Abstract
This study evaluated the biodiversity of 28 clinical and 24 environmental Mycobacterium isolates from Rio de Janeiro, Brazil, by using hsp65 sequences, with the aim of contributing to a better understanding of the genetic diversity and usefulness of this marker. An extensive phylogenetic analysis was performed. The nucleotide diversity was similar between clinical (0.06508) and environmental (0.06221) isolates.
Nontuberculous mycobacteria (NTM) are ubiquitous environmental microorganisms that can be found in a variety of ecosystems (2). The genus Mycobacterium comprises a wide range of organisms, including obligate parasites which cause serious human and animal diseases, opportunistic pathogens, and saprophytic species (1). Human activities likely influence the distribution and prevalence of mycobacteria. Mycobacteria are capable of inducing biofilm formation, which helps them to persist in a flowing system in spite of their slow growth. Biofilms may be important sources of NTM and may be responsible for pseudo-infections and pseudo-outbreaks as well as diseases and disease outbreaks. The rapid detection of pseudo-infections and diseases due to NTM is important and requires the use of molecular techniques. Telenti et al. (15) developed a rapid method based on evaluation of the gene encoding the 65-kDa heat shock protein by PCR. The 65-kDa protein contains epitopes that are common to various species of mycobacteria (13). Therefore, we assessed the feasibility of using hsp65 sequencing to identify mycobacteria and to analyze their genetic variability. In this study, we report a phylogenetic analysis of clinical mycobacteriology laboratory and environmental isolates.
The clinical isolates used in this study were provided by the Laboratory of Mycobacteria, Federal University of Rio de Janeiro, and were isolated from different places. Fifty-two isolates were investigated to determine the species (28 clinical isolates and 24 environmental isolates) (Table 1). Isolates were cultured in solid Lowenstein-Jensen medium, and conventional identification procedures were carried out according to the methods of Kent and Kubicae (4). DNA samples were extracted according to the cetyltrimethylammonium bromide protocol of Van Embden and colleagues (17), and PCR assays were performed according to the method of Telenti et al. (15). Samples were purified with MicroSpin S-400 columns (Amersham Biosciences) and a QIAquick PCR purification kit (Qiagen). Sequencing was performed with a DYEnamic ET dye terminator kit (MegaBace; Amersham Biosciences) and read with a MegaBace1000 (Amersham Biosciences) automated system. All chromatograms were checked using the CHROMAS 1.45 program, and the sequences were aligned using Clustal_X 1.83 (16), with manual adjustments using the BioEdit 7.0.9 program. The substitution model used for phylogenetic reconstructions was estimated with Modeltest 3.7 (11), using the minimum theoretical information criterion and the Bayesian information criterion, as suggested by Posada and Buckley (10). Isolates were identified by comparing unknown sequences to reference databases by a FASTA BLAST search (see the supplemental material). Genetic diversity parameters, such as haplotype and nucleotide diversity (6), were estimated employing DnaSP 4.0 software (Table 2). Phylogenetic trees were reconstructed by the maximum likelihood (ML) and neighbor-joining methods, using the program PAUP* 4.0b10 (D. Swofford, Sunderland, MA). The branch confidence values were estimated using 1,000 bootstrap replicates. We inferred ML trees with a heuristic nearest-neighbor interchange search option. The neighbor-joining analysis used the ML distance in the evolutionary model selected by the model test. Nocardia sp. and Corynebacterium sp. were used as outgroup species, and Mycobacterium tuberculosis was used as a more closely related species (Fig. 1).
TABLE 1.
Environmental and clinical strains
| Strain | GenBank accession no. |
|---|---|
| Environmental strains | |
| Swine source 255 | EU343669 |
| Swine source 259 | EU343670 |
| Swine source 260 | EU343671 |
| Water 262 | EU343672 |
| Water 263 | EU343673 |
| Water 264 | EU343674 |
| Water 269 | EU343675 |
| Soil 299 | EU343676 |
| Soil 301 | EU343677 |
| Bovine feces 308 | EU343678 |
| Bovine feces 314 | EU343679 |
| Bovine feces 315 | EU343680 |
| Bovine feces 318 | EU343681 |
| Bovine feces 328b | EU343682 |
| Bovine feces 328c | EU343683 |
| Bovine feces 345 | EU343684 |
| Bovine feces 346 | EU343685 |
| Swine source 358 | EU343686 |
| Swine source 359 | EU343687 |
| Swine source 369 | EU343688 |
| Swine source 370 | EU343689 |
| Swine source 371 | EU343690 |
| Soil 395 | EU343691 |
| Bovine feces 417 | EU343692 |
| Clinical strainsa | |
| 423 | EU343693 |
| 424 | EU343694 |
| 427 | EU343695 |
| 428 | EU343696 |
| 430 | EU343697 |
| 432 | EU343698 |
| 434 | EU343699 |
| 436 | EU343700 |
| 438 | EU343701 |
| 440 | EU343702 |
| 442 | EU343703 |
| 445 | EU343704 |
| 446 | EU343705 |
| 447 | EU343706 |
| 448 | EU343707 |
| 450 | EU343708 |
| 451 | EU343709 |
| 452 | EU343710 |
| 454 | EU343711 |
| 456 | EU343712 |
| 457 | EU343713 |
| 458 | EU343714 |
| 462 | EU343715 |
| 463 | EU343716 |
| 465 | EU343717 |
| 466 | EU343718 |
| 467 | EU343719 |
| 468 | EU343720 |
From induced sputum.
TABLE 2.
Genetic diversity parameters
| Sample group | No. of sequences | No. of variable sites | Total no. of mutations | No. of haplotypes | Haplotype diversity value | Nucleotide diversity (Pi) | Avg no. of nucleotide differences (K) |
|---|---|---|---|---|---|---|---|
| Clinical strains | 28 | 73 | 91 | 19 | 0.947 | 0.06508 | 23.88360 |
| Environmental strains | 24 | 85 | 106 | 17 | 0.920 | 0.06221 | 22.82971 |
| Clinical and environmental strains | 52 | 95 | 128 | 34 | 0.963 | 0.06590 | 24.18627 |
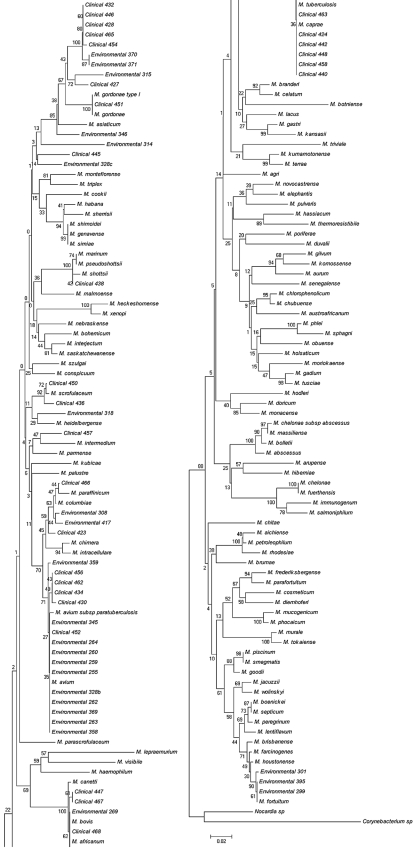
FIG. 1.
Consensus tree for 200 bootstraps showing the phylogenetic relationships among environmental and clinical isolates from several places in the region surrounding Rio de Janeiro, Brazil, and sequences of known mycobacterial species. The tree is based on a comparison of a 368-bp sequence from the mycobacterial hsp65 gene, using the neighbor-joining method. Bootstrap values of >70% are indicated. The distance between two strains is the sum of the branch lengths between them.
The clinical isolates are from Rio de Janeiro University Hospital, a reference hospital for mycobacterial detection, with a great number coming from long-term human immunodeficiency virus-positive patients. No incidences of hsp65 gene amplification, sequencing failure, or interprimer sequence variation were encountered in the 368 sites studied. The genetic variations of the hsp65 gene sequences between clinical and environmental samples were similar, which suggests an increasing relationship among some species from the environment and those infecting humans. Clinical sample 451 was clearly identified as M. gordonae, being grouped with 100% bootstrap support with M. gordonae ATCC 14470. Another clinical sample (438) was identified as M. shottsii, since it presented 100% identity with M. shottsii ATCC 700981. Samples used in the present study showed great variability, since we found many slow-growing isolates and also rapidly growing isolates in clinical and ambient samples. In general, the great majority of the samples showed slow growth. Many isolates were grouped with M. avium, M. intracellulare, M. scrofulaceum ATCC 19981, and the M. tuberculosis complex. A possible explanation for this fact is that environmental mycobacteria are normal inhabitants of a wide variety of environmental reservoirs, including natural and municipal water, soil aerosol, protozoans, animals, and humans. Environmental mycobacteria also have extraordinary starvation survival, persisting in tap water despite low nutrient levels (8, 12, 14).
Although water does not represent the only source of M. avium complex in humans, it is possible that it might be the primary source (3). Human activities probably have a great influence on the distribution and prevalence of mycobacteria. The treatment of drinking water supplies with chlorine or other disinfectants (e.g., ozone) leads to selection for environmental mycobacteria (7). This fact could explain why many clinical and environmental species in the present study grouped with M. avium. In this investigation, two clinical isolates grouped with M. scrofulaceum, maybe because of implementation of a clean water method, similar to the one that occurred in the United States in 1975, when increased chlorination rates may have led to a strong reduction of M. scrofulaceum in the water. Additionally, the epidemiology of infection by environmental mycobacteria is poorly understood due to a lack of data regarding the primary reservoirs of different mycobacterial species (5, 12).
A great number of the environmental isolates collected were identified by slow growth and were grouped with clinical isolates. This result could be related to the previously discussed question of adaptive value. There are many situations in which human and mycobacterial distributions can overlap geographically and environmentally, resulting in human exposure and in an impact on mycobacterial ecology. Humans are exposed to mycobacteria in water through drinking, swimming, and bathing. Contamination has been facilitated mainly in hospitals, where patients with reduced immunity are more exposed. Previous studies which correlate environmental parameters with the isolation of environmental mycobacteria were performed with acidic, organic, rich material and stagnant water reservoirs (1). However, we could see that infection by NTM can almost exclusively be associated with environmental mycobacteria that have adapted to humans. Our investigation provides evidence that hsp65 sequencing has the potential of being an accurate, reliable, and effective means for identifying clinical and environmental Mycobacterium isolates. It has the advantages over biochemical test profiles of being rapid and trustworthy. Moreover, the results of sequencing can be used to correlate the specimens between themselves and to give support in their identification.
Our results show that the hsp65 sequences from reference strains of mycobacteria provide a basis for determining systematic phylogenetic relationships. The phylogenetic analysis suggests that slow growth evolved recently in mycobacteria and, as discussed above, possibly has a great adaptive value (9). This study also shows the possibility that species correlate with each other and, moreover, the possible entry ports of mycobacteria in the artificial human environment. With regard to the information about DNA polymorphism obtained with the clinical and environmental isolates individually, we observed that it was greater in clinical than in environmental samples. This could be explained by an adaptation of the environmental species to the artificial human environment, probably through biofilms. It could also help us to understand why so many infections caused by mycobacteria have been reported recently. Currently, it is very important to understand associations between species of mycobacteria in Brazil because infections by these microorganisms have been increasing and causing outbreaks in hospitals, where the port of entry for infection is mainly surgery patients, and are becoming a serious and delicate problem for public health.
Supplementary Material
Acknowledgments
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ).
We are grateful to Denis Broock Rosemberg and Laura Utz for critical reviews of the manuscript.
Footnotes
Published ahead of print on 3 September 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Bland, C. B., J. M. Ireland, E. Lozano, M. E. Alvarez, and T. P. Primm. 2005. Mycobacterial ecology of the Rio Grande. Appl. Environ. Microbiol. 715719-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falkinham, J. O., III. 2002. Nontuberculous mycobacteria in the environment. Clin. Chest Med. 23529-551. [DOI] [PubMed] [Google Scholar]
- 3.Goslee, S., and E. Wolinsky. 1976. Water as a source of potentially pathogenic mycobacteria. Am. Rev. Respir. Dis. 113287-292. [DOI] [PubMed] [Google Scholar]
- 4.Kent, P. T., and G. P. Kubicae. 1985. Public health mycobacteriology. A guide for the level III laboratory. Centers for Disease Control, Atlanta, GA.
- 5.Moore, J. S., M. Christensen, R. W. Wilson, R. J. Wallace, Jr., Y. Zang, D. R. Nash, and B. Shelton. 2000. Mycobacterial contamination of metalworking fluids: involvement of a possible new taxon of rapidly growing mycobacteria. AIHAJ 61205-213. [DOI] [PubMed] [Google Scholar]
- 6.Nei, M. 1987. Molecular evolutionary genetics. Columbia University Press, New York, NY.
- 7.Norton, C. D., and M. W. LeChevallier. 2000. A pilot study of bacteriological population changes through potable water treatment and distribution. Appl. Environ. Microbiol. 66268-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nyka, W. 1974. Studies on the effect of starvation on mycobacteria. Infect. Immun. 9843-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitulle, C., M. Dorch, J. Kazda, J. Wloters, and E. Stackebrandt. 1992. Phylogeny of rapidly growing member of the genus Mycobacterium. Int. J. Syst. Bacteriol. 42337-343. [DOI] [PubMed] [Google Scholar]
- 10.Posada, D., and T. R. Buckley. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53793-808. [DOI] [PubMed] [Google Scholar]
- 11.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14817-818. [DOI] [PubMed] [Google Scholar]
- 12.Primm, T. P., C. A. Lucero, and J. O. Falkinham III. 2004. Health impacts of environmental mycobacteria. Clin. Microbiol. Rev. 1798-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schinnick, T. 1987. The 65-kilodalton antigen of Mycobacterium tuberculosis. J. Bacteriol. 1691080-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smeulders, M. J., J. Keer, A. Speight, and H. D. Williams. 1999. Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 181270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Telenti, A., F. Marehesi, M. Balz, E. C. Bottger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Soolingen, D., P. E. W. De Hass, P. W. M. Hermans, and J. D. A. Van Embden. 1994. RFLP analysis of mycobacteria. National Institute of Public Health and Environmental Protection, Bilthoven, The Netherlands.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.