Abstract
Epstein-Barr virus (EBV) DNA levels in whole-blood samples of 54 pediatric patients receiving T-cell-depleted haploidentical hematopoietic stem cell transplantation (HSCT) in 2003 to 2007 were retrospectively compared with EBV DNA loads in peripheral blood mononuclear cells (PBMC). Determination of EBV DNA in whole blood missed 1 of 19 patients (5.2%), who tested positive for EBV DNA in PBMC. The analytical sensitivity of EBV DNA detection in whole-blood samples relative to that in PBMC was 94.7%. Regression analysis showed a significant correlation between DNA levels in PBMC and whole blood (r = 0.81; P < 0.001). Relative to that in PBMC, the appearance of EBV DNA in whole blood was delayed in 9/18 patients (median, 49 days; range, 6 to 226 days), while peak levels and clearance were reached simultaneously. Following peak levels, EBV DNA showed a slower decline in whole blood than in PBMC. In conclusion, (i) EBV DNA levels in PBMC were significantly correlated with those in whole blood; (ii) a differential kinetics of EBV DNA in the two blood compartments was observed; and (iii) monitoring of EBV DNA levels in whole blood appears to be a valuable alternative to PBMC in the follow-up of pediatric recipients of haploidentical T-cell-depleted HSCT.
Epstein-Barr virus (EBV)-associated posttransplantation lymphoproliferative disorders (PTLD) represent a life-threatening complication for recipients of solid-organ transplantation (SOT) and hematopoietic stem cell transplantation (HSCT) (2, 11, 16). The heterogeneous clinical manifestations of PTLD are mostly due to B-lymphocyte proliferation driven and sustained by EBV latency products (5, 11, 22). In immunocompetent individuals, uncontrolled EBV-driven B-cell proliferation is prevented mainly by virus-specific cytotoxic T lymphocytes (CTL), while in transplant recipients, impaired CTL control of EBV infection due to pharmacological immune suppression or physical removal of T lymphocytes favors the development of PTLD (11, 16).
Recipients of T-cell-depleted, HLA-haploidentical HSCT are exposed to the highest risk of developing PTLD (8), because they cannot benefit from adoptive transfer of virus-specific CTL, and a timely diagnosis of EBV-driven lymphoproliferation is mandatory for prompt therapeutic intervention in this severe complication. Preemptive treatment of PTLD in HSCT includes infusion of an anti-CD20 monoclonal antibody (MAb), tapering of immune-suppressive regimens (whenever possible), and adoptive infusion of donor-derived EBV-specific CTL in patients at risk for PTLD (6, 26).
The EBV DNA load in blood is considered to reflect EBV-induced cell proliferation, and quantitative determination of EBV DNA levels in peripheral blood mononuclear cells (PBMC), plasma, or whole blood has been proposed as a means to identify SOT or HSCT recipients at risk for developing PTLD (1, 4, 7, 10, 12, 14-17, 19-21, 24, 25). While it is accepted that EBV DNA levels in the blood of patients with EBV-related PTLD are significantly higher than those in healthy EBV-seropositive individuals or transplant recipients without PTLD (4, 17, 19, 24), it is still not clear which might be the specimen of choice and what is the threshold EBV DNA level predictive for PTLD. Levels of cell-bound EBV DNA can be regarded as an indirect measure of EBV-driven B-cell proliferation (16, 22), while it is still unclear how levels of EBV DNA in plasma (containing only cell-free EBV DNA) or whole blood (containing both cell-associated and cell-free EBV DNA) correlate with EBV DNA levels in PBMC and with the development of PTLD.
From a technical standpoint, the recent introduction of real-time PCR techniques and automated extraction procedures, as well as the availability of international quality control panels, represented an advance toward a more reproducible quantification of EBV DNA. Quantification of EBV DNA in plasma or whole blood could represent a technical simplification of diagnostic procedures, since PBMC separation before DNA extraction and PCR amplification would be avoided, and several groups have adopted EBV DNA quantification in either specimen for the identification of patients at risk of developing PTLD after SOT or HSCT (1-4, 7-12, 14, 15, 17, 19-21, 24, 25).
Previous studies comparing EBV DNA determination in PBMC, whole blood, and plasma using real-time PCR assays showed higher analytical sensitivity for PBMC and whole blood; however, the impact on clinical management of the use of either material remains controversial (1, 10, 12, 21, 23-25). A possible reason for the discrepancies between the analytical and clinical performances could relate to a differential kinetics of EBV DNA in the different blood compartments.
The aim of this retrospective study was to compare the kinetics of EBV DNA in whole blood versus PBMC in pediatric patients receiving T-cell-depleted, HLA-haploidentical HSCT.
MATERIALS AND METHODS
Real-time PCR for EBV DNA quantification.
Since 2003, an original real-time PCR technique has been developed and utilized for monitoring EBV DNA in HSCT recipients. Briefly, a 59-nucleotide fragment of the BamH1 C/W region of the EBV genome was amplified using primers EBVSMF (5′-AGCGGGTCTATGGTTGGCT-3′) and EBVSMR (5′-GCTTATTCCTCTTTTCCCCTCTAAA-3′). The amplified DNA fragment was cloned into a plasmid vector (TA Cloning kit; Invitrogen, Carlsbad, CA) to generate an external quantification standard referred to as pEBV. Real-time PCR was performed in the presence of a 6-carboxyfluorescein-labeled MGB TaqMan probe (EBVSM [5′-CGCTGCTGCTATC-3′]) using a 7300 real-time PCR system (Applied Biosystems, Foster City, CA) and standard reagents (TaqMan Universal Master Mix; Applied Biosystems, Warrington, Cheshire, United Kingdom). The thermal profile was as follows: 1 cycle at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 60 s.
Following automatic extraction (EasyMag; Biomerieux, Lyon, France) of 2 × 105 PBMC, half of the eluate volume (corresponding to 1 × 105 PBMC) was used as input DNA in each real-time PCR. In addition, aliquots of 100 μl of whole blood stored at −80°C in the presence of 900 μl of EasyMag lysis buffer (Biomerieux) were submitted to automatic extraction, and 1/10 of the eluates (corresponding to 10 μl of whole blood) was used as input DNA in each real-time PCR.
Serial amounts of pEBV (106 to 100 input copies) were amplified in parallel with the clinical samples to generate a standard curve with threshold cycles from each log10 plasmid dilution, where threshold cycles from clinical samples were automatically interpolated to determine EBV DNA amounts.
Finally, the presence of inhibitors was evaluated by coamplification of both external standards and clinical samples with heterologous DNA (TaqMan exogenous internal positive control, VIC; Applied Biosystems, United Kingdom) using specific primers and a specific probe (23).
Patients and samples.
Pediatric patients receiving T-cell-depleted HSCT from HLA-haploidentical relatives in the period between 15 April 2003 and 31 December 2007 were included in the study.
For all patients, EBV DNA levels in PBMC were prospectively monitored with the newly developed real-time PCR assay once a week for at least the first 3 months after transplantation. Whole-blood samples stored at −80°C as routine backup specimens for the diagnosis and monitoring of other viral infections (such as human cytomegalovirus, human herpesvirus 6, human polyomavirus, and adenovirus) were used for retrospective evaluation of EBV DNA levels. All samples were collected, stored, thawed, and tested homogeneously.
Patients received preemptive treatment for EBV-associated PTLD according to the results of prospective determination of EBV DNA levels in PBMC, as previously reported (6). In detail, upon confirmed detection of >1,000 EBV DNA copies/1 × 105 PBMC, patients were given an anti-CD20 MAb (375 mg/m2/weekly for 2 consecutive weeks). In the presence of persistent or relapsed EBV DNAemia in PBMC or clinical symptoms suggestive of PTLD, patients were rescued with EBV-specific CTL generated from the donor (6).
Statistical analysis.
The Shapiro-Wilk test was used to test the normal distribution of quantitative variables. If they were normally distributed, means and standard deviations were used to summarize the results; otherwise, the median and range (minimum and maximum) were used. Detection of EBV DNA in PBMC was used as the “gold standard” to calculate the specificity and sensitivity (with 95% confidence intervals [95% CI]) of EBV DNA detection in whole blood or plasma. Pearson's r coefficient was used to evaluate correlations between log copies of EBV DNA in PBMC, whole blood, and plasma. A P value of <0.05 was considered statistically significant. All tests were two-sided. Statistica, version 6.0 (2006; Statsoft, Inc., Tulsa, OK), was used for statistical computations.
RESULTS
Analytical performance of the newly developed real-time PCR.
Repeated real-time PCR experiments on serial dilutions of predetermined amounts of pEBV showed that 10 input copies of the plasmid added to a background of extracted DNA corresponding to 1 × 105 cells could be amplified in 100% of the several dozens of replicates analyzed. The lower detection limit for EBV DNA in clinical samples was defined as 10 copies/1 × 105 PBMC. Samples showing no real-time PCR signal and the few samples (<10% of replicates) with positive real-time PCR signals below the lower limit of detection were scored as containing <10 EBV DNA copies/1 × 105 PBMC.
Real-time PCR sensitivity increased by 1 log10 unit when pEBV was added to a background of extracted DNA corresponding to 10 μl of whole blood or plasma. In fact, 1 input plasmid copy could be amplified in 100% of the several dozens of replicates analyzed. Thus, the lower detection limit for EBV DNA in clinical samples was defined as 1 copy/10 μl of whole blood. Results were then normalized to milliliters by multiplication by a factor of 100. Samples with no real-time PCR signal and the few samples (<10% of replicates) with positive real-time PCR signals below the lower detection limit were scored as containing <100 EBV DNA copies/ml of whole blood.
Patients and samples.
In the period from 15 April 2003 through 31 December 2007, EBV DNA levels in the PBMC of 54 pediatric haploidentical HSCT recipients were monitored by real-time PCR once a week for at least 3 months after the allograft. Forty-three patients (53.4% males) received transplantation due to underlying malignant disease (median age, 10.5 years; range, 3.8 to 22.1 years), and 11 patients (54.5% males) were originally affected by a nonmalignant disorder (median age, 3.3 years; range, 0.3 to 10.0 years).
For 32/54 (59.2%) patients, PBMC tested negative for EBV DNA over the entire follow-up period, while 22 (40.7%) patients showed at least one positive PBMC sample. Stored whole-blood samples were available for 41/54 (75.9%) patients (of whom 19 were EBV DNA positive and 22 were EBV DNA negative by detection in PBMC), while no samples were available for 13/54 (24.0%) patients (of whom 3 were EBV DNA positive and 10 were EBV DNA negative by detection in PBMC).
Overall, EBV DNA levels were determined in 378 PBMC and 320 whole-blood samples from patients testing positive for EBV DNA in PBMC. A comparison was also performed between 264 PBMC and whole-blood samples from patients testing negative for EBV DNA in PBMC. The median number of paired PBMC and whole-blood samples analyzed per patient was 19.5 (range, 6 to 29).
Detection and quantification of EBV DNA in PBMC and whole-blood samples.
PBMC from 19/41 (46.3%) patients were positive for EBV DNA, while whole-blood samples were positive for 18/41 (43.9%) patients. Thus, one patient positive for EBV DNA in PBMC was scored as negative for EBV DNA in whole blood. All whole-blood samples from the 22 patients testing negative for EBV DNA in PBMC were also negative for EBV DNA. Thus, with respect to EBV DNA determination in PBMC, detection of EBV DNA in whole blood could diagnose EBV infection in haploidentical HSCT recipients with 94.7% sensitivity (95% CI, 74.0 to 99.9%) and 100% specificity (95% CI, 84.5 to 100.0%).
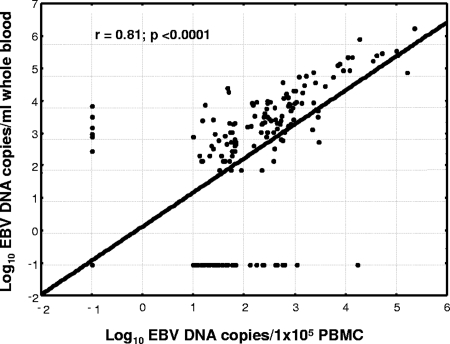
Considering all samples analyzed, 119 were positive and 420 were negative for EBV DNA both in PBMC and in whole blood; 6 samples were negative for EBV DNA in PBMC and positive for EBV DNA in whole blood; and 39 samples were positive for EBV DNA in PBMC and negative for EBV DNA in whole blood. Thus, compared to that in PBMC, EBV DNA determination in whole-blood samples showed 75.3% sensitivity (95% CI, 67.8 to 81.8%) and 98.6% specificity (95% CI, 96.9 to 99.5%). A significant correlation between EBV DNA levels in PBMC and those in whole blood (r = 0.81; P < 0.0001) was demonstrated by regression analysis (Fig. 1). EBV DNA positivity in whole-blood samples according to the stratification of PBMC DNA levels is detailed in Table 1.
FIG. 1.
Regression analysis of EBV DNA levels in PBMC and whole blood of pediatric haploidentical HSCT recipients.
TABLE 1.
EBV DNA positivity in whole-blood samples according to the stratification of EBV DNA levels in PBMC
| EBV DNA copy no./1 × 105 PBMC | No. of samples with the following EBV DNA copy no./ml of whole blood:
|
Total no. of samples | ||||
|---|---|---|---|---|---|---|
| <100 | 100-999 | 1,000-9,999 | 10,000-99,999 | ≥100,000 | ||
| <10 | 420 | 1 | 4 | 1 | 0 | 426 |
| 10-99 | 28 | 23 | 13 | 2 | 0 | 66 |
| 100-999 | 9 | 5 | 38 | 8 | 0 | 60 |
| 1,000-9,999 | 1 | 1 | 6 | 10 | 2 | 20 |
| ≥10,000 | 1 | 0 | 1 | 10 | 0 | 12 |
| Total | 459 | 30 | 62 | 31 | 2 | 584 |
Kinetics of EBV DNA in PBMC and whole-blood samples of patients receiving haploidentical HSCT.
The time to first appearance of EBV DNA in PBMC and whole blood was determined. Among the 19 patients with detectable EBV infections, the appearance of EBV DNA in PBMC anticipated that in whole blood for 9 (36.8%) patients (median time difference, 49 days; range, 6 to 226 days); EBV DNA appeared simultaneously in PBMC and whole blood for 9 patients; and, as mentioned above, EBV DNA was absent from the whole blood of 1 patient but present in that patient's PBMC. Overall, peak DNA levels in whole blood (median, 28,600 copies/ml; range, 1,700 to 3,280,000 copies/ml) and peak DNA levels in PBMC (median, 3,943 copies/1 × 105 PBMC; range, 25 to 226,875 copies/1 × 105 PBMC) were coincident (the median separation of peaks was 0 days; range, −6 to +7 days).
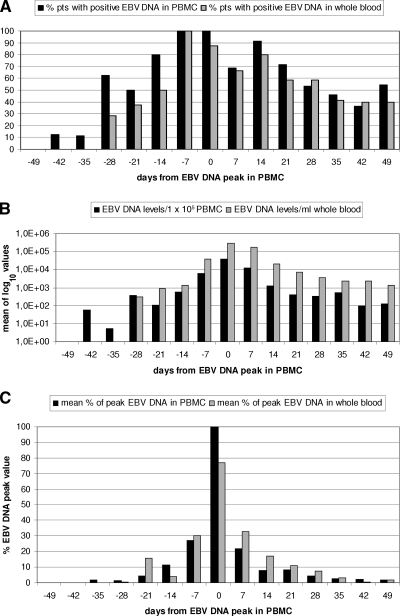
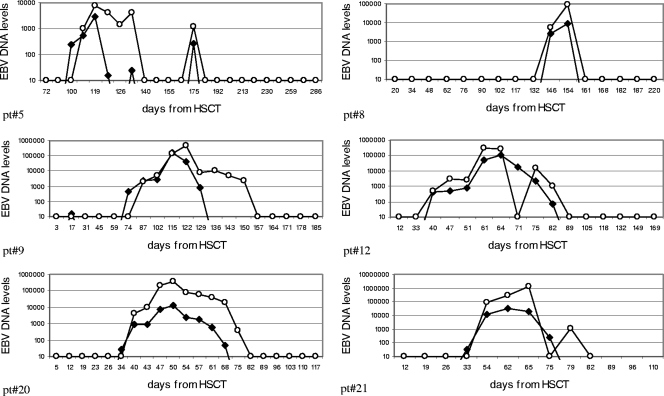
To compare the kinetics of EBV DNA in the two specimen types, EBV DNA in PBMC was used as a reference. For each patient, T0 corresponded to the time of the EBV DNA peak in PBMC. EBV DNA levels in PBMC and whole blood from the 19 positive patients were then stratified over 7 weeks before and after the PBMC peak time. As shown in Fig. 2A, the percentages of positive patients were similar for the two types of specimens at each time point, except for the earliest time points, when EBV DNA was positive in PBMC only. Mean log10 copies of EBV DNA followed similar kinetics in the two types of specimens, with delayed positivity in whole-blood samples, as already mentioned (Fig. 2B). Finally, when the increase and decrease in EBV DNA levels were evaluated with respect to individual peak values over time, a similar trend was shown, with minor differences (Fig. 2C). In more detail, peak EBV DNA levels in whole blood were mostly coincident with peak levels in PBMC, while delayed appearance and clearance of EBV DNA were observed in whole blood. In fact, besides a slower decline in EBV DNA levels, 6/19 (31.5%) patients had one whole-blood-positive sample after the first negative PBMC sample. This trend could be better appreciated in individual cases (Fig. 3).
FIG. 2.
Kinetics of EBV DNA in PBMC and whole-blood samples of pediatric haploidentical HSCT recipients. (A) Percentages of patients (pts) positive for EBV DNA in PBMC (filled bars) and in whole blood (shaded bars) with respect to the time of peak levels in PBMC. (B) Mean log10 copies of EBV DNA in PBMC (filled bars) and in whole blood (shaded bars) with respect to the time of peak levels in PBMC. (C) EBV DNA levels expressed as mean percentages of peak EBV DNA levels in PBMC (filled bars) and in whole blood (shaded bars) with respect to the time of peak levels in PBMC.
FIG. 3.
Representative EBV DNA kinetics in PBMC (filled circles) and whole-blood (open circles) samples of six pediatric haploidentical HSCT recipients. EBV DNA levels in PBMC are expressed as the number of EBV DNA copies per 1 × 105 cells, while EBV DNA levels in whole blood are expressed as the number of EBV DNA copies per milliliter. pt, patient.
Among the 19 patients positive for EBV DNA in PBMC, 13 (68.4%) with median EBV DNA levels of 15,248 copies/105 PBMC (range, 1,429 to 226,875 copies/105 PBMC) were given 17 courses of preemptive treatment of PTLD. The relevant median EBV DNA levels in the whole blood of these 13 children were 172,150 copies/ml (range, 2,000 to 3,280,000 copies/ml). Preemptive treatment of PTLD consisted of infusions of the anti-CD20 MAb for five patients, adoptive transfer of donor-derived EBV-specific CTL for two patients, and a combination of both for six patients. EBV DNA was cleared from both PBMC and whole blood after a median time of 28 days (range, 14 to 78 days).
DISCUSSION
EBV-driven PTLD is a severe and potentially fatal complication in high-risk transplant populations, such as recipients of T-cell-depleted, HLA-haploidentical HSCT. Treatment options for this severe condition are limited and, in the case of EBV-specific CTL, not widely available. Many groups are actively working on the identification of virologic markers predicting the development of PTLD with the objective of optimizing treatment options. However, biological and technical issues contribute to the difficulty of reaching a consensus. From a biological standpoint, it should be emphasized that EBV-driven lymphoproliferation is sustained by expression in infected cells of latency-associated antigens and is not associated with the lytic (active) phase of infection (22). While the determination of cell-associated EBV DNA loads could be regarded as a marker of EBV-induced cell proliferation, levels of cell-free EBV DNA could be expressions of either virus production or the release of episomal DNA from apoptotic cells, or both. The EBV lytic cycle is induced following differentiation of latently infected memory B cells in plasma cells, and virus progeny is released into body fluids (13, 22). In the plasma of patients with AIDS-related lymphomas and immunosuppressed/transplanted patients, cell-free EBV DNA was detected mostly as naked DNA fragments released from apoptotic cells, while a mixture of naked and encapsidated DNAs was reported in infectious mononucleosis patients (18). On the other hand, EBV DNA in whole blood comes from both EBV-positive cells and released virions or DNA.
In the present study, a cohort of patients at high risk for PTLD has been considered for retrospective comparison of EBV DNA levels in whole blood and PBMC. Positivity for EBV DNA in PBMC approached 50% of the 54 pediatric patients considered. Stored whole-blood samples were available for the majority of patients either positive or negative for EBV DNA in PBMC. Thus, the subset of patients analyzed could be considered representative of the entire cohort.
At the patient level, the determination of EBV DNA in whole blood was comparable to that in PBMC, since only a single PBMC-positive patient tested negative for EBV DNA in whole blood (94.7% sensitivity). At the sample level, the sensitivity of EBV DNA detection in whole blood was lower than that in PBMC (73.4%). In agreement with previous studies (9, 23), a significant correlation between EBV DNA levels in the two types of specimens was observed. However, a fair proportion of whole-blood samples were also negative in the presence of relatively high levels of EBV DNA in PBMC (as many as 10,000 copies/1 × 105 PBMC). These discrepant results could be explained by a differential kinetics of positivization of the two virologic assays. In fact, in half the patients, the onset of EBV DNA positivity was simultaneous in PBMC and whole blood, while in the other half there was a delay of about 7 weeks from the detection of EBV DNA in PBMC to its detection in whole blood. On the other hand, peak DNA levels were reached simultaneously, and the times of DNA clearance were comparable. However, after reaching a peak, EBV DNA levels showed a slower decline in whole blood than in PBMC before disappearing, and in about one-third of the patients, a whole-blood sample remained positive despite the detection of the first negative PBMC sample. Whether the delayed clearance of EBV DNA from whole blood can be considered a result of B-cell lysis following administration of the anti-CD20 MAb and/or EBV-specific CTL or was due to an as yet unknown biological event remains to be determined. Our results confirm and extend a previous observation (limited to only four HSCT recipients) of a similar kinetics of EBV DNAemia in PBMC and whole blood (12).
Treatment with the anti-CD20 MAb and an EBV-specific CTL infusion was started in the presence of median EBV DNA levels in PBMC of 15,248 copies/1 × 105 cells, which corresponded to 172,150 copies/ml of whole blood. Since treatment with the anti-CD20 MAb and/or virus-specific donor-derived CTL was successful for all patients, we can hypothesize that the corresponding EBV DNA level in whole blood could have been safely used for guiding preemptive treatment of EBV infection in recipients of T-cell-depleted, HLA-haploidentical HSCT. However, any proposal of cutoff values would require the standardization of EBV DNA quantification techniques and validation in prospective clinical trials.
In conclusion, (i) EBV DNA levels in PBMC were significantly correlated with those in whole blood; (ii) a differential kinetics of EBV DNA in the two blood compartments was observed; and (iii) monitoring of EBV DNA levels in whole blood appears to be a valuable alternative to determination of EBV DNA levels in PBMC in the follow-up of haploidentical T-cell-depleted HSCT pediatric recipients. Prospective studies comparing EBV DNA cutoff values in PBMC and whole blood for preemptive therapy of EBV-associated PTLD in high-risk HSCT recipients are needed.
Acknowledgments
We thank the technical staff for excellent technical assistance.
This work was supported by Ministero della Salute Ricerca Finalizzata grant 28C5/3 and Ricerca Corrente grant 80541 and by grants from the Ministero della Salute, Bandi Oncologia 2006, and from the Associazione Italiana Ricerca sul Cancro.
Footnotes
Published ahead of print on 17 September 2008.
REFERENCES
- 1.Aalto, S. M., E. Juvonen, J. Tarkkanen, L. Volin, H. Haario, T. Ruutu, and K. Hedman. 2007. Epstein-Barr viral load and disease prediction in a large cohort of allogeneic stem cell transplant recipients. Clin. Infect. Dis. 451305-1309. [DOI] [PubMed] [Google Scholar]
- 2.Bakker, N. A., G. W. van Imhoff, E. A. M. Verschuuren, and W. J. van Son. 2007. Presentation and early detection of post-transplant lymphoproliferative disorder after solid organ transplantation. Transpl. Int. 20207-218. [DOI] [PubMed] [Google Scholar]
- 3.Bakker, N. A., E. A. Verschuuren, N. J. Veeger, W. van der Bij, G. W. van Imhoff, C. G. Kallenberg, and B. G. Hepkema. 2008. Quantification of Epstein-Barr virus-DNA load in lung transplant recipients: a comparison of plasma versus whole blood. J. Heart Lung Transplant. 277-10. [DOI] [PubMed] [Google Scholar]
- 4.Baldanti, F., P. Grossi, M. Furione, L. Simoncini, A. Sarasini, P. Comoli, R. Maccario, R. Fiocchi, and G. Gerna. 2000. High levels of Epstein-Barr virus DNA in blood of solid-organ transplant recipients and their value in predicting posttransplant lymphoproliferative disorders. J. Clin. Microbiol. 38613-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capello, D., D. Rossi, and G. Gaidano. 2005. Post-transplant lymphoproliferative disorders: molecular basis of disease histogenesis and pathogenesis. Hematol. Oncol. 2361-67. [DOI] [PubMed] [Google Scholar]
- 6.Comoli, P., S. Basso, M. Zecca, D. Pagliara, F. Baldanti, M. E. Bernardo, W. Barberi, A. Moretta, M. Labirio, M. Paulli, M. Furione, R. Maccario, and F. Locatelli. 2007. Preemptive therapy of EBV-related lymphoproliferative disease after pediatric haploidentical stem cell transplantation. Am. J. Transplant. 71648-1655. [DOI] [PubMed] [Google Scholar]
- 7.Comoli, P., M. Labirio, S. Basso, F. Baldanti, P. Grossi, M. Furione, M. Viganò, R. Fiocchi, G. Rossi, F. Ginevri, B. Gridelli, A. Moretta, D. Montagna, F. Locatelli, G. Gerna, and R. Maccario. 2002. Infusion of autologous Epstein-Barr virus (EBV)-specific cytotoxic T cells for prevention of EBV-related lymphoproliferative disorder in solid organ transplant recipients with evidence of active virus replication. Blood 992592-2598. [DOI] [PubMed] [Google Scholar]
- 8.Curtis, R. E., L. B. Travis, P. A. Rowlings, G. Socié, D. W. Kingma, P. M. Banks, E. S. Jaffe, G. E. Sale, M. M. Horowitz, R. P. Witherspoon, D. A. Shriner, D. J. Weisdorf, H.-J. Kolb, K. M. Sullivan, K. A. Sobocinski, R. P. Gale, R. N. Hoover, J. F. Fraumeni, Jr., and H. J. Deeg. 1999. Risk of lymphoproliferative disorders after bone marrow transplantation: a multi-institutional study. Blood 942208-2216. [PubMed] [Google Scholar]
- 9.Fafi-Kremer, S., K. Brengel-Pesce, G. Barguès, M.-J. Bourgeat, O. Genoulaz, J.-M. Seigneurin, and P. Morand. 2004. Assessment of automated DNA extraction coupled with real-time PCR for measuring Epstein-Barr virus load in whole blood, peripheral mononuclear cells and plasma. J. Clin. Virol. 30157-164. [DOI] [PubMed] [Google Scholar]
- 10.Gärtner, B. C., H. Schäfer, K. Marggraff, G. Eisele, M. Schäfer, K. Roemer, H.-J. Laws, M. Sester, U. Sester, H. Einsele, and N. Mueller-Lantzsch. 2002. Evaluation of use of Epstein-Barr viral load in patients after allogeneic stem cell transplantation to diagnose and monitor posttransplant lymphoproliferative disease. J. Clin. Microbiol. 40351-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottschalk, S., C. M. Rooney, and H. E. Heslop. 2005. Post-transplant lymphoproliferative disorders. Annu. Rev. Med. 5629-44. [DOI] [PubMed] [Google Scholar]
- 12.Hakim, H., C. Gibson, J. Pan, K. Srivastava, Z. Gu, M. J. Bankowski, and R. T. Hayden. 2007. Comparison of various blood compartments and reporting units for the detection and quantification of Epstein-Barr virus in peripheral blood. J. Clin. Microbiol. 452151-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laichalk, L. L., and D. A. Thorley-Lawson. 2005. Terminal differentiation into plasma cells initiates the replicative cycle of Epstein-Barr virus in vivo. J. Virol. 791296-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meerbach, A., P. Wutzler, R. Häfer, F. Zintl, and B. Gruhn. 2008. Monitoring of Epstein-Barr virus load after hematopoietic stem cell transplantation for early intervention in post-transplant lymphoproliferative disease. J. Med. Virol. 80441-454. [DOI] [PubMed] [Google Scholar]
- 15.Oertel, S., R. U. Trappe, K. Zeidler, N. Babel, P. Reinke, M. Hummel, S. Jonas, M. Papp-Vary, M. Subklewe, B. Dörken, H. Riess, and B. Gärtner. 2006. Epstein-Barr viral load in whole blood of adults with posttransplant lymphoproliferative disorder after solid organ transplantation does not correlate with clinical course. Ann. Hematol. 85478-484. [DOI] [PubMed] [Google Scholar]
- 16.Preiksaitis, J. K. 2004. New developments in the diagnosis and management of posttransplantation lymphoproliferative disorders in solid organ transplant recipients. Clin. Infect. Dis. 391016-1023. [DOI] [PubMed] [Google Scholar]
- 17.Riddler, S. A., M. C. Breining, and J. L. McKnight. 1994. Increased levels of circulating Epstein-Barr virus (EBV)-infected lymphocytes and decreased EBV nuclear antigen antibody response are associated with the development of posttransplant lymphoproliferative disease in solid-organ transplant recipients. Blood 84972-984. [PubMed] [Google Scholar]
- 18.Ryan, J. L., H. Fan, L. J. Swinnen, S. A. Schichman, N. Raab-Traub, M. Covington, S. Elmore, and M. L. Gulley. 2004. Epstein-Barr virus (EBV) DNA in plasma is not encapsidated in patients with EBV-related malignancies. Diagn. Mol. Pathol. 1361-68. [DOI] [PubMed] [Google Scholar]
- 19.Savoie, A., C. Perpete, L. Carpentier, J. Joncas, and C. Alfieri. 1994. Direct correlation between the load of Epstein-Barr virus-infected lymphocytes in the peripheral blood of pediatric transplant patients and risk of lymphoproliferative disease. Blood 832715-2722. [PubMed] [Google Scholar]
- 20.Stevens, S. J., E. A. Verschuuren, S. A. Verkuijlen, A. J. Van Den Brule, C. J. Meijer, and J. M. Middeldorp. 2002. Role of Epstein-Barr virus DNA load monitoring in prevention and early detection of post-transplant lymphoproliferative disease. Leuk. Lymphoma 43831-840. [DOI] [PubMed] [Google Scholar]
- 21.Stevens, S. J., I. Pronk, and J. M. Middeldorp. 2001. Toward standardization of Epstein-Barr virus DNA load monitoring: unfractionated whole blood as preferred clinical specimen. J. Clin. Microbiol. 391211-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorley-Lawson, D. A., and A. Gross. 2004. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med. 3501328-1337. [DOI] [PubMed] [Google Scholar]
- 23.Wadowsky, R. M., S. L. M. Green, S. A. Webber, and D. Rowe. 2003. Measurement of Epstein-Barr virus DNA loads in whole blood and plasma by TaqMan PCR and in peripheral blood lymphocytes by competitive PCR. J. Clin. Microbiol. 415245-5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner, H. J., L. Fischer, W. Jabs, M. Holbe, K. Pething, and P. Bucsky. 2002. Longitudinal analysis of Epstein-Barr viral load in plasma and peripheral blood mononuclear cells of transplanted patients by real-time polymerase chain reaction. Transplantation 74656-664. [DOI] [PubMed] [Google Scholar]
- 25.Wagner, H. J., M. Wessel, W. Jabs, F. Smets, L. Fischer, G. Offner, and P. Bucsky. 2001. Patients at risk for development of posttansplant lymphoproliferative disorder: plasma versus peripheral blood mononuclear cells as material for quantification of Epstein-Barr viral load by using real-time quantitative polymerase chain reaction. Transplantation 721012-1019. [DOI] [PubMed] [Google Scholar]
- 26.Weinstock, D. M., G. G. Ambrossi, C. Brennan, T. E. Kiehn, and A. Jakubowski. 2006. Preemptive diagnosis and treatment of Epstein-Barr virus-associated post transplant lymphoproliferative disorder after hematopoietic stem cell transplant: an approach in development. Bone Marrow Transplant. 37539-546. [DOI] [PubMed] [Google Scholar]