Abstract
The development and validation of a one-step, single-tube, real-time accelerated reverse-transcription loop-mediated isothermal amplification (RT-LAMP) for the detection of the L RNA segment of Rift Valley fever virus (RVFV) are described. The assay was performed at a constant temperature (63°C), with a real-time follow-up using a LightCycler and a double-stranded-DNA-intercalating fluorochrome. The assay is highly sensitive and comparable to real-time RT-PCR, with a detection limit of ∼10 RNA copies per assay. However, the RT-LAMP assay is much faster than traditional RT-PCR and generates results in <30 min for most diluted samples. The specificity of the primers was established using other, related arboviruses as well as virus-containing and virus-free sera. The RT-LAMP assay reported here is thus a valuable tool for the rapid detection of RVFV in field diagnostic laboratories.
Rift Valley fever virus (RVFV) is a zoonotic mosquito-borne virus of the genus Phlebovirus in the family Bunyaviridae. Its tripartite negative-strand RNA genome is composed of a large segment (L), encoding the viral transcriptase, a medium segment (M), coding for the two external glycoproteins (GN and GC), and an S segment, which codes for the nucleocapsid protein (N) and a nonstructural protein (NSs) (22). First identified in Kenya in 1931 (7), RVFV is now considered an endemic zoonotic agent in sub-Saharan Africa and Madagascar (5). The virus causes explosive outbreaks in animals and humans (5) and has been observed in Egypt, in Mauritania, and more recently in the Arabian Peninsula (2).
The virus is transmitted to animals mainly by hematophagous mosquitoes of many genera (Aedes, Anopheles, Culex, Eretmapodites, and Mansonia) during seasons of high rainfall (6). Domestic animals are sensitive to RVFV infection and amplify the virus to high titers (26). Among cattle, sheep, goats, pigs, and camels, infection causes fever and anorexia. Although fewer than 10% of infected animals die, abortions are frequently observed among infected pregnant animals. (26).
Human infections are due mainly to direct exposure to infected material from sick animals during handling or slaughter (26) and sometimes to infected-mosquito bites. Consumption of raw milk from infected animal is also considered a risk. In most human cases, the disease is characterized by a brief self-limited febrile illness lasting 2 to 5 days, which progresses in 1 to 2% of patients to serious complications, including retinitis, blindness, encephalitis, hepatitis, or hemorrhagic syndromes that may lead to death in 10 to 20% of the cases (2).
RVFV diagnosis relies on specific-antibody detection, virus isolation (3, 5), and genome amplification (8, 21). Protocols have been developed that specifically detect RVFV genomes through technology such as real-time quantitative PCR (RT-PCR) (8). Among the alternative methods for nucleic acid amplification is loop-mediated isothermal amplification (LAMP), developed in 2000 (13, 24). LAMP amplifies specific sequences on nucleic acids using a set of six primers and relies on the strand displacement activity of the DNA polymerase. It is performed at a constant temperature (60 to 65°C), without cyclic denaturation of the template. RNA can be amplified simply by the addition of avian myeloblastosis virus reverse transcriptase to the reaction mix, keeping the same reaction conditions as for DNA amplification (24). High amplification rates are observed, leading to the production of large amounts of double-stranded DNA (dsDNA) and leading as well to production of a white precipitate of magnesium pyrophosphate that can be observed with the naked eye. When primers are properly designed, the reaction is as specific and sensitive as traditional PCR or RT-PCR and is, moreover, faster (12). LAMP or RT-LAMP is thus efficient, fast, and inexpensive, and since isothermal reaction conditions are easy to provide, LAMP is of particular interest for field diagnosis of tropical diseases.
Currently, primers sets have been published for the detection of various arboviruses, such as dengue virus serotypes 1, 2, 3, and 4 (17), Chikungunya virus (14), West Nile virus (16), and Japanese encephalitis virus (25), thus further contributing to the ease of field trials of this assay.
The aims of this study were to develop and validate a reverse-transcription LAMP method (RT-LAMP) for RVFV RNA amplification with a real-time follow-up and to evaluate the potential of this method for diagnostic purposes.
MATERIALS AND METHODS
Cell culture.
C6/36 cells (Aedes albopictus larva cells) were grown at 28°C in Leibowitz's L15 medium (BioWhittaker Europe, Verniers, Belgium) supplemented with 1% l-glutamine, 2% tryptose phosphate broth, and fetal calf serum (5%, final). Vero cells were grown at 37°C in MEM 199 (Invitrogen) supplemented with 5% fetal calf serum.
Virus strains.
RVFVs of the four topotypes were used: RVFV (IMTSSA/H-2067, human serum, Chad, 2005) (5), Senegal (ArSEN84, ArD38661), Madagascar (AnMg990), Egypt (ZH548, HEgy93), Kenya (HKen98), and RVFV clone 13 (note that for three of these topotypes, we used two different viral strains, for a total of seven viral strains corresponding to the four topotypes). Other arboviruses were also used: Toscana (IMTSSA/H-4906, human LCR, France, 1998) (20), Sandfly Sicilian (Sabin strain), Sandfly Naples (Sabin strain), Belterra (Brazil, 1977), Punta Toro (Adames strain; kindly provided by D. H. L. Bishop), dengue virus strains 1, 2, 3, and 4 (dengue virus strain 1, IMTSSA/H-658, isolated from human serum, Cambodia, 1998; dengue virus strain 2, IMTSSA/H-1164, human serum, Somalia, 1994; dengue virus strain 3, IMTSSA/H-1731, human serum [18], Guadeloupe, 1994; dengue virus strain 4, IMTSSA/H-812, human serum, Indonesia, 1994), yellow fever virus (17D), Japanese encephalitis virus (Nakayama), West Nile virus (Tunisia, 1997), Saint Louis encephalitis virus (MSI-7), tick-borne encephalitis virus (Langat), and Chikungunya (IMTSSA/H-6368, human serum, Réunion, 2005) (1). The viruses were propagated in C6/36 cells or Vero cells as described previously (5).
Clinical samples and infected sera.
Serum samples from patients with clinical suspicion of arbovirus infections were sent to the virological laboratory at the Institut de Médecine tropicale du Service de Santé des Armées. Control sera from healthy donors were also tested. All the samples were transported and stored at −80°C until use. Since we lacked human and animal RVFV-positive sera, we reconstituted sera and made them virus positive by mixing 200 μl of fetal calf serum with RVFV. Sera were then processed as described below.
RNA extraction.
Viral RNA was extracted from 140 μl of infected cell supernatants or (patient) sera by using a High Pure viral RNA kit or an RNeasy minikit (Qiagen) following the manufacturer's protocol. The eluted RNA was stored at −70°C until used.
Primer design.
The complete RVFV L segment sequences available in the GenBank database (see the legend to Fig. 1) were aligned using CLUSTAL software (23) to identify conserved regions (Fig. 1). Potential target regions were analyzed with the LAMP primer design software, and specific primers were automatically designed (www.http://primerexplorer.jp/elamp4.0.0/index.html). A set of six primers comprising two outer, two inner, and two loop primers was selected. FIP contained F1C (complementary to F1), a TTTT spacer, and the F2 sequence. BIP contained the B1C sequence (complementary to B1), a TTTT spacer, and the B2 sequence. All the described primers (Table 1) were ordered as standard desalted primers (Eurogentec, Angers, France).
FIG. 1.
Alignment of RVFV L segments (nucleotides 6117 to 6331) and positions of LAMP primers. The ends of the primers from which the elongation starts are indicated by arrows. Accession numbers for the strains aligned (top to bottom) are DQ375434.1, DQ375429.1, DQ375432.1, DQ375421.1, DQ375419.1, DQ375423.1, DQ375416.1, DQ375411.1, DQ375406.1, DQ375412.1, DQ375414.1, DQ375425.1, DQ375401.1, DQ375433.1, DQ375396.1, DQ375397.1, DQ375399.1, DQ375427.1, and DQ375428.1.
TABLE 1.
LAMP primers for RVFV amplificationa
| Primer | Type | Sequence (5′-3′)b |
|---|---|---|
| F3-RVF | Forward outer | TTTCTAAGATGGGGAGAGC |
| B3-RVF | Reverse outer | AAATCTAAGCCTGACTTTGC |
| FIP-RVF | Forward inner (F1c-TTTT-F2) | TGTAGAGTTCTGAAGAGAACCTTTGTTTTTAGGAAAGTTCTAGAGACAGG |
| BIP-RVF | Reverse inner (B1c-TTTT-B2) | GAGACCAGAAGAGAGCATTAGGATTTTTTGGTCTTAGCCTAGCATGT |
| LFP-RVF | Forward loop | TCTTTGCTGGAGCACCGA |
| BLP-RVF | Backward loop | ATCTGGAGTTATATGAGGAGACAGA |
Primers were designed using Primer Explorer 4 (http://primerexplorer.jp/elamp4.0.0/index.html) and modified to optimize annealing of the ends of the primers from which the elongation starts on different RVFV strains, according to the sequence alignment in Fig. 1.
Boldface characters indicate TTTT spacer.
RT-LAMP.
The RT-LAMP reaction was carried out in a 20-μl total reaction mixture volume containing 50 pmol each of the inner primers FIP and BIP, 5 pmol each of the outer primers F3 and B3, 20 pmol each of the loop primers loop F and loop B, 1.4 mM of each deoxynucleoside triphosphate, 0.6 M betaine (Sigma Aldrich, St. Quentin-Fallavier, France), 0.1 U of avian myeloblastosis virus RTase (Promega, Charbonnières les Bains, France), 6.5 U of Bst DNA polymerase (large fragment; New England Biolabs), 1× Thermopol buffer (Biolabs), and the appropriate amounts of target RNA. For field applicability, the mixture was incubated at 63°C in a heating block (Biometra, Goettingen, Germany) or a water bath. For real-time monitoring of the RT-LAMP reaction, the reaction mixture was incubated at 63°C in a LightCycler 1.0 (Roche Diagnostics, Meylan, France) with 1 μl of a 0.01-μg/ml solution of ethidium bromide (EtBr; Bio-Rad laboratories, Marne la Coquette, France) with fluorescence reading on channel 2 or 1 μl of 1/1,000 SYBR solution (Roche) with fluorescence reading on channel 1. The run was set up as follows: 60 cycles of 1 min at 63°C, with fluorescence reading at the end of each of these cycles, followed by a standard melting curve analysis by heating for 2 s at 95°C and 2 s at 60°C and then increasing to 95°C with 0.1°C/s ramping with continuous fluorescence monitoring, before a final cooling step at 40°C for 10 s. Interestingly, EtBr led to a better detection signal than the SYBR green classically used with the LightCycler under the reaction conditions used here. For both fluorochromes, the amplification curves were not sigmoidal, contrary to classical real-time PCR or even LAMP when a turbidimeter (650 nm) is used to detect magnesium pyrophosphate precipitate generation. The curves instead look like a hat: a fluorescence increase followed by a fluorescence decrease. We assume that during the RT-LAMP reaction, the synthesis of dsDNA leads to a fluorescence increase due to the fluorochrome binding. At the same time, the magnesium pyrophosphate precipitation leads to a turbidity increase that masks the fluorescence. This masking effect might be the most significant at the end of the reaction, leading to a fluorescence diminution detection by the optic sensor. Nevertheless, we managed to get sigmoid curves with EtBr using the F2/F3 LightCycler fluorescence report mode, allowing a subsequent threshold cycle (CT) calculation using the “second derivative maximum” setting.
Analysis of RT-LAMP products.
For agarose gel analysis, RT-LAMP products were incubated at 63°C, electrophoresed using a 2% agarose gel in Tris-borate buffer (Interchim, Montlucon, France) with EtBr, and visualized using a transilluminator (Bio-Rad laboratories). For naked-eye visualization following amplification, the tubes were examined for white turbidity or for the presence of a white pellet after a pulse spin. The positivity of the real-time RT-LAMP assay was determined as an increase of fluorescence above the baseline value and the time for positivity given by the CT value (second derivative maximum, 1 cycle corresponding to 1 min of reaction).
Real-time Taqman RT-PCR.
The RVFV Taqman assay developed by Garcia et al. (8) was used as a reference. Briefly, the reaction was conducted in a 20-μl assay with 10 μl of buffer, 0.8 μl of enzyme mix from a Superscript III one-step RT-PCR system with Platinum Taq (Invitrogen), 0.5 μM of the forward primer S432 (5′-ATG ATG ACA TTA GAA GG GA-3′), 0.5 μM of the reverse primer NS3m (5′-ATG CTG GGA AGT GAT GAG-3′), and 0.5 μM probe CRSSAr (5′-ATT GAC CTG TGC CTG TTGCC-3′) (8) during 30 min of RT at 50°C, 2 min of denaturation at 95°C, 50 cycles of 5 s at 95°C, and annealing/elongation for 1 min before fluorescence reading.
RESULTS
Sensitivity and specificity.
The L segment of the RVFV genome was used to design primers, as it encodes the viral polymerase and was thus more prone to contain conserved sequences than the S and M segments. We initially attempted to locate an ∼200-nucleotide sequence fully conserved among all the RVFV L-segment genomes. However, we were unable to identify a stretch of nucleotides that met these criteria. Therefore, primers were designed by Primerexplorer software based on a conserved segment of the L gene (nucleotides 6115 to 6404) and modified to fit the viral sequences. A limited number of mismatches (one or two, in the middle or at the 5′ ends of the primers) was thought to be acceptable; particular caution was exercised to avoid template and primer 3′-end mismatches (where elongation by the Bst DNA polymerase is initiated) (Fig. 1). A one-step, single-tube, real-time RT-LAMP assay was standardized with the selected primers under the conditions described above. A 282-nucleotide segment was then amplified (Table 1). The specificity was determined using RNA extracts of infected C6/36 and Vero cells. The reaction was monitored by naked-eye visualization, agarose gel deposition, and real-time follow-up. A total of ∼106 copies of viral RNA were tested in each sample. The seven RVFV strains were assayed, including the four RVFV subtypes: Chad, Senegal, Madagascar, Egypt (two strains), Kenya, and the vaccine candidate clone 13. All were positively amplified using this RT-LAMP protocol. The RVFV-specific RT-LAMP primers demonstrated a high degree of specificity for RVFV by yielding negative results for all other tested phleboviruses (Toscana, Sandfly Sicilian, Sandfly Naples, Belterra, and Punta Toro), flaviviruses (dengue virus types 1, 2, 3, and 4, yellow fever virus, Japanese encephalitis virus, West Nile virus, Saint Louis encephalitis virus, and tick-borne encephalitis virus), and an alphavirus (Chikungunya virus) (Table 2).
TABLE 2.
Species specificity of RVFV RT-LAMPa
| Virus (strain and/or source; reference) | RT-LAMP detection |
|---|---|
| RVFV (IMTSSA/H-2167, human serum, Chad, 2005) (5) | + |
| RVFV (Senegal) | + |
| RVFV (Madagascar) | + |
| RVFV (Egypt) | + |
| RVFV (Kenya) | + |
| RVFV (vaccine candidate clone 13) | + |
| RVFV (wild-type strain ZH548) | + |
| Toscana (IMTSSA/H-4906, human LCR, France, 1998) (19) | − |
| Sandfly Naples (Sabin strain) | − |
| Sandfly Sicilian (Sabin strain) | − |
| Belterra (Brazil, 1977) | − |
| Punta Toro (Adames strain, kindly provided by D. H. L. Bishop) | − |
| Dengue virus 1 (IMTSSA/H-658, isolated in human serum, Cambodia, 1998) | − |
| Dengue virus 2 (IMTSSA/H-1164, human serum, Somalia, 1994) | − |
| Dengue virus 3 (IMTSSA/H-1731, human serum, Guadeloupe, 1994) (19) | − |
| Dengue virus 4 (IMTSSA/H-812, human serum, Indonesia, 1994) | − |
| Yellow fever virus (17D) | − |
| Japanese encephalitis virus (Nakayama) | − |
| West Nile virus (Tunisia 1997) | − |
| Saint Louis encephalitis virus (MSI-7) | − |
| Tick-borne encephalitis virus (Langat) | − |
| Chikungunya virus (IMTSSA/H-6368, human serum, Réunion, 2005) (1) | − |
RNAs extracted from supernatants of cells infected with different arboviruses were assayed for detection using RVFV RT-LAMP; ∼106 copies of each viral RNA were used for each assay.
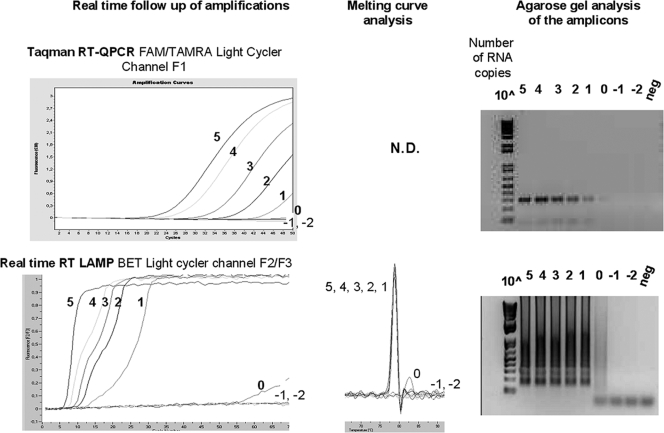
RVFV RT-LAMP and one-step Taqman RT-PCR (8) were compared by testing 10-fold serial dilutions of a viral RNA extracted from infected cells supernatant. Samples containing 105 to 10−2 RNA copies/assay of RVFV (strain Chad) were amplified by both Taqman RT-PCR (8) and RVFV RT-LAMP. The amplification was either real-time monitored or end point controlled by naked-eye visualization and agarose gel deposition. The RT-LAMP was as sensitive as the quantitative RT-PCR, with a detection limit of ∼101 RVFV RNA copies per assay (Fig. 2). The same analyses were performed on the other RVFV strains of this study: for all strains tested (Senegal, Kenya, Egypt [HEgy93 and ZH548], Madagascar, and clone 13), the detection limit corresponded to ∼101 RVFV RNA copies per assay (data not shown). This limit corresponded to that of Taqman RT-PCR (8). Note that clone 13 could not be amplified by the Taqman system, as described by others (8), since the primers are located in a genomic region that is not present in that attenuated strain. However, RT-LAMP amplification of clone 13 was not impaired, since the primers are in a genomic segment present in all strains.
FIG. 2.
Sensitivity of RVFV LAMP and real-time Taqman PCR (8). RVFV RNA extracted from supernatants of infected cells was serially diluted, and samples containing 105 to 10−2 copies were assayed in parallel using Taqman RT-PCR and our RT-LAMP assay. Real-time follow-up on a LightCycler (using channel F2/F3) and melting curve analysis for LAMP are reported. Reaction products were analyzed on agarose gels with ethidium bromide staining.
Rapidity and quantitation.
The RT-LAMP results indicated that the time required to detect a signal was 7.9 min when ∼105 RNA copies were present to 18.0 min when ∼10 copies of RNA were present, as compared to the Taqman RT-PCR CTs of 18.8 (corresponding to 30 min for RT plus 16.6 min for PCR) to 39.9 (30 min for RT plus 35 min for PCR) (Table 3). A standard curve showing the relationship between the concentrations of RVFV ranging from 101 to 105 RNA copies/assay and the time to positivity (CT) was built for the RT-LAMP (Fig. 3). A relationship was found between the number of RNA copies and the time to signal detection in the range of 100 to 100,000 copies, with a correlation coefficient (R2) of 0.95. No correlation (R2 < 0.8) was found in the range of 10 to 100,000 copies.
TABLE 3.
Comparison of RVFV Taqman CT and LAMP times for detection using real-time follow-upa
| Estimated no. of RNA copies |
CT
|
|
|---|---|---|
| Taqman RT-PCR (no. of cycles) | RT-LAMP (min) | |
| 100,000 (105) | 18.2 | 7.9 |
| 10,000 (104) | 20.3 | 8.5 |
| 1,000 (103) | 27.4 | 10.5 |
| 100 (102) | 33.4 | 12.4 |
| 10 (101) | 39.9 | 18.0 |
| ∼1 (10°) | − | − |
| ∼0.1 (10−1) | − | − |
| ∼0.01 (10−2) | − | − |
RT-PCR Taqman and RT-LAMP were performed with a LightCycler instrument following procedures described in Materials and Methods. The CT was determined by the fit points method at the end of the run. Note that for the Taqman PCR, the difference from CT to CT between dilutions is not 3, since efficiency of the reaction is not 2 (slope is −4 instead of −3,3) (8).
FIG. 3.
Standard curve for the RVFV-specific RT-LAMP. Time for LAMP positivity (CT estimated by the LightCycler) was plotted against initial number of RNA copies, on 10-fold serial dilutions of RVFV viral RNA. A logarithmic regression curve was constructed to evaluate the relationship between the two elements. The equation and R2 coefficient are presented for the range of 100 to 100,000 copies, since no significant relationship was found for smaller amounts (R2 < 0.8). Results are from one experiment representative of three different assays.
Evaluation of the RT-LAMP assay in serum samples.
We could not obtain any human or animal sera positive for the presence of RVFV. Nevertheless, to evaluate the reliability of our assay as a first diagnosis system in the field, we screened sera negative for RVFV and used reconstituted sera. Human sera in which other arboviruses had been detected by RT-PCR at different concentrations were used as negative controls. Fifty different sera were assayed: 9 positive for Toscana virus, 19 positive for dengue virus strain 1 virus, 3 positive for dengue virus strain 2, 1 positive for West Nile virus, 8 positive for Chikungunya virus, and 10 negative for all other viruses tested. None of these sera gave a positive signal after 60 min of RT-LAMP assay using a real-time follow-up with the LightCycler (Table 4). For positive samples, we used the strategy of reconstituted sera: we mixed the different RVFV strains with sterile fetal calf serum before RNA extraction to mimic a serum from a viremic animal. Similarly, we mixed the RVFV strains with five different human sera that were negative for RVFV, in order to mimic sera from viremic patients. All RVFV subtypes identified so far were used at a concentration corresponding to the LAMP detection limit as determined above. In all of these samples, the RVFV genome was positively amplified (Table 4); as expected, the presence of serum in the sample before the RNA extraction did not interfere with the RT-LAMP amplification process.
TABLE 4.
Evaluation of RT-LAMP assay in serum samplesa
| Sample | No. assayed | No. positive (all strains) |
|---|---|---|
| Human sera (arbovirus positive, RVFV negative) | 40 | 0 |
| Human sera (arbovirus negative) | 10 | 0 |
| Reconstituted animal sera | 7 | 7 |
| Reconstituted human sera | 7 | 7 |
Samples were either human sera negative for RVFV or reconstituted animal or human sera made positive for RVFV through individual mixing with each of the seven RVFV strains used in this study.
DISCUSSION
The outbreaks of RVFV in Africa and the Arabian Peninsula have highlighted the necessity of developing a surveillance plan for humans and animals in these regions. The data presented here may help predict and prevent future large-scale epidemics because of the speed and ease of reliably diagnosing Rift Valley fever at early stages, when control measures can be implemented before a large-scale outbreak occurs. The diagnosis of RVFV infection from clinical samples is currently based on specific immunoglobulin M or G detection, virus isolation, or genomic detection (3, 5, 8, 21). Although the classical methods are effective, they are time-consuming. Various nucleic acid amplification techniques allow rapid, specific, and sensitive detection. However, current nucleic acid-based detection methods are difficult to adapt for field use.
The RVFV RT-LAMP developed in this work is an easy, rapid, specific, and sensitive assay. A one-step, single-tube, real-time RT-LAMP assay was optimized to allow RVFV L-segment detection in less than 30 min, with a limit of detection of ∼10 RNA molecules/assay. A low limit of detection is characteristic of the LAMP method (13). The unmatched characteristics of the RT-LAMP, compared to quantitative RT-PCR (2 to 3 h) (8), also offer the advantage of continuous amplification under isothermal conditions that can be provided with a simple water bath. Other isothermal amplification techniques, such as nucleic acid sequence-based amplification (4) and the self-sustained sequence reaction, are reported to be less specific, probably due to the low stringency (reaction temperature is ∼40°C), and require either a precision instrument or a complex method for the detection of the amplified products due to the poor specificity of target sequence selection (4). Nevertheless, when performing LAMP under “field-like” conditions with a simple water bath, we noticed, as others have done (18), that two aspects should be considered. First, special care should be taken to avoid any contamination with previously amplified LAMP products. Due to the high amplification rate, any trace of LAMP DNA concatemers will be easily reamplified and lead to false-positive results. Thus, end-point reading options that do not require opening reaction tubes (turbidity or fluorescence reading) should be preferred to agarose gel analysis. Second, reagents should be kept on ice until heating of the samples to 60°C, in order to prevent nonspecific start of the reaction and primer multimer amplification. For that reason, it would be relevant to develop a “hot start” LAMP, which already exists for PCR, to facilitate processing of the reaction in field conditions.
The real-time monitoring of LAMP offers the fastest and most sensitive protocol. The RT-LAMP reaction can be achieved by the follow-up of the ethidium bromide fluorescence detected in the F2 channel of the LightCycler or by using any other dsDNA-intercalating fluorescent molecule and apparatus. The use of this in-house-developed protocol allows LightCycler-equipped laboratories to test the potential of the LAMP technique for molecular diagnosis.
The RT-LAMP assay requires six primers that recognize eight distinct regions of the target and, when properly designed, prevent cross-reactions (Table 2). Due to the setting of these primers in a highly conserved RVFV genetic region, we assumed that the RT-LAMP assay would amplify all the RVFV genomes in the genetic clusters. Accordingly, all the RVFV strains we assayed, representing all the virus subtypes identified so far, led to efficient amplification in the RT-LAMP assay. We did not get any RVFV-positive human or animal sera, so we chose to simulate viremic sera by mixing RVFV with bovine or human serum before RNA extraction. The RVFV RT-LAMP assay was successfully performed using such reconstituted positive samples. The RVFV-negative sera were found to be negative by using the RVFV-specific RT-LAMP. Therefore, we obtained no interference between components of the serum and the RT-LAMP process. This expected result is compatible with the literature, since LAMP and RT-LAMP have already been shown to be well-suited tools for the detection of pathogens in diagnosis samples, including viruses, bacteria, parasites, and fungi (see for instance references 9, 14, and 16).
The poor quantitative parameters of the assay (R2 = 0.95; the imprecise estimation of the CT value) render the RVFV RT-LAMP inappropriate as a precise quantification tool, in contrast to Taqman RT-PCR. Kuhara and collaborators reported the same limited relationship between the LAMP CT and herpesvirus 8 DNA concentration (11). Similarly, other authors have shown the real-time follow up of LAMP or RT-LAMP, using specifically designed turbidimeters (LA-100 and LA-200; Eiken Company). The nonregular spacing between the amplification curves from serial dilutions of the target nucleic acid points out the nonlinearity of the CT-concentration relationship over a wide concentration range (10, 14, 15). Nevertheless, for diagnostic purposes, exact quantification of the pathogen load is not necessary. A semiquantitative result, such as high, medium, or low viral titers, will provide clinically useful information. In this regard, the real-time RT-LAMP follow-up can easily be achieved with a LightCycler or a turbidimeter.
RT-LAMP is a new technique with potential for continued development. The RT-LAMP method appears to be well adapted for molecular diagnosis in laboratories, considering its high specificity, high speed, and low cost. Specific turbidimeters are available for the RT-LAMP real-time follow-up (Eiken Company). The ability to visually assess the white magnesium pyrophosphate precipitate at the end point of the RT-LAMP assay is a distinct advantage in the development of this method for field diagnosis of infectious diseases. As interest in techniques allowing precise and rapid diagnosis of infectious agents under sometime difficult conditions is increasing, the characteristics and performance of the RT-LAMP deserve attention.
Acknowledgments
We are indebted to Jon Davis for reviewing the manuscript.
This work was funded by the French Armed Forces Medical Service and the French Délégation Générale pour l'Armement (contrat d'objectif 08co402).
The opinions and assertions contained herein are those of the authors and are not to be construed as official or reflecting the views of the French Armed Forces Medical Service or the French Army at large.
Footnotes
Published ahead of print on 17 September 2008.
REFERENCES
- 1.Bessaud, M., C. N. Peyrefitte, B. A. Pastorino, F. Tock, O. Merle, J. J. Colpart, J. S. Dehecq, R. Girod, C. Jaffar-Bandjee, P. J. Glass, M. Parker, H. Tolou, and M. Grandadam. 2006. Chikungunya virus strains, Reunion island outbreak. Emerg. Infect. Dis. 121604-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bird, B. H., M. L. Khristova, P. E. Rollin, T. G. Ksiazek, and S. T. Nichol. 2007. Complete genome analysis of 33 ecologically and biologically diverse Rift Valley fever virus strains reveals widespread virus movement and low genetic diversity due to recent common ancestry. J. Virol. 812805-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2000. Outbreak of Rift Valley fever virus—Saudi Arabia, August-October, 2000. Morbid. Mortal. Wkly. Rep. 49:905-908. [PubMed] [Google Scholar]
- 4.Chan, A. B., and J. D. Fox. 1999. NASBA and other transcription-based amplification methods for research and diagnostic microbiology. Rev. Med. Microbiol. 10185-196. [Google Scholar]
- 5.Durand, J. P., M. Bouloy, L. Richecoeur, C. N. Peyrefitte, and H. Tolou. 2003. Rift Valley fever virus infection among French troops in Chad. Emerg. Infect. Dis. 9751-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faye, O., M. Diallo, D. Diop, O. E. Bezeid, H. Ba, M. Niang, I. Dia, S. A. Ould Mohamed, K. Ndiaye, D. Diallo, P. O. Ly, B. Diallo, P. Nabeth, F. Simon, B. Lo, and O. M. Diop. 2007. Rift Valley fever virus outbreak with East-Central African virus lineage in Mauritania, 2003. Emerg. Infect. Dis. 131016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Findlay, G. M., and R. Daubney. 1931. The virus of Rift Valley fever or enzootic hepatitis. Lancet ii1350-1351. [Google Scholar]
- 8.Garcia, S., J.-M. Crance, A. Billecocq, A. Peinnequin, A. Jouan, M. Bouloy, and D. Garin. 2001. Quantitative real-time PCR detection of Rift Valley fever virus and its application to evaluation of antiviral compounds. J. Clin. Microbiol. 394456-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han, E. T., R. Watanabe, J. Sattabongkot, B. Khuntirat, J. Sirichaisinthop, H. Iriko, L. Jin, S. Takeo, and T. Tsuboi. 2007. Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J. Clin. Microbiol. 452521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imai, M., A. Ninomiya, H. Minekawa, T. Notomi, T. Ishizaki, P. Van Tu, N. T. K. Tien, M. Tashiro, and T. Odagiri. 2007. Rapid diagnosis of H5N1 avian influenza virus infection by newly developed influenza H5 hemagglutinin gene-specific loop-mediated isothermal amplification method, J. Virol. Methods 141173-180. [DOI] [PubMed] [Google Scholar]
- 11.Kuhara, T., T. Yoshikawa, M. Ihira, D. Watanabe, Y. Tamada, H. Katano, Y. Asano, and Y. Matsumoto. 2007. Rapid detection of human herpesvirus 8 DNA using loop-mediated isothermal amplification, J. Virol. Methods 14479-85. [DOI] [PubMed] [Google Scholar]
- 12.Nagamine, K., T. Hase, and T. Notomi. 2002. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16223-229. [DOI] [PubMed] [Google Scholar]
- 13.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parida, M. M., S. R. Santhosh, P. K. Dash, N. K. Tripathi, V. Lakshmi, N. Mamidi, A. Shrivastva, N. Gupta, P. Saxena, and J. Pradeep. 2007. Rapid and real-time detection of Chikungunya virus by reverse transcription loop-mediated isothermal amplification assay J. Clin. Microbiol. 45351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parida, M. M., S. R. Santhosh, P. K. Dash, N. K. Tripathi, P. Saxena, S. Ambuj, A. K. Sahni, P. V. Lakshmana Rao, and K. Morita. 2006. Development and evaluation of reverse transcription-loop-mediated isothermal amplification assay for rapid and real-time detection of Japanese encephalitis virus. J. Clin. Microbiol. 4414172-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parida, M., G. Posadas, S. Inoue, F. Hasebe, and K. Morita. 2004. Real-time reverse transcription-loop-mediated isothermal amplification for rapid detection of West Nile virus. J. Clin. Microbiol. 42257-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parida, M., K. Horioke, H. Ishida, P. K. Dash, P. Saxena, A. M. Jana, M. A. Islam, S. Inoue, N. Hosaka, and K. Morita. 2005. Rapid detection and differentiation of dengue virus serotypes by real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 432895-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paris, D. H., M. Imwong, A. M. Faiz, M. Hasan, E. B. Yunus, K. Silamut, S. J. Lee, N. P. Day, and A. M. Dondorp. 2007. Loop-mediated isothermal PCR (LAMP) for the diagnosis of falciparum malaria. Am. J. Trop. Med. Hyg. 77972-976. [PubMed] [Google Scholar]
- 19.Peyrefitte, C. N., B. A. Pastorino, M. Bessaud, P. Gravier, F. Tock, P. Couissinier-Paris, J. Martial, P. Huc-Anaïs, R. Césaire, M. Grandadam, and H. J. Tolou. 2005. Dengue type 3 virus, Saint Martin, 2003-2004. Emerg. Infect. Dis. 11757-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peyrefitte, C. N., I. Devetakov, B. A. Pastorino, L. Villeneuve, M. Bessaud, P. Stolidi, J. Depaquit, L. Segura, P. Gravier, F. Tock, F. Durand, J. P. Vagneur, H. J. Tolou, and M. Grandadam. 2005. Toscana virus and acute meningitis, France. Emerg. Infect. Dis. 11778-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sall, A. A., J. Thonnon, O. K. Sene, A. Fall, M. Ndiaye, B. Baudez, C. Mathiot, and M. Bouloy. 2001. Single-tube and nested reverse transcriptase polymerase chain reaction for detection of Rift Valley fever virus in human and animal sera. J. Virol. Methods 19185-92. [DOI] [PubMed] [Google Scholar]
- 22.Schmaljohn, C. 1996. Bunyaviridae: the viruses and their replication, p. 1447-1471. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott Raven Publishers, Philadelphia, Pa.
- 23.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomita, N., Y. Mori, H. Kada, and T. Notomi. 2008. Loop-mediated isothermal amplification (LAMP) of the genome sequences and simple visual detection of products. Nat. Protoc. 3:877-882. [DOI] [PubMed] [Google Scholar]
- 25.Toriniwa, H., and T. Komiya. 2006. Rapid detection and quantification of Japanese encephalitis virus by real-time reverse transcription-loop-mediated isothermal amplification. Microbiol. Immunol. 50:379-387. [DOI] [PubMed] [Google Scholar]
- 26.Zeller, H., and M. Bouloy. 2000. Infections by viruses of the families Bunyaviridae and Filiviridae. Rev. Sci. Tech. Off. Int. Epizoot. 19:79-91. [DOI] [PubMed] [Google Scholar]