A recent study reported microsatellite subtyping of isolates from cases in five drinking water outbreaks (3) but lacked patient specific exposure data, which prevented the demonstration of the full value of subtyping in outbreak investigation. We report the use of multilocus microsatellite subtyping to investigate an outbreak of drinking-water-associated cryptosporidiosis in the northwest of England which affected residents during April and May 2000 and in which exposure data were available.
The outbreak was in an area where there hade been multiple outbreaks over previous years (4). Some 238 cases were identified in the affected areas, though not all were thought to be related to the primary outbreak. The typing methods used have been extensively described in our previous papers (1, 2). A total of 99 strains were typed, of which 10 strains of Cryptosporidium hominis and 78 of Cryptosporidium parvum were typeable at all three loci. The distribution of multilocus fragment types (MLFTs) for these typeable strains is shown in Table 1. For C. parvum there were 17 different MLFTs detected; the most common type (P36) accounted for 37% (29) of strains, and the second (P5) accounted for 19% of strains. Ten of the detected types (P34 to P45) were distinct from strain types identified in the previously reported study of sporadic infections (2). All but four of the C. parvum strains had the ML1 242 allele previously shown to be associated with zoonotic infection (2).
TABLE 1.
Distribution of Cryptosporidium hominis and Cryptosporidium parvum MLFTs
| Cryptosporidium species (n) | MLFT | No. of cases | Allele no.
|
||
|---|---|---|---|---|---|
| ML1 | ML2 | gp60 | |||
| C. hominis (10) | H1 | 10 | 233 | 180 | 371 |
| C. parvum (78) | P1 | 6 | 242 | 229 | 341 |
| P2 | 2 | 242 | 229 | 338 | |
| P5 | 15 | 242 | 231 | 341 | |
| P7 | 1 | 242 | 231 | 338 | |
| P8 | 4 | 242 | 233 | 341 | |
| P14 | 6 | 242 | 231 | 356 | |
| P19 | 1 | 242 | 235 | 338 | |
| P34 | 5 | 242 | 207 | 338 | |
| P36 | 29 | 242 | 229 | 344 | |
| P37 | 1 | 242 | 211 | 344 | |
| P38 | 1 | 227 | 197 | 368 | |
| P39 | 2 | 242 | 231 | 362 | |
| P40 | 1 | 242 | 221 | 341 | |
| P41 | 1 | 242 | 235 | 341 | |
| P42 | 1 | 245 | 229 | 338 | |
| P44 | 1 | 227 | 197 | 335 | |
| P45 | 1 | 239 | 231 | 341 | |
Illness associated with C. parvum reached a peak in week 4, and both P36 and P5 had the greatest number of associated cases in that week. There was no significant difference between the number of weekly cases of the most common MLFT (P36) and those of all other C. parvum MLFTs (Wilcoxon signed-rank test; Z = −1.552, P = 0.121).
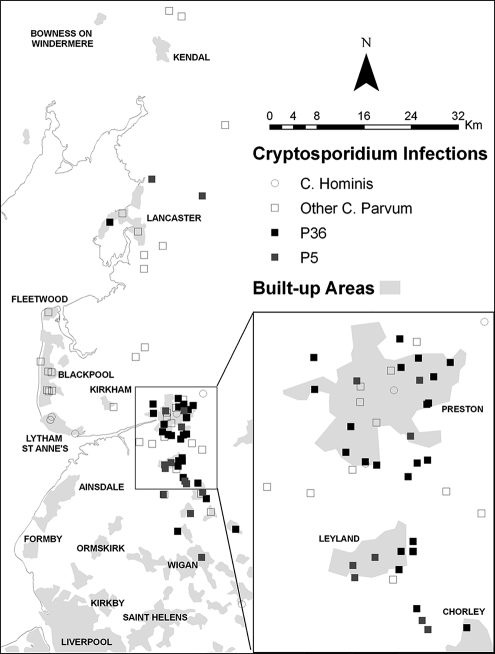
Figure 1 shows the geographical distribution of the two main strains and other C. parvum strains. Nearly all P36 strains were strongly clustered in the area around Preston and Chorley, the main focus for the outbreak and the area most dependent on water from the implicated supply. When comparing the distances of each of the P36 strains to their geographic centroid with that for other strains of C. parvum, this clustering of P36 was highly significant (Mann-Whitney U test, Z = 4.580, P = 0.000004).
FIG. 1.
Geographical distribution of P36, P5, and other C. parvum infection strains.
The value of typing in this outbreak was the demonstration that most strains were of the zoonotic ML1 242 allele. Subtyping enabled identification of a particularly common type but one that represented only 37% of strains. Subtyping also demonstrated the lack of this type in areas not reliant on the affected water supply. It currently remains unclear whether the variation in subtypes identified represented different lineages or evolution of strains during the outbreak. The issue of the stability of MLFTs in natural infections needs to be investigated in more detail. In this context it is worth noting that a further nine strains differed from P36 at only one locus and so could potentially be derived from P36 with just a single mutation. Microsatellite typing of Cryptosporidium isolates associated with waterborne outbreaks can provide useful additional epidemiological information especially when linked to individual exposure data.
Acknowledgments
We thank Kristin Elwin, Guy Robinson, and other staff of the UK Cryptosporidium Reference Unit for help with Cryptosporidium species determination and for the provision of additional subtyping data.
This project was funded by the Department for Environment, Food and Rural Affairs and managed by the Drinking Water Inspectorate.
Footnotes
Published ahead of print on 3 September 2008.
REFERENCES
- 1.Chalmers, R. M., S. J. Hadfield, C. J. Jackson, K. Elwin, L. Xiao, and P. Hunter. 2008. Geographic linkage and variation in Cryptosporidium hominis. Emerg. Infect. Dis. 14496-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter, P. R., S. J. Hadfield, D. Wilkinson, I. R. Lake, F. C. D. Harrison, and R. M. Chalmers. 2007. Subtypes of Cryptosporidium parvum in humans and disease risk. Emerg. Infect. Dis. 1382-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leoni, F., M. E. Mallon, H. V. Smith, A. Tait, and J. McLauchlin. 2007. Multilocus analysis of Cryptosporidium hominis and Cryptosporidium parvum isolates from sporadic and outbreak-related human cases and C. parvum isolates from sporadic livestock cases in the United Kingdom. J. Clin. Microbiol. 453286-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naumova, E. N., J. Christodouleas, P. R. Hunter, and Q. Syed. 2005. Temporal and spatial variability in cryptosporidiosis recorded by the surveillance system in North West England in 1990-1999. J. Water Health 3185-196. [PubMed] [Google Scholar]