Abstract
Detection of galactomannan antigen (GMA) in serum is the standard assay for the diagnosis of invasive aspergillosis (IA) in high-risk patients with hematological disorders. Detection of Aspergillus DNA in serum has been proposed, but its sensitivity is lower than that of GMA when small serum volumes (SSV) are used. In this study, we investigated whether extraction of DNA from large serum volumes (LSV) improves diagnostic yield. In a 13-month prospective study, we compared the performances of twice-weekly screening of serum for GMA by an enzyme immunoassay and weekly screening for Aspergillus fumigatus DNA by a real-time PCR (RT-PCR) assay of 1.0 ml (LSV) or 100 μl (SSV) of serum. We included 124 patients (138 treatment episodes), with 17 episodes of EORTC (European Organization for Research and Treatment of Cancer)/MSG (Mycoses Study Group)-documented IA. In all, 1,870 samples were screened for GMA. The sensitivity (Se), specificity (Sp), and positive and negative predictive values (PPV and NPV, respectively) of GMA for IA were 88.2%, 95.8%, 75%, and 98.3%, respectively. We screened 938 samples for Aspergillus DNA by using LSV; 404 of these samples were also tested with SSV. The Se, Sp, PPV, and NPV of RT-PCR were 100%, 96.7%, 81%, and 100%, respectively, with LSV and 76.5%, 96.7%, 81.3%, and 95.6%, respectively, with SSV. DNA detection gave a positive result when performed on LSV in two cases of IA where the GMA assay result remained negative. Furthermore, in four IA cases, DNA was detected earlier than GMA. The use of LSV for extraction improved the performance of the RT-PCR, which appears highly sensitive and specific for the early diagnosis of IA in high-risk patients with hematological disorders.
Invasive aspergillosis (IA) is currently the most frequent fungal infection in patients with hematological malignancies (18). Despite highly active new antifungal drugs, mortality remains as high as 50 to 70% (4, 23). Early initiation of effective antifungal treatment is essential in order to improve the outcomes for these patients. In this context, high-risk patients with hematological disorders should benefit from efficient, noninvasive diagnostic strategies allowing early diagnosis of IA. The screening of galactomannan antigen (GMA) in serum is currently the only indirect microbiological assay approved for the diagnosis of IA in such patients (2). However, the GMA assay yields a number of false-positive results, owing to cross-reactivity between Aspergillus spp. and other fungi (25) or to detection of circulating GMA resulting from contamination by certain antibiotics or parenteral nutrition preparations (3, 10, 20-22, 24). In addition, high rates of false-positive results for galactomannan (GM) antigenemia have been reported recently for allogeneic hematopoietic stem cell transplantation (HSCT) recipients during the first 100 days following transplantation, or for those with chronic gastrointestinal graft-versus-host disease (GvHD) (1). Thus, the diagnostic value of GM antigenemia detection should be interpreted with caution for these patients, in conjunction with the results of other diagnostic procedures.
Screening for circulating DNA of Aspergillus spp. by PCR has shown potential in the definitive diagnosis of IA, especially in combination with antigen testing (17, 26). However, in most studies, the sensitivity of DNA detection was lower than that of GMA screening, possibly because small serum volumes (SSV) (100 to 200 μl) were used for DNA extraction (14, 15, 17). The current availability of automated nucleic acid extraction techniques, such as the MagNA Pure LC (Roche Diagnostics) apparatus, which allows safe DNA extraction from larger volumes of serum (i.e., 1 ml), prompted us to test this hypothesis. We performed a prospective study of adult patients with hematological disorders who were at high risk for IA in order to determine the diagnostic contribution of weekly screening of large serum volumes (LSV) for Aspergillus fumigatus DNA in comparison with conventional serum volumes. We then compared the contributions of these molecular diagnostic approaches with that of GM antigenemia assessed on a biweekly basis.
MATERIALS AND METHODS
Study design.
This study was conducted prospectively from February 2006 to March 2007 (13 months) in the adult hematology and bone marrow transplant unit at Necker-Enfants Malades hospital, a tertiary-care university hospital (Paris, France). All adult patients receiving allogeneic or autologous HSCT, or intensive (induction, consolidation, or salvage) chemotherapy for hematological malignancies, and who were routinely monitored for biweekly GM detection were included in the study. A treatment episode was defined as a single cycle of chemotherapy (with or without HSCT). IA was classified as proven, probable, or possible, according to EORTC (European Organization for Research and Treatment of Cancer)/MSG (Mycoses Study Group) definitions (2).
Patient management.
Empirical antifungal therapy (amphotericin B deoxycholate, liposomal amphotericin B, or caspofungin in cases of renal dysfunction) was started 2 days after the onset of fever in cases of antibiotic-resistant fever, as recommended for neutropenic patients (12).
Diagnostic procedures included daily physical examinations, weekly bacterial and fungal stool and urine cultures, weekly chest radiography, and, as stated, twice-weekly tests for GM detection. When pulmonary IA was suspected, sputum samples were also collected for fungal cultures. When possible, a computed-tomography scan of the chest was performed early on, followed by bronchoalveolar lavage (BAL).
Detection of circulating GMA.
The GM assay was performed, as recommended by the manufacturer, on samples collected twice weekly (Mondays and Thursdays) using the Platelia Aspergillus enzyme immunossay (Bio-Rad Laboratories, Marnes-la-Coquette, France). Serum samples with an index of ≥0.5 were retested the following day and were considered positive if the GM index was again ≥0.5 (16).
Extraction of DNA from serum.
Samples for the PCR assay were collected weekly (on Mondays). DNA was extracted from serum using the MagNA Pure LC apparatus (Roche). For DNA extraction from SSV, 100 μl of serum was processed with the instrument's Total Nucleic Acid External Lysis protocol and MagNA Pure LC DNA isolation kit III (Bacterial, Fungal) (Roche Diagnostics).
For DNA extraction from LSV, 1 ml of serum was processed with the instrument's Total Nucleic Acid Isolation Large Volume Serum protocol (Roche Diagnostics) in conjunction with the MagNA Pure LC Total Nucleic Acid Isolation Kit-Large Volume.
RT-PCR assay.
A. fumigatus DNA was amplified by real-time PCR (RT-PCR) with a Thermocycler/ABI Prism 7300 sequence detector (Applied Biosystems) as described previously (6). The target was a 67-bp DNA fragment specific to the multicopy gene encoding the 28S rRNA of A. fumigatus. The sensitivity of RT-PCR (expressed as the mean minimum number of cycles necessary to detect A. fumigatus DNA) was 34.3 cycles (1 copy), as determined previously (6). A sample was considered positive only when the crossing point value was ≤41 cycles.
Statistical analysis.
Our objective was to compare the diagnostic contributions of the GMA and RT-PCR assays in a setting where these tests were performed twice weekly and weekly, respectively, to monitor the subsequent development of IA. Thus, the positivity and negativity of a test were defined for each treatment episode. For the GMA assay, an episode was considered positive when at least two consecutive samples were positive; for RT-PCR, an episode was considered positive when at least one sample was positive. To calculate the sensitivity (Se), specificity (Sp), positive predictive value (PPV), and negative predictive value (NPV) of each test, we used the EORTC/MSG criteria (except the GM results) for diagnosis of IA (2).
The date of diagnosis of IA (proven, probable, or possible) was defined for a given patient as the day on which the first clinical, radiological, and/or microbiological EORTC/MSG criteria, other than a GM-positive result, appeared.
RESULTS
During the 13-month period of this prospective study, 124 adult patients, corresponding to 138 treatment episodes, were considered at high risk for IA; 17 of these patients developed IA (1 proven, 14 probable, and 2 possible). The cumulative incidence of proven and probable IA, per patient or per treatment episode, was 12.1% or 10.1%, respectively. The incidence reached 13.7% or 12.3%, respectively, when possible cases were also taken into account. The characteristics of IA patients are shown in Table 1.
TABLE 1.
Characteristics of 17 adult patients with proven, probable, or possible IA
| Patient no. | Age (yr) | Gendera | SCT | Primary disease or risk factorb | IA classificationc | Days posttransplantation | GvHD | Neutropenia (<0.5 × 109/liter) | Chemotherapy during the preceding 4 wks | Steroid use (>1 mg/ kg/day) during the preceding 3 wks | Outcome at 3 mo after diagnosis |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 51 | F | Allogeneic | AML | Probable | 21 | No | Yes | Yes | No | Survival |
| 2 | 68 | F | No | CLL | Probable | No | No | No | No | Death | |
| 3 | 27 | M | No | AML | Probable | No | Yes | Yes | No | Death | |
| 4 | 55 | M | Allogeneic | Myeloma | Probable | 70 | Yes | Yes | No | Yes | Death |
| 5 | 31 | M | Allogeneic | AML | Probable | 28 | Yes | Yes | No | Yes | Death |
| 6 | 54 | F | Allogeneic | CLL | Probable | 180 | Yes | Yes | No | Yes | Death |
| 7 | 69 | F | No | CLL | Probable | No | Yes | Yes | No | Survival | |
| 8 | 52 | F | Allogeneic | AML | Probable | 124 | Yes | Yes | No | No | Survival |
| 9 | 56 | M | Allogeneic | Aplasia | Probable | Yes | Yes | No | Yes | Death | |
| 10 | 34 | M | No | AML | Probable | No | Yes | Yes | No | Survival | |
| 11 | 77 | F | No | ALL | Probable | No | Yes | Yes | No | Death | |
| 12 | 53 | F | No | Myeloma | Probable | No | Yes | Yes | No | Death | |
| 13 | 79 | M | No | ALL | Probable | No | No | No | No | Death | |
| 14 | 57 | F | Allogeneic | CLL | Proven | 340 | Yes | No | No | Yes | Death |
| 15 | 63 | M | No | AML | Probable | No | Yes | Yes | No | Death | |
| 16 | 47 | M | No | AML | Possible | No | Yes | Yes | No | Survival | |
| 17 | 18 | F | Allogeneic | AML | Possible | 150 | No | Yes | Yes | Yes | Death |
M, male; F, female.
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CLL, chronic lymphoid leukemia.
According to European Organization for Research and Treatment of Cancer criteria. Criteria are from Ascioglu et al. (2).
Comparative diagnostic contribution of detection of circulating A. fumigatus DNA from SSV or LSV.
The performances of the RT-PCRs using SSV or LSV procedures for DNA extraction were compared for 43% (404/938) of the samples available for DNA analysis. These were from 107 treatment episodes for 105 patients, including the 17 IA cases (median number of serum samples per episode, 3; range, 1 to 14). Circulating DNA was detected in 16 treatment episodes (38 samples) by use of SSV and in 20 treatment episodes (60 samples) by use of LSV. A total of 109 serum samples from the 17 patients with IA (17 treatment episodes) were tested using both methods. Thirty-five samples (from 13 treatment episodes) were positive by the SSV procedure compared to 57 (from 17 treatment episodes) by the LSV procedure (P = 0.004). The RT-PCR using SSV was repeatedly negative in four treatment episodes (cases 1, 4, 10, and 14 [Table 2]) with IA. In all, three serum samples from three episodes, not related to IA, yielded false-positive DNA results by using LSV and SSV. The Se, Sp, PPV, and NPV of RT-PCR were 100%, 96.7%, 85%, and 100%, respectively, when DNA was extracted from LSV and 76.5%, 96.7%, 81.3%, and 95.6%, respectively, when SSV were used. Thus, increasing the volume of serum for DNA extraction to detect IA cases allowed a higher Se and NPV without affecting the Sp and PPV.
TABLE 2.
Diagnosis of IA and its documentation
| Patient no. | Criteria for IA according to EORTCa
|
Date of diagnosisb | Result for the following assay:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Host factors | Clinical evidence
|
Microbiological evidence
|
GMA
|
RT-PCR
|
||||||||
| Major | Minor | Culture | GM in CSF or BAL | LSV
|
SSV
|
|||||||
| No. of positive samples | Date of first positive sample | No. of positive samples | Date of first positive sample | No. of positive samples | Date of first positive sample | |||||||
| 1 | Yes | C | S, N, P | NP | NP | 3/14/2006 | 2 | 2/20/2006 | 2 | 2/27/2006 | 0 | |
| 2 | Yes | H | S, N, P | A. fumigatus from BAL | BAL | 12/4/2006 | 3 | 12/22/2006 | 1 | 12/22/2006 | 1 | 12/22/2006 |
| 3 | Yes | H | S, N | Negative | NP | 2/12/2006 | 6 | 2/12/2006 | 2 | 2/12/2006 | 2 | 2/12/2006 |
| 4 | Yes | H | S, N, P | A. fumigatus from BAL | ? | 1/11/2007 | 2 | 1/8/2007 | 1 | 1/13/2007 | 0 | |
| 5 | Yes | H | S, N, P | Negative | NP | 2/10/2006 | 2 | 2/5/2007 | 4 | 1/29/2007 | 2 | 2/5/2007 |
| 6 | Yes | No | N, I | Negative | NP | 4/24/2006 | 9 | 3/20/2006 | 5 | 4/4/2006 | 3 | 4/4/2006 |
| 7 | Yes | No | N, I | Negative | NP | 6/1/2006 | 2 | 7/3/2006 | 1 | 7/3/2006 | 1 | 7/3/2006 |
| 8 | Yes | H, C | S, I, P | Negative | Negative | 1/23/2007 | 2 | 2/14/2007 | 4 | 1/24/2007 | 1 | 2/19/2007 |
| 9 | Yes | H | S, P | Negative | NP | 4/11/2006 | 26 | 4/3/2006 | 14 | 4/3/2006 | 14 | 4/3/2006 |
| 10 | Yes | No | S, I, N, P | Negative | NP | 8/17/2006 | 2 | 9/6/2006 | 3 | 9/4/2006 | 0 | |
| 11 | Yes | No | S, N | NP | NP | 4/27/2006 | 2 | 4/25/2006 | 4 | 4/19/2006 | 1 | 4/25/2006 |
| 12 | Yes | No | S, I, N, P | Negative | NP | 11/16/2006 | 7 | 11/8/2006 | 4 | 11/8/2006 | 3 | 11/8/2006 |
| 13 | Yes | H | S, I, P | A. fumigatus from BAL | NP | 2/24/2007 | 3 | 2/19/2007 | 5 | 2/19/2007 | 2 | 2/21/2007 |
| 14 | Yes | No | S, P | A. fumigatus from sputum and skin | CSF | 1/9/2007 | 4 | 12/28/2006 | 3 | 1/2/2007 | 0 | |
| 15 | Yes | H | S | Negative | NP | 2/21/2006 | 5 | 2/16/2006 | 4 | 2/16/2006 | 3 | 2/20/2006 |
| 16 | Yes | No | N, I | Negative | NP | 10/10/2006 | 0 | 1 | 10/2/2006 | 1 | 10/10/2006 | |
| 17 | Yes | No | N, P | Negative | NP | 4/4/2006 | 1 | 3/28/2006 | 1 | 3/28/2006 | 1 | 3/28/2006 |
Criteria are from Table 2 in Ascioglu et al. (2). CSF, cerebrospinal fluid; H, halo sign; C, air-crescent sign or cavity within area of consolidation; S, symptoms of lower respiratory tract infection; I, any new infiltrate not fulfilling major criteria; N, multiple nodular lesions in the lung, P, pleural effusion; NP, not performed.
Defined as the first day on which clinical and/or microbiological criteria were met.
Comparative values of detection of circulating A. fumigatus DNA (from LSV) and GMA for the diagnosis of IA.
In all, 1,870 serum samples from 138 treatment episodes for 124 patients were screened for GMA (median number of serum samples per episode, 10; range, 2 to 55). Circulating GMA was detected in 35 patients (113 positive serum samples). Among the 17 patients with IA, 15 had at least 2 consecutive positive GMA samples (median number of consecutive positive GMA samples, 9; range, 3 to 23). These 15 patients were therefore considered positive for GMA by EORTC criteria. One patient with IA (case 17) had only 1 positive serum sample out of 39 tested and was thus considered negative for GMA. Another IA patient (case 16) had a total of 42 serum samples analyzed, all of which were negative for GMA. Nineteen patients without IA had at least one positive GMA result (7.6% of their serum samples [34/444]) (median number of positive assays, 1; range, 1 to 9). Among these, five had at least two consecutive positive GMA samples (median, 2; range, 2 to 9). They were therefore considered positive for GMA.
A total of 938 sera from 138 treatment episodes for 124 patients were screened for A. fumigatus DNA by RT-PCR using the LSV procedure (median number of serum samples per episode, 5; range, 1 to 32). Circulating A. fumigatus DNA was detected in 21 treatment episodes (64 positive samples). All 17 patients with IA had at least one positive RT-PCR result, with 28.9% (59/205) of their serum samples positive for A. fumigatus DNA (median number of positive samples, 3; range, 1 to 14). Four patients who did not meet the EORTC/MSG criteria for IA also had one positive RT-PCR result each (1 positive sample per patient; overall, 4 positive RT-PCR results out of 36 sera). Two patients with false-positive RT-PCR results were also positive for GMA (3 of 4 and 1 of 23 serum samples, respectively), and two had one RT-PCR-positive sample each but tested negative for GMA (0 of 3 and 0 of 23 serum samples, respectively).
Sensitivity, specificity, PPV, and NPV were 100%, 96.7%, 81%, and 100%, respectively, for RT-PCR using LSV and 88.2%, 95.8%, 75.%, and 98.3%, respectively, for GMA. Thus, the overall performance of RT-PCR using LSV was consistently higher than that of GMA (Table 3).
TABLE 3.
Sensitivity, specificity, and predictive valuesa of weekly RT-PCR and biweekly GM detection for the diagnosis of IA in 138 treatment episodes for 124 high-risk patients with hematological disorders
| Assay | Se | Sp | PPV | NPV |
|---|---|---|---|---|
| RT-PCR | ||||
| LSV | 100 (90.5.-100) | 96.7 (92.1-98.3) | 81.0 (58.9-89.7) | 100 (98.4-100) |
| SSVb | 76.5 (50.7-86.9) | 96.7 (91.1-98.3) | 81.3 (55.6-89.7) | 95.6 (89.5-97.8) |
| GM | 88.2 (65.5-93.1) | 95.8 (90.8-97.8) | 75.0 (51.2-85.8) | 98.3 (94.4-99.0) |
Data are percentages (95% confidence intervals).
Evaluated in 107 treatment episodes for 105 patients.
Timing of RT-PCR and GM positivity compared with other clinical diagnostic criteria.
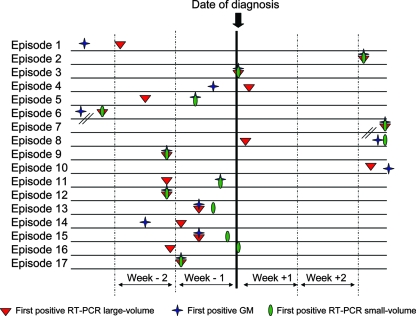
The dates of diagnosis and the dates on which the first positive test results for A. fumigatus DNA and GM were obtained are presented in Table 2. As shown in Fig. 1 and summarized in Table S1 in the supplemental material, RT-PCR performed on LSV yielded positive results either earlier than GMA (in four cases: cases 5, 8, 10, and 11) or simultaneously (in eight cases: cases 2, 3, 7, 9, 12, 13, 15, and 17). Similarly, positive results were obtained later by RT-PCR with SSV than by RT-PCR with LSV in six cases (median, 7 days; range, 7 to 25 days). No correlation was observed between the extent of pulmonary disease and the rate of positive tests by either method (data not shown).
FIG. 1.
Time interval between the day of diagnosis of IA (data from 17 episodes) and the day of the first positive result by the RT-PCR or GM assay. Week −2, days 15 through 8 before diagnosis; week −1, days 7 through 1 before diagnosis; week +1, the day of diagnosis through day 7 after diagnosis; week +2, days 8 through 15 after diagnosis.
Analysis of patients with circulating A. fumigatus DNA who did not meet the EORTC/MSG criteria for IA.
Patients were considered false positives if they tested positive (two consecutive serum samples positive for GMA and/or a single serum sample positive for A. fumigatus DNA) without meeting the EORTC/MSG criteria for IA at the time of sampling or at any time during follow-up. In all, eight patients were considered false positives for the GMA and/or RT-PCR assay. In four cases, only the GMA assay was positive; in three cases, only RT-PCR was positive; and in one case, both GMA and RT-PCR were positive.
Among the four patients with false-positive RT-PCR results, two had undergone allogeneic SCT and had grade III acute GvHD requiring intensive immunosuppression; another was neutropenic following autologous SCT for multiple myeloma. A. fumigatus DNA was detected in one instance in each case. The two patients with GvHD were placed on prophylactic posaconazole, and the third patient recovered rapidly from neutropenia. None subsequently developed clinical criteria for IA.
DISCUSSION
We show here that a slight technical adaptation in the way DNA was extracted, i.e., a 10-fold increase in the extraction volume, significantly increased the performance of RT-PCR in the detection of A. fumigatus DNA in high-risk patients with hematological malignancies. Indeed, increasing the starting serum volume for DNA extraction from 100 μl to 1 ml increased the sensitivity and NPV of the test to 100% without affecting the other performance parameters. This gain in sensitivity allowed the detection of smaller amounts of circulating DNA in the sera of high-risk patients. In six cases, the positivity of RT-PCR performed on large volumes preceded both the positive results revealed in small volumes and those identified with GMA, permitting an earlier diagnosis. In four cases, RT-PCR yielded negative results with small volumes but positive results when performed on large volumes. The use of an automated DNA extraction system made the extraction procedures easier and faster and provided a higher degree of safety with respect to avoiding DNA contamination, which may also explain, at least in part, why the number of false-positive results was low in our study.
We have also observed a high performance of prospective weekly screening of circulating DNA versus that of twice-weekly screening of GMA for the diagnosis of aspergillosis in high-risk patients with hematological disorders. The Se, Sp, PPV, and NPV for the detection of A. fumigatus DNA in at least a single sample were all higher than those for the detection of GM in two consecutive samples. A cutoff of two consecutive positive PCR results has been suggested to define a PCR-positive episode (11, 26), but we believe that this criterion is too stringent and could delay diagnosis. Indeed, because our assay was highly specific (96.7%), the diagnostic value of a single episode of DNA detection was higher than that of GM detection (PPV, 81 versus 75%; NPV, 100 versus 98.3%). However, 70% (12/17) of our IA cases had several consecutive positive PCR samples.
Several studies have prospectively evaluated the diagnostic contribution of a PCR to detect Aspergillus DNA in whole blood or serum by weekly screening for adult patients with hematological disorders who are at high-risk for IA (5, 9, 11, 13, 15). Their results have shown that the global performance of the assay, as performed, was too low to be of clinical interest but that the assay showed the ability to produce an earlier IA diagnosis when used in combination with GM detection. Different situations have been reported: PCR either had high sensitivity and NPV, while specificity and PPV were low (11, 13), or, conversely, high specificity and PPV with low sensitivity and NPV (9, 15). These discrepancies could be due to the different technical approaches used. Indeed, a major difference was the type of PCR method used in these studies, i.e., nested PCR (5, 11), PCR-enzyme-linked immunosorbent assay (9), or RT-PCR (13, 15). These different types of PCR are not equivalent in terms of contamination with previously amplified products; the nested PCR dramatically increases the risk of “false-positive results.” A second major difference was the type of blood sample used for the molecular detection of DNA. Different studies used whole blood (5, 11), plasma (15), or serum (9). A comparison of the efficiencies of the different specimen types showed that serum is an appropriate source for the diagnosis of IA by PCR and should be preferred to white blood cells (7, 8). However, in the studies that used serum, a small starting volume was used for DNA extraction, which may explain, in part, the lower performances reported in comparison with our results (6, 17).
Our RT-PCR assay also has some potential limitations. Because it was highly specific for A. fumigatus, the other species of Aspergillus were not detected. In contrast, GMA detects all Aspergillus species. However, this limitation is of little clinical consequence, because A. fumigatus is by far the species most frequently causing aspergillosis in adult oncohematology patients (19).
With high-risk patients, it is often difficult to distinguish false-positive from true results. Analysis of the case histories of three of the four patients with false-positive detection of circulating A. fumigatus DNA showed that they may have had abortive IA that failed to develop fully because of the introduction of prophylactic posaconazole in the first two cases and recovery from neutropenia in the third case. It is therefore possible that DNA detection for these patients was in fact the first sign of IA caused by A. fumigatus rather than a false-positive result. Recently, other investigators have also suggested that DNA detection by PCR is a more sensitive tool than current diagnostic procedures for distinguishing true from “false” cases of subclinical IA (13, 26). Although this hypothesis requires further confirmation by large-scale studies, our results will be of interest in helping to design the molecular diagnostic approach for IA in high-risk neutropenic patients that is currently most appropriate.
In summary, our results showed that the use of a larger volume of serum improves the performance of RT-PCR detection of A. fumigatus DNA and adds clinical value to GMA detection for high-risk adult patients with hematological disorders.
Supplementary Material
Acknowledgments
We thank Eric Abachin for fruitful discussions and Orene Greer for extensive review of the English language in the manuscript.
Footnotes
Published ahead of print on 24 September 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Asano-Mori, Y., Y. Kanda, K. Oshima, S. Kako, A. Shinohara, H. Nakasone, M. Kaneko, H. Sato, T. Watanabe, N. Hosoya, K. Izutsu, T. Asai, A. Hangaishi, T. Motokura, S. Chiba, and M. Kurokawa. 2008. False-positive Aspergillus galactomannan antigenaemia after haematopoietic stem cell transplantation. J. Antimicrob. Chemother. 61411-416. [DOI] [PubMed] [Google Scholar]
- 2.Ascioglu, S., J. H. Rex, B. de Pauw, J. E. Bennett, J. Bille, F. Crokaert, D. W. Denning, J. P. Donnelly, J. E. Edwards, Z. Erjavec, D. Fiere, O. Lortholary, J. Maertens, J. F. Meis, T. F. Patterson, J. Ritter, D. Selleslag, P. M. Shah, D. A. Stevens, and T. J. Walsh. 2002. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin. Infect. Dis. 347-14. [DOI] [PubMed] [Google Scholar]
- 3.Aubry, A., R. Porcher, J. Bottero, S. Touratier, T. Leblanc, B. Brethon, P. Rousselot, E. Raffoux, J. Menotti, F. Derouin, P. Ribaud, and A. Sulahian. 2006. Occurrence and kinetics of false-positive Aspergillus galactomannan test results following treatment with beta-lactam antibiotics in patients with hematological disorders. J. Clin. Microbiol. 44389-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes, P. D., and K. A. Marr. 2007. Risks, diagnosis and outcomes of invasive fungal infections in haematopoietic stem cell transplant recipients. Br. J. Haematol. 139519-531. [DOI] [PubMed] [Google Scholar]
- 5.Buchheidt, D., M. Hummel, D. Schleiermacher, B. Spiess, R. Schwerdtfeger, O. A. Cornely, S. Wilhelm, S. Reuter, W. Kern, T. Sudhoff, H. Morz, and R. Hehlmann. 2004. Prospective clinical evaluation of a LightCycler-mediated polymerase chain reaction assay, a nested-PCR assay and a galactomannan enzyme-linked immunosorbent assay for detection of invasive aspergillosis in neutropenic cancer patients and haematological stem cell transplant recipients. Br. J. Haematol. 125196-202. [DOI] [PubMed] [Google Scholar]
- 6.Challier, S., S. Boyer, E. Abachin, and P. Berche. 2004. Development of a serum-based TaqMan real-time PCR assay for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 42844-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa, C., J. M. Costa, C. Desterke, F. Botterel, C. Cordonnier, and S. Bretagne. 2002. Real-time PCR coupled with automated DNA extraction and detection of galactomannan antigen in serum by enzyme-linked immunosorbent assay for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 402224-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa, C., D. Vidaud, M. Olivi, E. Bart-Delabesse, M. Vidaud, and S. Bretagne. 2001. Development of two real-time quantitative TaqMan PCR assays to detect circulating Aspergillus fumigatus DNA in serum. J. Microbiol. Methods 44263-269. [DOI] [PubMed] [Google Scholar]
- 9.Florent, M., S. Katsahian, A. Vekhoff, V. Levy, B. Rio, J. P. Marie, A. Bouvet, and M. Cornet. 2006. Prospective evaluation of a polymerase chain reaction-ELISA targeted to Aspergillus fumigatus and Aspergillus flavus for the early diagnosis of invasive aspergillosis in patients with hematological malignancies. J. Infect. Dis. 193741-747. [DOI] [PubMed] [Google Scholar]
- 10.Hage, C. A., J. M. Reynolds, M. Durkin, L. J. Wheat, and K. S. Knox. 2007. Plasmalyte as a cause of false-positive results for Aspergillus galactomannan in bronchoalveolar lavage fluid. J. Clin. Microbiol. 45676-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halliday, C., R. Hoile, T. Sorrell, G. James, S. Yadav, P. Shaw, M. Bleakley, K. Bradstock, and S. Chen. 2006. Role of prospective screening of blood for invasive aspergillosis by polymerase chain reaction in febrile neutropenic recipients of haematopoietic stem cell transplants and patients with acute leukaemia. Br. J. Haematol. 132478-486. [DOI] [PubMed] [Google Scholar]
- 12.Hughes, W. T., D. Armstrong, G. P. Bodey, E. J. Bow, A. E. Brown, T. Calandra, R. Feld, P. A. Pizzo, K. V. Rolston, J. L. Shenep, and L. S. Young. 2002. 2002 guidelines for the use of antimicrobial agents in neutropenic patients with cancer. Clin. Infect. Dis. 34730-751. [DOI] [PubMed] [Google Scholar]
- 13.Jordanides, N. E., E. K. Allan, L. A. McLintock, M. Copland, M. Devaney, K. Stewart, A. N. Parker, P. R. Johnson, T. L. Holyoake, and B. L. Jones. 2005. A prospective study of real-time panfungal PCR for the early diagnosis of invasive fungal infection in haemato-oncology patients. Bone Marrow Transplant. 35389-395. [DOI] [PubMed] [Google Scholar]
- 14.Kami, M., T. Fukui, S. Ogawa, Y. Kazuyama, U. Machida, Y. Tanaka, Y. Kanda, T. Kashima, Y. Yamazaki, T. Hamaki, S. Mori, H. Akiyama, Y. Mutou, H. Sakamaki, K. Osumi, S. Kimura, and H. Hirai. 2001. Use of real-time PCR on blood samples for diagnosis of invasive aspergillosis. Clin. Infect. Dis. 331504-1512. [DOI] [PubMed] [Google Scholar]
- 15.Kawazu, M., Y. Kanda, Y. Nannya, K. Aoki, M. Kurokawa, S. Chiba, T. Motokura, H. Hirai, and S. Ogawa. 2004. Prospective comparison of the diagnostic potential of real-time PCR, double-sandwich enzyme-linked immunosorbent assay for galactomannan, and a (1→3)-β-d-glucan test in weekly screening for invasive aspergillosis in patients with hematological disorders. J. Clin. Microbiol. 422733-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maertens, J. A., R. Klont, C. Masson, K. Theunissen, W. Meersseman, K. Lagrou, C. Heinen, B. Crepin, J. Van Eldere, M. Tabouret, J. P. Donnelly, and P. E. Verweij. 2007. Optimization of the cutoff value for the Aspergillus double-sandwich enzyme immunoassay. Clin. Infect. Dis. 441329-1336. [DOI] [PubMed] [Google Scholar]
- 17.Millon, L., R. Piarroux, E. Deconinck, C. E. Bulabois, F. Grenouillet, P. Rohrlich, J. M. Costa, and S. Bretagne. 2005. Use of real-time PCR to process the first galactomannan-positive serum sample in diagnosing invasive aspergillosis. J. Clin. Microbiol. 435097-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pagano, L., M. Caira, A. Nosari, M. T. Van Lint, A. Candoni, M. Offidani, T. Aloisi, G. Irrera, A. Bonini, M. Picardi, C. Caramatti, R. Invernizzi, D. Mattei, L. Melillo, C. de Waure, G. Reddiconto, L. Fianchi, C. G. Valentini, C. Girmenia, G. Leone, and F. Aversa. 2007. Fungal infections in recipients of hematopoietic stem cell transplants: results of the SEIFEM B-2004 study—Sorveglianza Epidemiologica Infezioni Fungine Nelle Emopatie Maligne. Clin. Infect. Dis. 451161-1170. [DOI] [PubMed] [Google Scholar]
- 19.Perfect, J. R., G. M. Cox, J. Y. Lee, C. A. Kauffman, L. de Repentigny, S. W. Chapman, V. A. Morrison, P. Pappas, J. W. Hiemenz, and D. A. Stevens. 2001. The impact of culture isolation of Aspergillus species: a hospital-based survey of aspergillosis. Clin. Infect. Dis. 331824-1833. [DOI] [PubMed] [Google Scholar]
- 20.Pfeiffer, C. D., J. P. Fine, and N. Safdar. 2006. Diagnosis of invasive aspergillosis using a galactomannan assay: a meta-analysis. Clin. Infect. Dis. 421417-1427. [DOI] [PubMed] [Google Scholar]
- 21.Sulahian, A., S. Touratier, and P. Ribaud. 2003. False positive test for Aspergillus antigenemia related to concomitant administration of piperacillin and tazobactam. N. Engl. J. Med. 3492366-2367. [DOI] [PubMed] [Google Scholar]
- 22.Surmont, I., and W. Stockman. 2007. Gluconate-containing intravenous solutions: another cause of false-positive galactomannan assay reactivity. J. Clin. Microbiol. 451373. (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Upton, A., K. A. Kirby, P. Carpenter, M. Boeckh, and K. A. Marr. 2007. Invasive aspergillosis following hematopoietic cell transplantation: outcomes and prognostic factors associated with mortality. Clin. Infect. Dis. 44531-540. [DOI] [PubMed] [Google Scholar]
- 24.Viscoli, C., M. Machetti, P. Cappellano, B. Bucci, P. Bruzzi, M. T. Van Lint, and A. Bacigalupo. 2004. False-positive galactomannan Platelia Aspergillus test results for patients receiving piperacillin-tazobactam. Clin. Infect. Dis. 38913-916. [DOI] [PubMed] [Google Scholar]
- 25.Wheat, L. J., and T. J. Walsh. 2008. Diagnosis of invasive aspergillosis by galactomannan antigenemia detection using an enzyme immunoassay. Eur. J. Clin. Microbiol. Infect. Dis. 27245-251. [DOI] [PubMed] [Google Scholar]
- 26.White, P. L., C. J. Linton, M. D. Perry, E. M. Johnson, and R. A. Barnes. 2006. The evolution and evaluation of a whole blood polymerase chain reaction assay for the detection of invasive aspergillosis in hematology patients in a routine clinical setting. Clin. Infect. Dis. 42479-486. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.