Abstract
We used real-time PCR to examine the persistence of Bordetella pertussis DNA in serial nasopharyngeal aspirates from 22 children treated for pertussis. After 5 days of treatment, PCR was positive for all 21 assessable patients. After 14 and 21 days, PCR was still positive for 83% (10/12) and 66% (4/6) of assessable patients, respectively. One patient was tested 1 month after treatment initiation, and B. pertussis DNA was still detectable. Quantitative analysis showed that the DNA concentration diminished during treatment in all except one case. The PCR cycle threshold at which B. pertussis DNA became detectable increased by a mean of 1.7 cycles per day (range, 0.86 to 3.68 cycles per day). Real-time PCR can thus be used to diagnose pertussis in young children for up to 3 weeks after treatment initiation. Its potential value for assessing the treatment outcome remains to be determined.
Despite the high rate of coverage with an effective vaccine for more than 40 years, pertussis remains endemic in France (1, 6). Pertussis can occur in adolescents and adults vaccinated during childhood and can then be transmitted to infants who are too young to be vaccinated or who have been only partially vaccinated (11). Bordetella pertussis is still one of the leading bacterial causes of death among very young infants (5). Rapid and sensitive diagnostic methods are needed to guide treatment and to limit transmission. Real-time PCR targeting the IS481 locus in nasopharyngeal aspirates is considered the “gold standard” method by a European consensus group (8). The changes in the bacterial DNA load from the time of diagnosis to the time of posttherapeutic follow-up have not been studied in this setting. We have previously reported on the case of a patient in whom B. pertussis DNA persisted for more than 7 weeks after treatment initiation (3). In the present study, using the IS481 real-time PCR assay, we assessed the persistence of B. pertussis DNA in serial nasopharyngeal aspirates from 22 children treated for pertussis.
MATERIALS AND METHODS
Patients and specimens.
Children hospitalized in the Hôpital Robert Debré, a pediatric hospital, for pertussis between January 2005 and March 2008 were included in the study if they met the following criteria: they had a PCR-based diagnosis of pertussis before treatment initiation and at least one B. pertussis DNA PCR assay of a nasopharyngeal aspirate obtained after treatment initiation was performed. Nasopharyngeal secretions were obtained by aspiration and were immediately placed at −20°C until DNA extraction.
Culture.
When sufficient sample volume was available, we inoculated charcoal agar plates (Oxoid, France). Suspected B. pertussis colonies were presumptively identified from their phenotypic characteristics, before they were sent to the Bordetella National Reference Centre for confirmation and further analysis.
DNA extraction and real-time PCR.
Nasopharyngeal secretions were fluidized with an equal volume of Mucomyst solution (Bristol-Myers, Rueil Malmaison, France). DNA was extracted with an EZ1 BioRobot apparatus (Qiagen S.A., Courtaboeuf, France) by use of the EZ1 DNA tissue kit (Qiagen S.A.). Extraction was performed with 200 μl of specimen, and the extract was eluted into a 100-μl volume. The DNA extracts were stored at −80°C. The B. pertussis real-time PCR was based on the IS481 target, as described previously (3, 10). Briefly, the PCR was performed with a reaction mixture of 50 μl consisting of 25 μl of iQ Supermix (Bio-Rad, Marnes la Coquette, France), 0.2 μM each primer, 0.2 μM Molecular Beacon fluorogenic probe, and 5 μl of template. The thermal profile consisted of 15 min at 95°C, followed by 50 cycles of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C. Detection and analysis were performed with an iQ Cycler apparatus (Bio-Rad).
The quality of the nasopharyngeal aspirates, the quality of the nucleic acid extraction step, and the presence of PCR inhibitors were controlled for by amplification of the human β2-microglobulin gene in each run with primers B2M-TR-1 (5′-GCAAGGACTGGTCTTTCTATC-3′) and B2M-TR-2 (5′-TACACAACTTTCAGCAGCTTACA-3′) and the Molecular Beacon probe B2M-TR-Sde (5′-6-carboxyfluorescein-CGTGCCCTGCCGTGTGAACCATGTGACTTTGGCACG-Black Hole Quencher 1-3′). The primer and probe concentrations and the PCR thermal profile were identical to those used for the IS481 real-time PCR. In our experience with this technique, 90% of patients have a β2-microglobulin cycle threshold (CT) of between 20 and 26. A CT value above 26 was therefore considered to denote an aspirate with too few epithelial cells, a poor extraction procedure, or the presence of inhibitors.
Quality controls.
The real-time PCR diagnosis of pertussis in our laboratory is regularly evaluated through an external quality control program managed by the Bordetella National Reference Centre. During the study period, nine control evaluations with a total of 40 samples were conducted. The sensitivity and specificity of this technique in our hands were 100% and 97.5%, respectively. The sensitivity was determined with serial dilutions of B. pertussis Tohama I DNA (1,000 fg/μl to 0.01 fg/μl) and was found to be 0.02 CFU/μl. Quantification was linear from 1,000 fg/μl to 0.1 fg/μl of purified B. pertussis DNA (R2 = 0.99).
RESULTS
Between January 2005 and March 2008, 44 children were admitted to Robert-Debré Hospital with a diagnosis of pertussis, as determined by real-time PCR. For 22 of these children, one or more real-time PCR assays of the B. pertussis DNA in nasopharyngeal aspirates after treatment initiation were also performed: 10, 7, and 5 patients provided one, two, and three supplementary samples for testing, respectively. All 37 samples gave β2-microglobulin CT values between 20 and 26, thus validating the B. pertussis PCR results.
Clinical and microbiological data for the 22 patients are shown in Table 1. Twenty patients were less than 6 months old, one was 1 year old, and one was 9 years old. Most were unvaccinated against pertussis or had received only one vaccine injection. Only one patient had coughed for more than 15 days before the diagnosis was made. Two patients were considered immunocompromised: patient 8 had a nephrosis syndrome that was treated with immunosuppressive drugs, and patient 14 had immunoglobulin G deficiency. All but two of the patients received josamycin for 14 days. One patient received azithromycin, and patient 8 received ciprofloxacin because macrolides and co-trimoxazole were incompatible with the drugs used to treat nephrosis.
TABLE 1.
Clinical and biological characteristics of 22 patients with pertussis
| Patient | Age at onset | Duration of cough before diagnosis (days) | Vaccination history | Culture result
|
Resultb of real-time PCR for Bordetella pertussis DNA collected at the following times (days) after treatment initiation:
|
|||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Initial sample | Subsequent samplea | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | ||||
| P8 | 9 yr | 10 | Four vaccines | Positive | Negative | + | + | |||||||||||||||||||||||||||||||
| P14 | 1 yr | 3 | Three vaccines | Not available | Not available | + | + | - | ||||||||||||||||||||||||||||||
| P12 | 5 mo | 10 | Unvaccinated | Not available | Not available | + | + | + | ||||||||||||||||||||||||||||||
| P1 | 4 mo | 7 | Unvaccinated | Not available | Not available | + | + | + | ||||||||||||||||||||||||||||||
| P15 | 4 mo | 4 | One vaccine | Positive | Negative | + | + | + | ||||||||||||||||||||||||||||||
| P18 | 3 mo | 28 | Unvaccinated | Not available | Not available | + | + | + | + | |||||||||||||||||||||||||||||
| P21 | 3 mo | 15 | One vaccine | Not available | Negative | + | + | + | ||||||||||||||||||||||||||||||
| P9 | 3 mo | 2 | One vaccine | Negative | Negative | + | − | |||||||||||||||||||||||||||||||
| P20 | 2 mo | 15 | Unvaccinated | Not available | Not available | + | + | |||||||||||||||||||||||||||||||
| P10 | 2 mo | 15 | Unvaccinated | Positive | Negative | + | + | |||||||||||||||||||||||||||||||
| P2 | 2 mo | 14 | One vaccine | Positive | Negative | + | + | + | + | |||||||||||||||||||||||||||||
| P22 | 2 mo | 12 | Unvaccinated | Positive | Negative | + | + | |||||||||||||||||||||||||||||||
| P11 | 2 mo | 8 | Unvaccinated | Not available | Not available | + | + | + | + | |||||||||||||||||||||||||||||
| P13 | 2 mo | 7 | Unvaccinated | Not available | Not available | + | + | |||||||||||||||||||||||||||||||
| P17 | 2 mo | 7 | Unvaccinated | Negative | Negative | + | + | |||||||||||||||||||||||||||||||
| P19 | 1 mo | 10 | Unvaccinated | Negative | Negative | + | + | |||||||||||||||||||||||||||||||
| P16 | 1 mo | 4 | Unvaccinated | Positive | Negative | + | + | + | ||||||||||||||||||||||||||||||
| P4 | 1 mo | 3 | Unvaccinated | Positive | Negative | + | + | |||||||||||||||||||||||||||||||
| P7 | 1 mo | 2 | Unvaccinated | Positive | Negative | + | + | |||||||||||||||||||||||||||||||
| P6 | 1 mo | 10 | Unvaccinated | Positive | Negative | + | + | + | ||||||||||||||||||||||||||||||
| P3 | 28 days | 14 | Unvaccinated | Not available | Not available | + | + | + | − | |||||||||||||||||||||||||||||
| P5 | 27 days | 11 | Unvaccinated | Positive | Negative | + | + | + | + | |||||||||||||||||||||||||||||
A subsequent sample for culture was obtained 5 to 14 days after treatment initiation.
+, positive result; −, negative result.
The intervals between the times of collection of the samples used for diagnosis and the times of collection of the subsequent samples are also shown in Table 1, along with the results of PCR and culture. Diagnostic cultures were performed for only 13 patients, and 11 of these were positive. After 5 days of treatment, all 21 assessable patients were still PCR positive. Ten (83%) of the 12 patients who were assessable 14 days after treatment initiation were still positive, and 4 (66%) of the 6 patients who were assessable at 21 days were still positive. One patient was tested 1 month after treatment initiation and was still PCR positive for B. pertussis DNA. All 11 initially culture-positive patients were culture negative when new samples were tested between 5 and 14 days after treatment initiation. All 22 patients were seen 1 month after discharge, and none had clinical signs of resurgent pertussis.
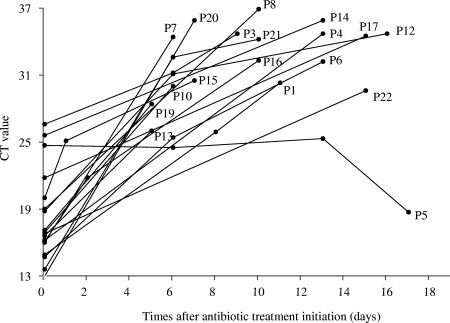
Real-time PCR was applied in the same run to all stored DNA extracts from 18 patients (the initial samples from patients 2, 11, and 18 were missing; and the second sample from patient 9 was PCR negative) (Fig. 1). Overall, the B. pertussis DNA level fell progressively during antibiotic therapy. Only one patient (patient 5) had a paradoxical response, with an increase in the B. pertussis DNA load during the 16 days of follow-up. After the data for this patient were excluded, the mean and median increases in the CT number were 1.7 cycles per day (range, 0.86 to 3.68 cycles per day) and 1.5 cycles per day, respectively, indicating a marked gradual fall in the B. pertussis DNA load during and after treatment in the vast majority of patients.
FIG. 1.
Time course of IS481 real-time PCR CT values applied to serial nasopharyngeal samples from 18 infants treated for pertussis.
DISCUSSION
To our knowledge this is the first study by real-time PCR of the persistence of B. pertussis DNA in the nasopharyngeal secretions of hospitalized patients treated for pertussis. Using this highly sensitive technique, we found that B. pertussis DNA persisted for long periods. Fourteen days after treatment initiation—the period of treatment with old macrolides—the PCR result was positive for 10 (87%) of the 12 assessable patients.
The significance of the persistence of B. pertussis DNA is unclear. Nasopharyngeal secretions became culture negative in the vast majority of patients after 5 days of macrolide therapy. None of our patients remained culture positive after the start of treatment. The persistence of B. pertussis DNA may correspond to the presence of antibiotic-damaged bacteria. Whether these bacteria are still viable is questionable, but it is noteworthy that there were no recurrences of pertussis in our study. However, pertussis treatment failure has been observed in some patients with compromised immunity (3, 9). Further studies, to search for specific B. pertussis mRNA, for instance, are needed to determine whether the DNA detected corresponds to that from viable but antibiotic-damaged bacteria.
One previous study examined the persistence of B. pertussis DNA during antibiotic therapy by means of conventional PCR with serial nasopharyngeal swabs from young infants (4). Among the nine patients from whom follow-up samples were available, six (66%) were positive after 5 days of erythromycin treatment. On day 10 of treatment, three (43%) of the seven assessable patients were still positive. Samples from only two patients were obtained after 14 days or more, and neither was positive. The rate of PCR positivity during pertussis therapy was lower than that in our study, possibly because real-time PCR is more sensitive than conventional PCR and also because we tested nasopharyngeal aspirates, which are now considered more suitable for use than swabs in this setting (8). In addition, josamycin was used to treat most of our patients, whereas erythromycin was used in the study based on conventional PCR.
In a prospective trial of azithromycin treatment for pertussis, Pichichero et al. found that all 29 patients with culture-proven pertussis were PCR negative 2 to 3 days after treatment (7). It is noteworthy that Pichichero et al. also used conventional PCR and that their patients were older than ours (all were >6 months old). Whether the use of a new macrolide had an influence on the prompt eradication of Bordetella DNA remains questionable. However, in our study, the only patient (patient 17) treated with azithromycin was still PCR positive 15 days after treatment initiation.
In our study, the CT number (the number at which PCR amplification products become detectable) increased by a mean of 1.7 cycles per day during macrolide therapy, reflecting a gradual fall in the B. pertussis DNA level. Serial real-time PCR might be useful for the assessment of treatment efficacy in patients with pertussis. Although it is rare, macrolide resistance has been described in B. pertussis (2). In culture-negative patients, real-time PCR repeated 5 to 7 days after treatment initiation might help to predict antibiotic resistance, and a stable or decreasing CT value during the first week could be interpreted as a reflection of treatment failure. However, we observed a paradoxical decrease in the CT value in one successfully treated patient (patient 5) whose isolate was sensitive to josamycin (data not shown). To check that this result was not due to the persistence of another Bordetella species, we performed a ptxA PCR with the different samples, and all were positive, indicating the true persistence of B. pertussis DNA (data not shown). Of note, this patient was the youngest in our series, and immature innate immunity might have contributed to the slow clearance of B. pertussis. Alternatively, this case might correspond to in vivo resistance, but unfortunately, no subsequent samples were cultured.
In conclusion, real-time PCR can be used to diagnose pertussis even 3 weeks after treatment initiation in infants less than 6 months of age. Whether or not this also applies to older children and adults remains to be determined. Serial real-time PCR might be useful for the prediction of treatment failure and comparison of efficacies of different antibiotics, although further studies are required.
Footnotes
Published ahead of print on 3 September 2008.
REFERENCES
- 1.Baron, S., E. Njamkepo, E. Grimprel, P. Begue, J. C. Desenclos, J. Drucker, and N. Guiso. 1998. Epidemiology of pertussis in French hospitals in 1993 and 1994: thirty years after a routine use of vaccination. Pediatr. Infect. Dis. J. 17412-418. [DOI] [PubMed] [Google Scholar]
- 2.Bartkus, J. M., B. A. Juni, K. Ehresmann, C. A. Miller, G. N. Sanden, P. K. Cassiday, M. Saubolle, B. Lee, J. Long, A. R. Harrison, Jr., and J. M. Besser. 2003. Identification of a mutation associated with erythromycin resistance in Bordetella pertussis: implications for surveillance of antimicrobial resistance. J. Clin. Microbiol. 411167-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonacorsi, S., C. Farnoux, P. Bidet, V. Caro, S. Aizenfisz, M. Benhayoun, Y. Aujard, N. Guiso, and E. Bingen. 2006. Treatment failure of nosocomial pertussis infection in a very-low-birth-weight neonate. J. Clin. Microbiol. 443830-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edelman, K., S. Nikkari, O. Ruuskanen, Q. He, M. Viljanen, and J. Mertsola. 1996. Detection of Bordetella pertussis by polymerase chain reaction and culture in the nasopharynx of erythromycin-treated infants with pertussis. Pediatr. Infect. Dis. J. 1554-57. [DOI] [PubMed] [Google Scholar]
- 5.Floret, D. 2001. Pediatric deaths due to community-acquired bacterial infection. Survey of French pediatric intensive care units. Arch. Pediatr. 8(Suppl. 4)705s-711s. [DOI] [PubMed] [Google Scholar]
- 6.Gilberg, S., E. Njamkepo, I. P. Du Chatelet, H. Partouche, P. Gueirard, C. Ghasarossian, M. Schlumberger, and N. Guiso. 2002. Evidence of Bordetella pertussis infection in adults presenting with persistent cough in a French area with very high whole-cell vaccine coverage. J. Infect. Dis. 186415-418. [DOI] [PubMed] [Google Scholar]
- 7.Pichichero, M. E., W. J. Hoeger, and J. R. Casey. 2003. Azithromycin for the treatment of pertussis. Pediatr. Infect. Dis. J. 22847-849. [DOI] [PubMed] [Google Scholar]
- 8.Riffelmann, M., C. H. Wirsing von Konig, V. Caro, and N. Guiso. 2005. Nucleic acid amplification tests for diagnosis of Bordetella infections. J. Clin. Microbiol. 434925-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinberg, J. M., and I. Srugo. 2002. Reoccurrence of culture-positive pertussis in an infant initially treated with azithromycin and steroids. Arch. Pediatr. Adolesc. Med. 1561057-1058. [PubMed] [Google Scholar]
- 10.Templeton, K. E., S. A. Scheltinga, A. van der Zee, B. M. Diederen, A. M. van Kruijssen, H. Goossens, E. Kuijper, E. C. Claas, K. Kosters, U. Reischl, J. Schmetz, M. Riffelmann, C. H. Wirsing von Konig, N. Lehn, G. N. Sanden, and M. J. Loeffelholz. 2003. Evaluation of real-time PCR for detection of and discrimination between Bordetella pertussis, Bordetella parapertussis, and Bordetella holmesii for clinical diagnosis. J. Clin. Microbiol. 414121-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wendelboe, A. M., E. Njamkepo, A. Bourillon, D. D. Floret, J. Gaudelus, M. Gerber, E. Grimprel, D. Greenberg, S. Halperin, J. Liese, F. Munoz-Rivas, R. Teyssou, N. Guiso, and A. Van Rie. 2007. Transmission of Bordetella pertussis to young infants. Pediatr. Infect. Dis. J. 26293-299. [DOI] [PubMed] [Google Scholar]