1, 15, 16, 23, 26, 27). This conclusion appears to be based solely on the abundance of this protein in virus-infected cells. Although 3AB uridylylation has been shown to occur in the presence of Mn2+ (30), we were unable to observe 3AB uridylylation in the presence of Mg2+ (lanes 3 and 4 of Fig. 2B), a condition supporting robust VPg uridylylation (lane 5 of Fig. 2B). Mn2+ is seldom the preferred divalent cation employed by polymerases in vivo and is known to substantially relax substrate specificity of PV 3Dpol (31, 32). In contrast to 3AB, both 3BC and 3BCD were uridylylated in the presence of Mg2+ (lanes 1 and 6 of Fig. 2B). These observations are consistent with a recent structure for the 3Dpol-VPg complex from foot-and-mouth disease virus that located the VPg amino terminus in the polymerase active site (33). 3AB would not be able to locate its VPg domain in the 3Dpol active site without reorganizing the active site in a manner that would be predicted to substantially reduce nucleotidyl transfer activity. Worth noting, foot-and-mouth disease virus 3BC is also uridylylated (34). These data are consistent with 3BC(D) being employed for uridylylation instead of VPg.
Inhibiting processing between picornaviral 3A and 3B proteins is lethal for genome replication (26); however, inhibiting processing between 3B and 3C is not (Gly-Gly mutant, Fig. 6) (35). The kinetics of Gly-Gly mutant RNA synthesis was temperature-sensitive and slow relative to wild type (Fig. 6 and Fig. 8, A and B). Importantly, 3BC-linked RNA was clearly made (Fig. 8C). 3BCD-linked RNA was not observed (data not shown). Together, these data support a model in which 3A-3B processing occurs to form 3BCD, which is subsequently uridylylated. Extension of 3BC(D)-pUpU leads to 3BC-RNA, which in turn leads to VPg-RNA. Therefore, proteolytic processing of the P3 precursor may represent an important mechanism for regulating the transition from initiation to elongation during RNA synthesis and the transition from genome replication to genome encapsidation. Infectious virus particles were not observed for the Gly-Gly mutant (data not shown). These data strongly support the ability of P3 precursors containing 3B at the amino terminus to be uridylylated in vivo and rule out the requirement for processed VPg.
We observed that the uridylylation efficiency of 3BC was ∼10-fold greater than VPg and that the uridylylation efficiency of VPg was ∼10-fold greater than 3BCD (Fig. 3). 3BC was a better oriI-binding protein than 3C (Fig. 5) as well as a better VPg donor than the VPg peptide (Fig. 4). These data suggest that 3B contributes to the RNA-binding affinity of 3C. It has been shown that 3AB is an RNA-binding protein and the RNA-binding determinant maps to 3B as 3A lacks demonstrable RNA binding activity (36, 37). The carboxyl-terminal two-thirds of picornaviral 3B proteins contain several basic amino acid residues (38). Therefore, 3B may interact with the phosphodiester backbone of oriI in or around the loop. 3B also confers increased binding affinity to 3CD as the Kd value for 3BCD binding to oriI is 10-fold lower than that reported by us previously for 3CD (Fig. 5) (3). The ability for 3C to contribute to the affinity of VPg binding to the uridylylation complex could be facilitated by 3BC-3C and/or 3BC-3D interaction. In this case, the 3B domain would direct binding to 3Dpol, and the 3C domain would increase retention time of 3B in the uridylylation complex by interacting with 3C and/or 3D domains of 3CD. The structure of PV 3CD identified numerous possibilities for 3C-3D interaction surfaces that are independent of those that contribute to formation of the (3C)2-3Dpol core of the uridylylation complex (39). Retention of 3BCD in the uridylylation complex may be enhanced over that observed for 3BC by using its 3D domain for additional interactions with components of the uridylylation complex.
Studies of oriI-templated VPg uridylylation in vitro are generally performed under steady-state conditions, i.e. the concentration of VPg is much greater than the other components of the reaction; multiple turnovers occur (2–5, 7–9, 11, 12, 14, 34, 38, 40, 41). The rate of VPg uridylylation under standard conditions is too slow to support the rate of replication observed in vivo (3, 21). Unfortunately, the rate-limiting step(s) for VPg uridylylation is not known, thus precluding an explanation for the “inefficiency” of the reaction. The existence of an optimal equilibrium dissociation constant of the 3C-containing protein-oriI complex (Fig. 5B) for the maximal rate of uridylylation suggests that both formation and collapse of the uridylylation complex contribute to the rate-limiting step. The observation of an inverse correlation between uridylylation efficiency and the K0.5 value for the VPg donor (Fig. 4) suggests that the kinetics of release of the VPg donor and/or its uridylylated products contribute to the rate-limiting step. We propose that the reduced affinity of 3C for oriI relative to 3C-containing precursors would cause formation and/or collapse of the (3C)2-oriI complex to both partially limit the rate of VPg uridylylation at concentrations of VPg well above its K0.5 value. We propose that the increased affinity of 3BC(D) for the uridylylation complex when serving as the VPg donor would cause 3BC(D) release to limit the rate of VPg uridylylation. Release of 3BCD would be slower than 3BC for the reason provided above.
Slow turnover in the steady state that is driven by product release is the paradigm for a processive, polymerase-catalyzed reaction (32). Consistent with product release limiting turnover of 3BC is the finding that 3BC uridylylation was more processive than VPg uridylylation (Fig. 2C). The inability to chase VPg-pU into VPg-pUpU mandates a processive reaction. Release of 3BCD-pUpU would be slower than 3BC due to the increased interaction of this protein with the uridylylation complex as discussed above. It would be most advantageous for release of the uridylylated product to be coupled to transfer to the 3′ end of plus- or minus-strand RNA. We envisage two possible mechanisms for this coupling. It is possible that binding of oriR in the 3′ end of plus-strand RNA or the functional equivalent in the 3′ end of minus-strand RNA stimulates release. Alternatively, it is possible that proteolysis of the uridylylated 3BCD protein bound to the 3′ end of plus- or minus-strand RNA to remove 3D stimulates release. We conclude that the lower-than-expected uridylylation efficiency observed in vitro may be biologically relevant.
Our studies were consistent with the existence of two independent pathways for P3 polyprotein processing (Fig. 7, B and D). The major pathway produced 3AB and 3CD. Impaired cleavage between 3B and 3C activated cleavage between 3C and 3D, yielding 3ABC and 3D (Fig. 7, B and C). 3CD is highly resistant to cleavage when present in its normal location, the cytoplasm and nucleoplasm (1). 3AB is targeted to membranes (1). If P3 is cleaved into 3AB and 3CD in the cytoplasm, followed by movement of 3AB to membranes, then inhibition of 3B-3C cleavage would take the intact precursor to membranes. Perhaps in this environment, the conformation of 3CD is altered in a manner that leads to 3C-3D cleavage. The minor pathway produced 3A and 3BCD; this pathway was not perturbed (Fig. 7, B and C). Under normal conditions, processed 3D is difficult to detect (Fig. 7, B and C), suggesting that processed 3D originates from the minor pathway. Only processed 3D would exhibit polymerase activity (39, 42). We propose that the minor pathway is involved in producing all of the proteins that participate directly in genome replication, including the VPg donor employed for uridylylation and RNA synthesis. The major pathway would then be involved in the virus-host functions of 3AB and 3CD (43–45). It is not clear what determines whether a P3 precursor is processed by the major or minor pathway. The major pathway could represent a default pathway that is bypassed by interaction of the P3 precursor with viral RNA. Additional studies will be required to address this question.
Semler and co-workers (46) have also suggested two independent processing pathways for the P3 precursor based on pulse-chase experiments. Their model is consistent with ours in that processed 3C and 3D do not originate from 3CD.
A recent study by Wimmer and co-workers (26) has shown that a P3 precursor encoding the uridylylation inactivating Y3F mutation in 3B (3AB*CD, where the asterisk denotes the mutant 3B) cannot be rescued by creating a 3BAB*CD precursor but can be rescued by creating a 3ABAB*CD precursor. This study revealed a function for Tyr-3 in the context of 3AB for formation of a complex with 3CD or 3Dpol that is required for some unknown aspect of genome replication. Given the requirement for two distinct functions for Tyr-3 in genome replication, this mutant allele could not be used to prove the VPg donor employed in vivo. However, cleavage between the amino-terminal 3AB of 3ABAB*CD was absolutely essential for genome replication, consistent with our hypothesis that a 3BCD precursor is employed in vivo. In the case of the Wimmer mutant, this precursor would be 3BAB*CD. A prediction of this hypothesis is that 3ABAB*CD incapable of releasing the amino-terminal 3AB should be complemented in trans by 3BAB*CD.
The ability to produce any RNA in the absence of 3CD is surprising. 3CD is thought to be required for most (if not all) polyprotein-processing events (1). However, P2 and P3 processing was as robust as wild type (Fig. 7). Perhaps precursors larger than 3CD function in vivo. This possibility would provide an alternative explanation for the very low cleavage efficiency of both 3C and 3CD in vitro (47). 3CD is more robust at P1 (capsid) precursor processing. Therefore, the inability to recover virus from mutated genomic RNA (data not shown) could be attributed to the absence of 3CD. 3CD is also implicated in the switch from translation to replication (48–50). The Gly-Gly mutant may be “switch-defective.” There was an incongruence between the kinetics of luciferase production (Fig. 6B) and the kinetics of RNA synthesis (Fig. 8) assessed by Northern analysis that was consistent with enhanced translation of the mutant replicon.
In conclusion, this study provides very compelling evidence for use of a 3B(VPg)-containing precursor instead of 3AB for uridylylation and RNA synthesis in vivo. A unique pathway appears to exist for processing of the P3 to produce 3B-containing precursors functioning in genome replication. Use of this 3B-containing precursor will ensure processive uridylylation, as well as permit regulated, coupled transfer of the uridylylated product to the 3′ end of either plus- or minus-strand RNA. This model represents a paradigm shift relative to existing models of picornavirus genome replication. Therefore, future studies must scrutinize this model and determine the extent to which it applies to picornaviruses other than poliovirus.
Supplementary Material
Acknowledgments
We thank Saikat Kumar B. Ghosh for purifying the 3AB protein, Zachary J. Reitman for purifying active PV 3C protease, and Mehul Suthar for construction of the pET26Ub-N-His plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grant AI053531 (NIAID research grant) (to C. E. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, additional references, and Table S1.
Footnotes
The abbreviations used are: PV, poliovirus; NTR, nontranslated region; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; MOPS, 4-morpholinepropanesulfonic acid; nt, nucleotide; CRE, cis-acting replication element.
I. G. Goodfellow, unpublished results.
References
- 1.Semler, B. L., and Wimmer, E. (eds) (2002) Molecular Biology of Picornaviruses, American Society for Microbiology, Washington, DC
- 2.Pathak, H. B., Ghosh, S. K., Roberts, A. W., Sharma, S. D., Yoder, J. D., Arnold, J. J., Gohara, D. W., Barton, D. J., Paul, A. V., and Cameron, C. E. (2002) J. Biol. Chem. 277 31551-31562 [DOI] [PubMed] [Google Scholar]
- 3.Pathak, H. B., Arnold, J. J., Wiegand, P. N., Hargittai, M. R., and Cameron, C. E. (2007) J. Biol. Chem. 282 16202-16213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen, M., Wang, Q., Yang, Y., Pathak, H. B., Arnold, J. J., Castro, C., Lemon, S. M., and Cameron, C. E. (2007) J. Virol. 81 12485-12495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen, M., Reitman, Z. J., Zhao, Y., Moustafa, I., Wang, Q., Arnold, J. J., Pathak, H. B., and Cameron, C. E. (2008) J. Biol. Chem. 283 875-888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paul, A. V., van Boom, J. H., Filippov, D., and Wimmer, E. (1998) Nature 393 280-284 [DOI] [PubMed] [Google Scholar]
- 7.Paul, A. V., Rieder, E., Kim, D. W., van Boom, J. H., and Wimmer, E. (2000) J. Virol. 74 10359-10370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerber, K., Wimmer, E., and Paul, A. V. (2001) J. Virol. 75 10979-10990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang, Y., Rijnbrand, R., McKnight, K. L., Wimmer, E., Paul, A., Martin, A., and Lemon, S. M. (2002) J. Virol. 76 7485-7494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mason, P. W., Bezborodova, S. V., and Henry, T. M. (2002) J. Virol. 76 9686-9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Ooij, M. J., Vogt, D. A., Paul, A., Castro, C., Kuijpers, J., van Kuppeveld, F. J., Cameron, C. E., Wimmer, E., Andino, R., and Melchers, W. J. (2006) J. Gen. Virol. 87 103-113 [DOI] [PubMed] [Google Scholar]
- 12.Rieder, E., Paul, A. V., Kim, D. W., van Boom, J. H., and Wimmer, E. (2000) J. Virol. 74 10371-10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paul, A. V., Yin, J., Mugavero, J., Rieder, E., Liu, Y., and Wimmer, E. (2003) J. Biol. Chem. 278 43951-43960 [DOI] [PubMed] [Google Scholar]
- 14.Yin, J., Paul, A. V., Wimmer, E., and Rieder, E. (2003) J. Virol. 77 5152-5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray, K. E., and Barton, D. J. (2003) J. Virol. 77 4739-4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morasco, B. J., Sharma, N., Parilla, J., and Flanegan, J. B. (2003) J. Virol. 77 5136-5144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thiviyanathan, V., Yang, Y., Kaluarachchi, K., Rijnbrand, R., Gorenstein, D. G., and Lemon, S. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 12688-12693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang, Y., Rijnbrand, R., Watowich, S., and Lemon, S. M. (2004) J. Biol. Chem. 279 12659-12667 [DOI] [PubMed] [Google Scholar]
- 19.Amero, C. D., Arnold, J. J., Moustafa, I. M., Cameron, C. E., and Foster, M. P. (2008) J. Virol. 82 4363-4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korneeva, V. S., and Cameron, C. E. (2007) J. Biol. Chem. 282 16135-16145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnold, J. J., and Cameron, C. E. (2000) J. Biol. Chem. 275 5329-5336 [DOI] [PubMed] [Google Scholar]
- 22.Barton, D. J., Black, E. P., and Flanegan, J. B. (1995) J. Virol. 69 5516-5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Racaniello, V. R. (2007) in Fields Virology (Knipe, D. M., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B., and Straus, S. E., eds) 4th Ed., pp. 795-838, Lippincott-Raven Publishers, Philadelphia
- 24.Koch, F., and Koch, G. (1985) The Molecular Biology of Poliovirus, pp. 311-371, Springer-Verlag, Inc., New York
- 25.Wong, I., and Lohman, T. M. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 5428-5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, Y., Franco, D., Paul, A. V., and Wimmer, E. (2007) J. Virol. 81 5669-5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao, X., and Wimmer, E. (1996) EMBO J. 15 23-33 [PMC free article] [PubMed] [Google Scholar]
- 28.Baltimore, D., Girard, M., and Darnell, J. E. (1966) Virology 29 179-189 [DOI] [PubMed] [Google Scholar]
- 29.van Ooij, M. J., Polacek, C., Glaudemans, D. H., Kuijpers, J., van Kuppeveld, F. J., Andino, R., Agol, V. I., and Melchers, W. J. (2006) Nucleic Acids Res. 34 2953-2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards, O. C., Spagnolo, J. F., Lyle, J. M., Vleck, S. E., Kuchta, R. D., and Kirkegaard, K. (2006) J. Virol. 80 7405-7415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnold, J. J., Ghosh, S. K., and Cameron, C. E. (1999) J. Biol. Chem. 274 37060-37069 [DOI] [PubMed] [Google Scholar]
- 32.Arnold, J. J., Gohara, D. W., and Cameron, C. E. (2004) Biochemistry 43 5138-5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrer-Orta, C., Arias, A., Agudo, R., Perez-Luque, R., Escarmis, C., Domingo, E., and Verdaguer, N. (2006) EMBO J. 25 880-888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nayak, A., Goodfellow, I. G., Woolaway, K. E., Birtley, J., Curry, S., and Belsham, G. J. (2006) J. Virol. 80 9865-9875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall, D. J., and Palmenberg, A. C. (1996) J. Virol. 70 5954-5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lama, J., Sanz, M. A., and Rodriguez, P. L. (1995) J. Biol. Chem. 270 14430-14438 [DOI] [PubMed] [Google Scholar]
- 37.Kusov, Y. Y., Morace, G., Probst, C., and Gauss-Muller, V. (1997) Virus Res. 51 151-157 [DOI] [PubMed] [Google Scholar]
- 38.Paul, A. V., Peters, J., Mugavero, J., Yin, J., van Boom, J. H., and Wimmer, E. (2003) J. Virol. 77 891-904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marcotte, L. L., Wass, A. B., Gohara, D. W., Pathak, H. B., Arnold, J. J., Filman, D. J., Cameron, C. E., and Hogle, J. M. (2007) J. Virol. 81 3583-3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nayak, A., Goodfellow, I. G., and Belsham, G. J. (2005) J. Virol. 79 7698-7706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boerner, J. E., Lyle, J. M., Daijogo, S., Semler, B. L., Schultz, S. C., Kirkegaard, K., and Richards, O. C. (2005) J. Virol. 79 7803-7811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson, A. A., and Peersen, O. B. (2004) EMBO J. 23 3462-3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doedens, J. R., and Kirkegaard, K. (1995) EMBO J. 14 894-907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wessels, E., Duijsings, D., Niu, T. K., Neumann, S., Oorschot, V. M., de Lange, F., Lanke, K. H., Klumperman, J., Henke, A., Jackson, C. L., Melchers, W. J., and van Kuppeveld, F. J. (2006) Dev. Cell 11 191-201 [DOI] [PubMed] [Google Scholar]
- 45.Belov, G. A., and Ehrenfeld, E. (2007) Cell Cycle 6 36-38 [DOI] [PubMed] [Google Scholar]
- 46.Lawson, M. A., and Semler, B. L. (1992) Virology 191 309-320 [DOI] [PubMed] [Google Scholar]
- 47.Parsley, T. B., Cornell, C. T., and Semler, B. L. (1999) J. Biol. Chem. 274 12867-12876 [DOI] [PubMed] [Google Scholar]
- 48.Gamarnik, A. V., and Andino, R. (1998) Genes Dev. 12 2293-2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verlinden, Y., Cuconati, A., Wimmer, E., and Rombau, B. (2002) Arch. Virol. 147 731-744 [DOI] [PubMed] [Google Scholar]
- 50.Perera, R., Daijogo, S., Walter, B. L., Nguyen, J. H., and Semler, B. L. (2007) J. Virol. 81 8919-8932 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.