Abstract
The Beijing genotype is a globally spread lineage of Mycobacterium tuberculosis. In Russia, these strains constitute half of the local population of M. tuberculosis; they are associated with multidrug resistance and show increased transmissibility. Here, we analyzed traditional and new markers for the rapid and simple genotyping of the Beijing strains. A representative sample of 120 Beijing genotype strains was selected from a local IS6110-restriction fragment length (RFLP) database at the St. Petersburg Pasteur Institute. These strains were subjected to variable-number tandem-repeat (VNTR) typing using 24 loci of a newly proposed format and three hypervariable (HV) loci (QUB-3232, VNTR-3820, and VNTR-4120). Ten of the 27 VNTR loci were monomorphic, while five loci, MIRU26, QUB-26, QUB-3232, VNTR-3820, and VNTR-4120, were the most polymorphic (Hunter Gaston index, >0.5). VNTR typing allowed us to differentiate between two large IS6110-RFLP clusters known to be prevalent across the entire country (clusters B0/W148 and A0) and identified in 27 and 23% of strains, respectively, in the Beijing genotype database. The B0/W148 strains were grouped closely in the VNTR dendrogram and could be distinguished by a characteristic signature of the loci MIRU26 and QUB-26. Consequently, this clinically important IS6110-RFLP variant, B0/W148, likely presents a successful clonal group within the M. tuberculosis Beijing lineage that is widespread in Russia. To conclude, the IS6110-RFLP method and VNTR typing using a reduced set of the most polymorphic loci complement each other for the high-resolution epidemiological typing of the M. tuberculosis Beijing genotype strains circulating in or imported from Russia.
The population structure of Mycobacterium tuberculosis in Russia is marked by a significant prevalence of the Beijing genotype, as demonstrated in many studies in different regions across the country. This situation is an important public health issue, as an association of the Beijing genotype with multidrug-resistant tuberculosis (MDR-TB) was found in several Russian settings (4, 29), whereas a global spread of these strains makes any world region vulnerable.
The Beijing genotype was first identified by the use of IS6110-restriction fragment length polymorphism (RFLP) and spacer oligonucleotide typing (spoligotyping) in strains collected in 1992 to 1994 in the Beijing area in China (39). These strains are endemic in east Asia, South Africa, and northern Eurasia (1, 9, 17). Mokrousov et al. (25) hypothesized that the primary dispersal of the Beijing genotype strains took place in China, whereas their initial introduction into northern Eurasia was historically recent and might have been associated with the expansion of the Mongol empire in the 13th to 15th centuries. Recently, the Beijing genotype strains have been found in countries as distant as Argentina (27), Malawi (8), and Australia (21), and new unexpected routes of their apparently secondary transmission are being uncovered (7).
The development and implementation of molecular tools for strain typing has greatly and critically aided the epidemiology of tuberculosis. However, in many world regions, especially those with a high rate of recent transmission and/or MDR-TB, local population structures are dominated by a single or a few genetic families of closely related and hard-to-discriminate strains. The gold standard method to identify Beijing strains is spoligotyping, while the best approach to further subtype them is still IS6110-RFLP typing. The latter method is rather time-consuming and cumbersome, and it requires the extraction and purification of large amounts of DNA. The alternative and also presently widely used approach is VNTR (variable number of tandem repeats) typing (6, 33, 34). The apparent advantage of the VNTR approach is its portability due to the easy digitalization of the generated profiles and, consequently, easy interlaboratory exchange and database management. The task of high-level discrimination long remained a challenge.
Our previous results clearly demonstrated that the 12-locus mycobacterial interspersed repetitive-unit (12-MIRU) method (34) cannot substitute for IS6110-RFLP analysis for the comprehensive epidemiological subtyping of the Beijing strains in Russia (26). The newly proposed 15- and 24-locus VNTR formats were shown to have a higher discriminatory power in a worldwide collection of M. tuberculosis strains (33) and also in Beijing family strains from Japan (11, 41) and China (12). Consequently, the aim of the present study was to investigate the power of the classical IS6110-RFLP typing and the new VNTR combinations for the differentiation of the Beijing family strains from the area of its likely long-term and endemic presence, Russia.
MATERIALS AND METHODS
Bacterial strains.
The mycobacteriology laboratory in the St. Petersburg Research Institute of Phthsiopulmonology serves as a reference center for northwest Russia (11 provinces; 1,700,000 km2; population of 13.5 million); additionally, the laboratory occasionally receives strains from other regions across Russia. A total of 1,130 M. tuberculosis clinical strains recovered from different adult pulmonary TB patients admitted to the hospitals of the St. Petersburg Research Institute of Phthisiopulmonology and the City Anti-Tuberculosis Dispensary were examined by genotyping techniques in the Laboratory of Molecular Microbiology in the St. Petersburg Pasteur Institute between 1996 and 2007. This sample included strains collected during population-based and cross-sectional studies (28-30). The patients originated mainly from St. Petersburg and northwest Russia, as well as from northern Russia, central Russia, Ural, Siberia, and the Far East.
Fifty-two percent of the studied strains were defined as the Beijing genotype by the use of spoligotyping and IS6110-RFLP typing, as recommended previously (17, 39). These strains (from an in-house IS6110-RFLP database) served to select two specific subsets that corresponded to the objectives of the study.
Study set 1.
Forty-eight M. tuberculosis Beijing genotype strains were selected for this study based on a fine analysis of the IS6110-RFLP database at the St. Petersburg Pasteur Institute. The IS6110-RFLP-based unweighted pair-group method of arithmetic averages (UPGMA) tree of all available profiles of the Beijing genotype strains was constructed. Subsequently, a selection of strains from different branches across the dendrogram was performed, and care was taken to include both similar and different patterns (dissimilarity of up to 30%). All selected strains were recovered from patients not found to be epidemiologically linked, in different years, and originating from different locations across Russia.
Forty-four of these strains previously served to evaluate the 12-MIRU format (26). The conclusions of that study were further confirmed in other Russian settings, including a population-based study in Samara (31) and convenience sample-based studies in Ural (15) and Irkutsk (23). For example, locus MIRU26 was confirmed to be the most variable locus (in the 12-MIRU format) in the Beijing genotype strains in all Russian settings (15, 23, 26, 31). Further, similar Hunter Gaston index (HGI) values for 12-MIRU (HGI12-MIRU) were found for Beijing genotype strains from Ural (0.64) (15), Samara (0.61) (31), Irkutsk (0.68) (23), and northwestern Russia (0.65) (26). These findings validated the representativeness of our sample to evaluate the diversity of new VNTR loci in the Russian Beijing genotype strains.
Subsequently, this collection served as a core panel to test the newly proposed VNTR loci in order to find those that are most informative and/or variable of the 24-locus format (33) and the three hypervariable (HV) loci (11).
Study set 2.
The second study set (n = 72) served to evaluate the capacity of the VNTR markers to discriminate among strains in large IS6110-RFLP clusters. It included strains with identical IS6110-RFLP profiles, B0 (n = 34) and A0 (n = 38), which were identified in 27 and 23% of the Beijing genotype strains in our database, respectively (28, 29), and also frequently described in other studies in the countries of the former Soviet Union (1, 3, 19, 37). All strains were recovered in 2003 to 2007 from patients not found to be epidemiologically linked and originating from St. Petersburg and other Russian regions, mainly northwest Russia.
DNA fingerprinting.
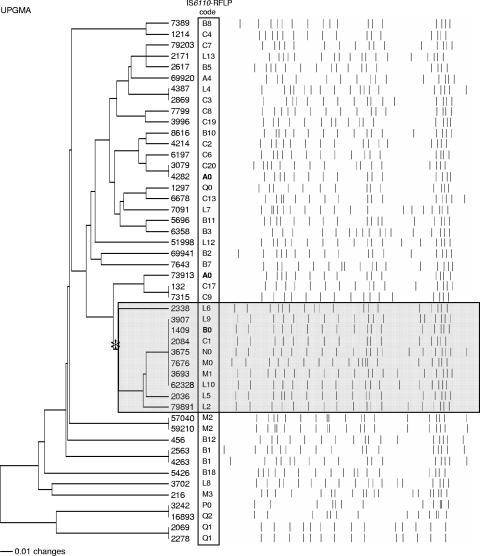
DNA isolation, IS6110-RFLP typing, and spoligotyping were performed as described previously (14, 38). The IS6110-RFLP patterns were compared using the GelCompar version 4.1 package (BVBA Applied Maths, St. Martens-Latem, Belgium) by UPGMA using the Dice coefficient (position tolerance, 2.50%). Before assigning a new code number to a profile, the profiles were visually reverified in the membranes to avoid a possible misalignment during computer processing (e.g., profiles C9 and C17 in Fig. 1 represent different patterns with a single band shift).
FIG. 1.
Twenty-seven VNTR locus-based UPGMA dendrogram and IS6110-RFLP profiles of the 48 M. tuberculosis Beijing genotype strains (study set 1). IS6110-RFLP profile codes are boxed. The B0 cluster (B0 and B0-like profiles) is shaded and marked by an asterisk.
MIRU-VNTR analysis was performed essentially as previously described (11, 33, 34); a 100-bp DNA ladder (GE Healthcare) was used as a molecular size marker. The H37Rv strain was run as an additional control of the performance of the method. To test the reproducibility of the results obtained for HV loci (11), the PCRs were repeated and initial results were confirmed.
The size analysis of the PCR fragments in 1.5% agarose gels and the assignment of the VNTR alleles were done using TotalLab TL100 software (Nonlinear Dynamics Ltd., United Kingdom) and by comparison with correspondence tables kindly provided by Philip Supply and Tomotada Iwamoto.
Statistical analysis.
The HGI was calculated as described previously (10) and was used to evaluate the discriminatory power of the typing methods and the allelic diversity of the VNTR loci. The PAUP* 4.0 package (36) and PARS program of Phylip 3.6 (5) were used to construct the most parsimonious dendrogram and minimum spanning tree, respectively, based on the VNTR digital profiles that were treated as categorical variables.
RESULTS
VNTR typing of strains with diverse IS6110-RFLP profiles (study set 1).
A total of 48 M. tuberculosis strains from St. Petersburg, Russia, identified as the Beijing genotype by spoligotyping, were used to evaluate a discriminatory capacity of the novel VNTR loci. These strains were selected from our IS6110-RFLP database as described in Materials and Methods. The strains differed in their RFLP profiles from a single band of variation of up to 30% dissimilarity. We evaluated the discriminatory power of different loci used alone and in combination (Tables 1 and 2; also see the table in the supplemental material), while IS6110-RFLP typing was considered the gold standard method. The studied 27 loci showed a remarkable difference in their allelic diversity in the studied Beijing genotype strains. Ten loci were found to be monomorphic, thus making the HGI difference between the 15- and 24-locus schemes negligible (0.905 and 0.906, respectively). The use of the three HV loci significantly improved discrimination compared to that of the 15-locus scheme (HGIs of 0.974 and 0.905, respectively). A comparison of the individual diversities of the studied loci showed that only one of the three HV loci, QUB-3232, was polymorphic enough, while two other HV loci were moderately polymorphic, similarly to the most variable loci of the 15-locus scheme, MIRU26 and QUB26 (Table 2). Indeed, a higher diversity of the HV loci was manifested as a greater number of alleles and a wider range of the number of repeats per locus (Table 2).
TABLE 1.
Discriminatory power of the typing methods in M. tuberculosis Beijing genotype strains from Russia, study set 1 (48 strains with mainly different IS6110-RFLP profiles)
| Methoda | No. of types | No. of unique isolates | No. of clustered isolates | No. of clusters | Range of cluster sizes (no. of strains) | HGI |
|---|---|---|---|---|---|---|
| IS6110-RFLP | 44 | 40 | 8 | 4 | 2 | 0.996 |
| 12-MIRU | 12 | 7 | 41 | 5 | 2-23 | 0.723 |
| 15-VNTR | 23 | 15 | 33 | 8 | 2-13 | 0.905 |
| 24-VNTR | 24 | 16 | 32 | 8 | 2-13 | 0.906 |
| 15 VNTR loci plus 3 HV loci | 34 | 25 | 23 | 9 | 2-7 | 0.974 |
| Reduced scheme A (11 loci) | 34 | 25 | 23 | 9 | 2-7 | 0.974 |
| Reduced scheme B (7 loci) | 29 | 18 | 30 | 11 | 2-8 | 0.963 |
| Reduced scheme C (5 loci) | 25 | 14 | 34 | 11 | 2-10 | 0.941 |
The 12-MIRU is from Supply et al. (29); 15- and 24-MIRUs are from Supply et al. (33); the three HV loci are from Iwamoto et al. (11). Reduced scheme A loci (HGI > 0.1) included QUB-11b, Mtub21, QUB-26, MIRU26, ETR-A, MIRU40, MIRU31, MIRU20, QUB-3232, VNTR-3820, and VNTR-4120. Reduced scheme B loci (HGI > 0.1; number of alleles, >3) included QUB-11b, Mtub21, QUB-26, MIRU26, QUB-3232, VNTR-3820, and VNTR-4120. Reduced scheme C loci (HGI > 0.5 or >3 alleles) included QUB-26, MIRU26, QUB-3232, VNTR-3820, and VNTR-4120.
TABLE 2.
Allelic diversity of the VNTR loci in 48 M. tuberculosis Beijing genotype strains with mainly different IS6110-RFLP profiles (study set 1)
| VNTR locus or combination (no. of loci) | VNTR alias | No. of alleles | No. of repeats (range) | Allelic diversity (HGI) |
|---|---|---|---|---|
| Discriminatory (15) | ||||
| 2163b | QUB-11b | 4 | 4-7 | 0.205 |
| 1955 | Mtub21 | 4 | 3-6 | 0.330 |
| 4052 | QUB-26 | 4 | 6-9 | 0.636 |
| 4156 | QUB-4156 | 2 | 2-4 | 0.082 |
| 2996 | MIRU 26 | 4 | 3-7 | 0.520 |
| 424 | Mtub04 | 1 | 4 | 0 |
| 2165 | ETR A | 3 | 2-4 | 0.158 |
| 802 | MIRU 40 | 3 | 1-4 | 0.122 |
| 3690 | Mtub39 | 1 | 3 | 0 |
| 3192 | MIRU 31; ETR E | 3 | 4-6 | 0.160 |
| 960 | MIRU 10 | 2 | 1-3 | 0.082 |
| 580 | MIRU 4; ETR D | 1 | 2 | 0 |
| 577 | ETR C | 2 | 2-4 | 0.042 |
| 1644 | MIRU 16 | 2 | 1-3 | 0.082 |
| 2401 | Mtub30 | 2 | 3-4 | 0.042 |
| Supplementary (9) | ||||
| 2347 | Mtub29 | 2 | 2-4 | 0.087 |
| 4348 | MIRU 39 | 1 | 3 | 0 |
| 3007 | MIRU 27; QUB-5 | 1 | 3 | 0 |
| 2461 | ETR B | 1 | 2 | 0 |
| 3171 | Mtub34 | 1 | 3 | 0 |
| 2531 | MIRU 23 | 1 | 5 | 0 |
| 2059 | MIRU 20 | 2 | 1-2 | 0.120 |
| 154 | MIRU 2 | 1 | 2 | 0 |
| 2687 | MIRU 24 | 1 | 1 | 0 |
| HV | ||||
| 3232 | QUB-3232 | 9 | 5-19 | 0.729 |
| 3820 | VNTR-3820 | 6 | 10-16 | 0.542 |
| 4120 | VNTR-4120 | 6 | 5-16 | 0.370 |
As shown in previous studies (11, 13, 33), PCR failure was found for some loci, namely QUB-26 and Mtub29 in strain 73913 and QUB-11b in strain 7315 (see the table in the supplemental material). These PCR failures were repeated even though the tests were performed under different PCR conditions. This might result from the decreased quality of DNA. On the other hand, as suggested by Iwamoto et al. (11), such negative data are useful for indicating the characteristics that could be used as reference information for genotyping. In this study, these cases were considered to be missing data in the calculation of the pairwise distances under phylogenetic analysis.
We further tested various combinations of loci in order to find one based on a reduced number of VNTR loci and close in discrimination power to that of the 27-locus and IS6110-RFLP typing methods. The applied criteria were the number of alleles and individual diversities (as HGI values) of the loci. Some combinations and rules to include/exclude loci are given in Table 1. Accordingly, the reduced scheme B (seven loci) appears to provide an acceptable balance between a limited number of loci and an HGI close to that of the best-discriminating scheme of VNTR using 15 loci and three HV loci (designated 15 + 3 VNTR). It should be noted that in all combinations, the use of the HV loci was critical, whereas the resulting HGI, while below that of the IS6110-RFLP typing, still was superior to those of the 15- and 24-locus formats (Table 1).
A comparison of the two methods with study set 1 (Fig. 1, Table 1) showed that IS6110-RFLP had more discriminatory capacity than VNTR by further subdividing VNTR clusters, even when all 27 loci were considered. 27-loci VNTR typing (i.e., 24 loci + 3 HV loci) identified eight clusters (Table 1, Fig. 1). The largest VNTR cluster included seven strains that remained nondifferentiated even when all loci were used; not surprisingly, these strains all had different but similar IS6110-RFLP profiles (Fig. 1). These strains had a characteristic upper double band and were defined as belonging to the B0 cluster, since they included profile B0 (27% in our database) and similar B0-like profiles (Fig. 1); a more detailed VNTR analysis of the enlarged set of the B0 cluster strains is presented below.
Three other 27-VNTR clusters (two strains each) also corresponded to IS6110-RFLP clusters, while four other 27-VNTR clusters (also two strains each) included strains with only slightly different IS6110-RFLP profiles (Fig. 1). This may be considered both a general agreement of the typing outcomes by the two methods and the utility of RFLP typing to differentiate among strains in VNTR clusters. On the other hand, in one case, strains with the identical IS6110-RFLP profile A0 were differentiated by three VNTR loci, including two HV loci (QUB-3232 and VNTR-3820) and one low-polymorphic MIRU20 locus (Fig. 1; also see the table in the supplemental material); a more detailed analysis of the enlarged collection of the A0 strains is presented below.
VNTR subtyping within the predominant IS6110-RFLP clusters (study set 2).
As mentioned above, two RFLP profiles, A0 and B0, have been identified in a high proportion of the Beijing genotype strains in St. Petersburg and Russia as a whole (1, 28, 29). Seventy-two strains representing these two profiles (A0, 38 strains; and B0, 34 strains) were subjected to the typing with 17 VNTR loci that were found to be variable in the analysis of the diverse collection of 48 strains (Table 2). In general, VNTR typing underlined both inter- and intracluster differentiation. In particular, 34 B0 strains were subdivided into 11 different types; 26 strains were clustered, whereas the largest cluster included 22 strains (HGI = 0.585). In contrast, no predominant VNTR cluster was found among the A0 strains that were subdivided into 12 different types (33 clustered strains; HGI = 0.860). The five loci previously found to be polymorphic were also the most variable in the combined sample of the A0 and B0 strains, while other tested loci showed negligible or null variation (Table 3).
TABLE 3.
Allelic diversity of the 17 VNTR loci in the 72 M. tuberculosis Beijing genotype strains of study set 2a
| VNTR locus | VNTR alias | No. of alleles | No. of repeats (range) | Allelic diversity (HGI) |
|---|---|---|---|---|
| 2163b | QUB-11b | 2 | 5-6 | 0.106 |
| 1955 | Mtub21 | 1 | 5 | 0 |
| 4052 | QUB-26 | 3 | 6-8 | 0.656 |
| 4156 | QUB-4156 | 2 | 1-2 | 0.028 |
| 2996 | MIRU 26 | 3 | 4-7 | 0.534 |
| 2165 | ETR A | 2 | 3-4 | 0.028 |
| 802 | MIRU 40 | 2 | 2-3 | 0.028 |
| 3192 | MIRU 31; ETR E | 2 | 5-6 | 0.028 |
| 960 | MIRU 10 | 1 | 3 | 0 |
| 577 | ETR C | 1 | 4 | 0 |
| 1644 | MIRU 16 | 3 | 1-4 | 0.082 |
| 2401 | Mtub30 | 2 | 2-4 | 0.055 |
| 2347 | Mtub29 | 4 | 1 | 0 |
| 2059 | MIRU 20 | 1 | 2 | 0 |
| 3232 | QUB-3232 | 5 | 12-18 | 0.655 |
| 3820 | VNTR-3820 | 6 | 12-19 | 0.716 |
| 4120 | VNTR-4120 | 3 | 6-10 | 0.478 |
The 17 loci were found to be polymorphic in 48 strains of study set 1 (Table 2). Study set 2 was composed of two IS6110-RFLP clusters, A0 (n = 38) and B0 (n = 34).
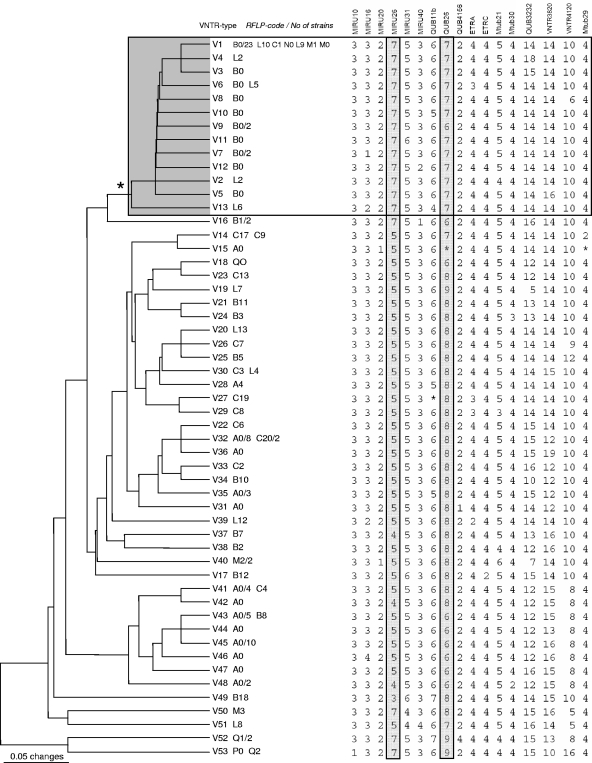
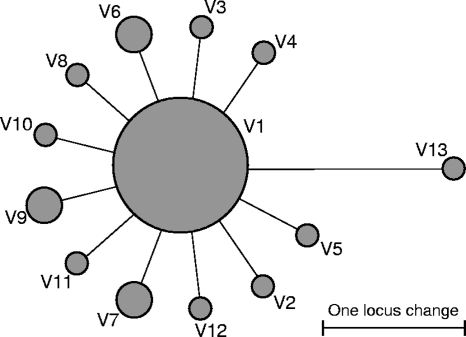
When both study sets 1 and 2 were pooled together (120 strains in total), a total of 53 different VNTR types were identified. To assess the relationships between these types, a dendrogram based on 17 variable loci was constructed (Fig. 2). It showed that B0 and B0-like strains grouped closely, while A0 strains were dispersed all over the dendrogram. In the VNTR-based minimum spanning tree of the B0-cluster strains, the largest VNTR type V1 was in the core position, while all other types could be directly derived from it, mainly by a single locus change (Fig. 3).
FIG. 2.
Seventeen VNTR locus-based UPGMA dendrogram of the combined sample of 120 M. tuberculosis Beijing genotype strains. The B0 cluster (B0 and B0-like profiles) is shaded. The asterisk to the left of the VNTR digital profile designates missing data due to PCR failure. The number of strains is shown if it is greater than one.
FIG. 3.
VNTR-based minimum spanning tree of the VNTR types within the B0 cluster. Circle sizes are roughly proportional to the number of strains. VNTR digital profiles are shown in Fig. 2.
DISCUSSION
This study was undertaken to evaluate new VNTR markers for genotyping within the M. tuberculosis strains of the Beijing genotype in Russia. Currently, these strains attract great attention worldwide, because they demonstrate some important pathogenic properties, such as high transmissibility (2), association with multiple-drug resistance (3, 4, 18, 29), and increased virulence in BCG-vaccinated mice (22). A close genetic relatedness of the Beijing strains makes an epidemiological investigation involving these strains a real challenge.
As multiband IS6110-RFLP profiles evolve faster than low-band profiles, a high HGI, close to the absolute of 1, is not an unexpected finding for the Beijing genotype strains in any setting (Russia, 0.996 [this study, set 1]; Japan, 0.9978 [11]; China, 0.999 [12]). However, the most recent (compared to the other areas in which Beijing strains are endemic) dissemination in Russia since approximately 100 years ago (25) explains the lower HGI for the 12-locus set in the Russian Beijing collection (0.723) (Table 1). On the contrary, in east Asia, where the Beijing strains have circulated for a much longer time, such as in Japan, a basic 12-MIRU set achieved a good discrimination among the Beijing genotype strains (0.941) while the use of the additional, especially HV, loci resulted in HGI values that were superior to that of the IS6110-RFLP typing (HGI15-MIRU + 3HV, 0.9980; HGIIS6110, 0.9978) (11). In fact, this finding of the low diversity of the Beijing strains across Russia may be considered a reflection of the recent dissemination of the Beijing genotype in this country.
On the other hand, we have noticed that the HGI for the 12-MIRU scheme was higher for the setting of Japan (0.941) than for that of Beijing, China (0.778). A Hong Kong study demonstrated a higher diversity of the Beijing genotype by both methods, whereas the HGIIS6110 (0.998) was higher than the HGI12-MIRU (0.882) (13). The Beijing genotype is historically endemic in central and southern China and Japan (11, 12, 24, 39), and it may be that this estimation for the Beijing city (12) was biased by a smaller sample size compared to those of the Hong Kong and Japan studies.
A comparison of the diversities of the individual loci showed that HV loci were indeed the most polymorphic in our study, although their HGI values were below the respective values in the Japan study (Table 4). A closer look at the loci included in the new 15- and 24-locus formats showed that some of them remained lowly polymorphic in every published setting in Russia and east Asia, e.g., loci MIRU2 and ETR-B. Locus MIRU24 remained monomorphic, which is in agreement with the previous observation that this locus is phylogenetically conserved and discriminates between ancestral/modern M. tuberculosis lineages with/without the TbD1 genome region (32). It may be mentioned that locus QUB-3232 was similarly and highly polymorphic both in our study and in the large population-based study in the Samara province in Russia (31), which may be considered an additional validation of the representativeness of our study panel (Table 4). Some other loci were lowly or less polymorphic in Russian settings but showed high or higher diversity in east Asian studies, e.g., loci QUB-11b, Mtub04, MIRU10, and Mtub30 (Table 4). A generally lower VNTR diversity of the Russian setting compared to those of the east Asian settings may be explained by the fact that the clonal expansion of the Beijing genotype strains in Russia began more recently.
TABLE 4.
VNTR loci diversity in Beijing genotype strains from different locations
| VNTR locus or combination (no. of loci) | VNTR alias | Locus diversitya in area of isolation (reference)
|
||||||
|---|---|---|---|---|---|---|---|---|
| St. Petersburg, Russia (this study, set 1) | Samara, Russia (31) | West Siberia, Russia (35) | Beijing, China (12) | Hong Kong, China (13) | Hong Kong, China (16) | Kobe, Japan (11) | ||
| Discriminatory (15) | ||||||||
| 2163b | QUB-11b | 0.205 | 0.210 | 0.651 | 0.669 | 0.618 | 0.772 | |
| 1955 | Mtub21 | 0.330 | 0.105 | 0.110 | 0.556 | 0.393 | ||
| 4052 | QUB-26 | 0.636 | 0.780 | 0.518 | 0.314 | 0.299 | 0.741 | |
| 4156 | QUB-4156 | 0.082 | 0.395 | 0.167 | 0.611 | |||
| 2996 | MIRU 26 | 0.520 | 0.445 | 0.353 | 0.200 | 0.383 | ||
| 424 | Mtub04 | 0 | 0.061 | 0.306 | 0.459 | |||
| 2165 | ETR-A | 0.158 | 0.046 | 0 | 0.232 | 0.188 | 0.201 | 0.147 |
| 802 | MIRU40 | 0.122 | 0.031 | 0.390 | 0.194 | 0.196 | 0.327 | |
| 3690 | Mtub39 | 0 | 0.016 | 0 | 0.171 | 0.186 | ||
| 3192 | MIRU31; ETR-E | 0.160 | 0.176 | 0 | 0.169 | 0.156 | 0.200 | 0.322 |
| 960 | MIRU10 | 0.082 | 0.046 | 0.144 | 0.377 | 0.419 | ||
| 580 | MIRU4; ETR-D | 0 | 0.046 | 0.120 | 0.072 | 0.019 | 0.086 | |
| 577 | ETRC | 0.042 | 0.016 | 0 | 0.094 | 0.057 | 0.165 | 0.022 |
| 1644 | MIRU16 | 0.082 | 0.016 | 0.068 | 0.058 | 0.310 | ||
| 2401 | Mtub30 | 0.042 | 0.068 | 0.403 | ||||
| Supplementary (9) | ||||||||
| 2347 | Mtub29 | 0.087 | 0.180 | 0.119 | 0.043 | |||
| 4348 | MIRU39 | 0 | 0.016 | 0.119 | 0.320 | 0.221 | ||
| 3007 | MIRU27; QUB-5 | 0 | 0 | 0.014 | 0.115 | |||
| 2461 | ETR-B | 0 | 0 | 0.014 | 0.064 | 0 | 0.033 | |
| 3171 | Mtub34 | 0 | 0.014 | 0.065 | ||||
| 2531 | MIRU23 | 0 | 0.016 | 0 | 0.014 | 0.176 | ||
| 2059 | MIRU20 | 0.120 | 0 | 0.014 | 0.022 | |||
| 154 | MIRU2 | 0 | 0.031 | 0 | 0 | |||
| 2687 | MIRU24 | 0 | 0 | 0 | 0 | |||
| HV | ||||||||
| 3232 | QUB-3232 | 0.729 | 0.621 | 0.804 | 0.880 | |||
| 3820 | VNTR-3820 | 0.542 | 0.800 | |||||
| 4120 | VNTR-4120 | 0.370 | 0.902 | |||||
An increase in the number of targeted VNTR loci is expected to achieve an increased discrimination that ultimately is close to that of IS6110-RFLP. Nevertheless, this raises a question about the practical utility of schemes based on, for example, more than 10 loci. A first-line screening may be reasonably limited to a few loci if they achieve a sufficiently high discrimination. As the population structures of the circulating M. tuberculosis strains vary across different world regions, these first-line/primary typing schemes may be country specific and include different sets of loci. For example, in Russia, such a scheme may target the MDR-associated Beijing genotype strains that constitute half of the circulating strains in this country. The most polymorphic loci in our study also were found to be polymorphic in other Russian settings (Table 4). This leads us to suggest that these most-polymorphic five/seven loci (schemes B and C, respectively, in Table 1) be used in the first-line screening of the M. tuberculosis Beijing genotype not only in northwest Russia but for the whole country. Nonetheless, further studies are required to validate the proposed reduced format. At the same time, several large VNTR types (V1, V32, V41, and V43) (Fig. 2) were further subdivided by the use of IS6110-RFLP, thus demonstrating its apparent utility as a secondary typing method of the VNTR clustered isolates. The reverse situation, i.e., VNTR subtyping within RFLP clusters, is discussed below.
The possibility to differentiate using VNTRs within IS6110 clusters has been shown in studies that were focused mainly on non-Beijing strains and low-band clusters. Regarding Beijing genotype strains, 24-locus VNTR typing showed a lower level of discrimination than IS6110-RFLP typing in Chinese strains, but still one RFLP cluster of two strains was discriminated by the ETR-A locus (12). A large-scale Japan study showed a slightly higher HGI for the 24-VNTR locus plus 3 HV locus method than for the IS6110-RFLP typing method (11). In that study, three RFLP clusters within Beijing genotype strains (two to four strains each) were further differentiated by VNTRs due to 2 to 10 variant loci. In this study, we addressed this issue by testing two large RFLP clusters of the Beijing strains (A0 and B0) that are found at high rates across Russia (1, 28).
Interestingly, 40 strains with RFLP profile A0 were distributed in different and distant branches of the VNTR dendrogram (Fig. 2). Whether this situation reflects a high stability of this particular IS6110-RFLP profile or results from the convergent evolution of the IS6110 elements remains to be addressed in further studies targeting other genome regions in these strains.
Unlike the case for A0 strains, 44 strains of the B0 cluster were related in their VNTR profiles and clearly diverged from other strains (Fig. 2). At the same time, the B0 cluster strains were marked by a specific combined signature of the two loci MIRU26 (seven copies in all strains) and QUB-26 (seven copies in 42/44 strains). Applying a rigorous definition (seven copies in MIRU26 and seven copies in QUB26), this would correctly identify 42 (95%) of 44 strains of the B0 cluster. The specificity to detect the B0 cluster would be 100%, since none of the other 76 strains had this two-locus signature (Fig. 2). At the same time, the application of the phylogenetic analysis based on the complete set of the polymorphic loci would correctly identify all strains of the B0 cluster (Fig. 2).
B0, also defined as W148 in the database of the Public Health Research Institute, Newark, NJ (1), is the most frequent Russian Beijing variant and is identified in approximately one-third of the Beijing strains in different geographic settings in Russia and countries of the former Soviet Union (1, 3, 4, 19, 28, 29, 31, 35, 37), as well as in the United States and Germany from recent immigrants from the former Soviet Union (1, 20). These strains were shown to have been involved in the nosocomial outbreak in St. Petersburg (30), they were found to be more virulent in mice (40) and macrophage models (E. Lasunskaia, S. Ribeiro, O. Manicheva, L. Lima Gomes, P. N. Suffys, I. Mokrousov, L. Ferrazoli, A. Kritski, T. Otten, T. L. Kipnis, B. Vishnevsky, and O. Narvskaya, submitted for publication), and they were associated with MDR-TB in newly diagnosed TB patients (28). This two-locus MIRU26/QUB26 signature apparently is specific for this clonal group in northwest Russia; its specificity for the country as a whole remains to be tested with large strain collections in different Russian regions. It may be noted that the same two-locus signature was found in the Beijing genotype strains with clearly distinct IS6110-RFLP profiles in Japan (11) and China (12). In this sense, our finding cannot be applied to all Beijing genotype strains worldwide. Nevertheless, in view of the increased transmissibility and hypervirulence of this particular B0/W148 Russian (northwest Russian) subvariant of the Beijing genotype (28, 29, 30, 40, and Lasunskaia et al., submitted), the availability of a simple molecular test for its detection has an apparent practical utility. The fact that these strains isolated in patients in different years and distant locations were grouped closely in the VNTR-based tree supports their true clonality. On the other hand, a star-like VNTR-based network with short edges linking the large core type with other types suggests a recent outburst dissemination of these strains in the survey area (Fig. 3). Consequently, this closely related group of strains, the B0 cluster, may indeed represent a successful clone that is widely and recently disseminated due to its specific pathogenic properties.
An important additional implication from our finding of the VNTR signature of the specific B0 cluster is that it opens new perspectives in the analysis of archival noncultured samples that have insufficient quantity/quality of DNA for IS6110-RFLP analysis but still are sufficient for PCR-based VNTR analysis. Consequently, this could help to trace back the origin of the B0 clonal group.
In conclusion, a related group of the Beijing genotype strains, the B0 cluster, appears to represent a successful clone that is widely and recently disseminated across Russia. The IS6110-RFLP method and VNTR typing using a reduced set of the most polymorphic loci critically complement each other for the high-resolution epidemiological typing of the M. tuberculosis Beijing genotype strains circulating in or imported from Russia.
Supplementary Material
Acknowledgments
We are grateful to Philip Supply and Tomotada Iwamoto for kindly providing the correspondence tables for VNTR copy number analysis.
This study was supported by NATO's Public Diplomacy Division in the framework of the Science for Peace program (grant SFP-982319, Detect Drug-Resistant TB) and by a research fellowship from the European Commission within the EU Marie Curie Mobility program to Igor Mokrousov.
Footnotes
Published ahead of print on 27 August 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 1045-52. [DOI] [PubMed] [Google Scholar]
- 2.Caminero, J. A., M. J. Pena, M. I. Campos-Herrero, J. C. Rodriguez, I. Garcia, P. Cabrera, C. Lafoz, S. Samper, H. Takiff, O. Afonso, J. M. Pavon, M. J. Torres, D. van Soolingen, D. A. Enarson, and C. Martin. 2001. Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. Am. J. Respir. Crit. Care Med. 1641165-1170. [DOI] [PubMed] [Google Scholar]
- 3.Cox, H. S., T. Kubica, D. Doshetov, Y. Kebede, S. Rüsch-Gerdess, and S. Niemann. 2005. The Beijing genotype and drug resistant tuberculosis in the Aral Sea region of Central Asia. Respir. Res. 6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drobniewski, F., Y. Balabanova, V. Nikolayevsky, M. Ruddy, S. Kuznetzov, S. Zakharova, A. Melentyev, and I. Fedorin. 2005. Drug-resistant tuberculosis, clinical virulence, and the dominance of the Beijing strain family in Russia. JAMA 2932726-2731. [DOI] [PubMed] [Google Scholar]
- 5.Felsenstein, J. 2004. PHYLIP (Phylogeny Inference Package) version 3.6b. Department of Genome Sciences, University of Washington, Seattle.
- 6.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 1441189-1196. [DOI] [PubMed] [Google Scholar]
- 7.García de Viedma, D., F. Chaves, J. Inigo, et al. 2006. New route of importation of Mycobacterium tuberculosis Beijing genotype. Emerg. Infect. Dis. 12169-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glynn, J. R., A. C. Crampin, H. Traore, M. D. Yates, F. D. Mwaungulu, B. M. Ngwira, S. D. Chaguluka, D. T. Mwafulirwa, S. Floyd, C. Murphy, F. A. Drobniewski, and P. E. Fine. 2005. Mycobacterium tuberculosis Beijing genotype, northern Malawi. Emerg. Infect. Dis. 11150-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpsons's index of diversity. J. Clin. Microbiol. 262465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwamoto, T., S. Yoshida, K. Suzuki, M. Tomita, R. Fujiyama, N. Tanaka, Y. Kawakami, and M. Ito. 2007. Hypervariable loci that enhance the discriminatory ability of newly proposed 15-loci and 24-loci variable-number tandem repeat typing method on Mycobacterium tuberculosis strains predominated by the Beijing family. FEMS Microbiol. Lett. 272282-283. [DOI] [PubMed] [Google Scholar]
- 12.Jiao, W. W., I. Mokrousov, G. Z. Sun, Y. J. Guo, A. Vyazovaya, O. Narvskaya, and A. D. Shen. 2008. Evaluation of new variable-number tandem repeat typing systems of Mycobacterium tuberculosis with Beijing genotype isolates from Beijing, China. J. Clin. Microbiol. 461045-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kam, K. M., C. W. Yip, K. L. Leung, K. L. Wong, W. M. Ko, and W. S. Wong. 2006. Optimization of variable number tandem repeat typing set for differentiating Mycobacterium tuberculosis strains in the Beijing family. FEMS Microbiol. Lett. 256258-265. [DOI] [PubMed] [Google Scholar]
- 14.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovalev, S. Y., E. Y. Kamaev, M. A. Kravchenko, N. E. Kurepina, and S. N. Skorniakov. 2005. Genetic analysis of Mycobacterium tuberculosis strains isolated in Ural region, Russian Federation, by MIRU-VNTR genotyping. Int. J. Tuberc. Lung Dis. 9746-752. [PubMed] [Google Scholar]
- 16.Kremer, K., B. K. Au, P. C. Yip, R. Skuce, P. Supply, K. M. Kam, and D. van Soolingen. 2005. Use of variable-number tandem-repeat typing to differentiate Mycobacterium tuberculosis Beijing family isolates from Hong Kong and comparison with IS6110 restriction fragment length polymorphism typing and spoligotyping. J. Clin. Microbiol. 43314-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kremer, K., J. R. Glynn, T. Lillebaek, S. Niemann, N. E. Kurepina, B. N. Kreiswirth, P. J. Bifani, and D. van Soolingen. 2004. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J. Clin. Microbiol. 424040-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krüüner, A., S. E. Hoffner, H. Sillastu, M. Danilovits, K. Levina, S. B. Svenson, S. Ghebremichael, T. Koivula, and G. Källenius. 2001. Spread of drug-resistant pulmonary tuberculosis in Estonia. J. Clin. Microbiol. 393339-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubica, T., R. Agzamova, A. Wright, M. A. Aziz, G. Rakishev, V. Bismilda, E. Richter, S. Rüsch-Gerdes, and S. Niemann. 2005. The Beijing genotype is a major cause of drug-resistant tuberculosis in Kazakhstan Int. J. Tuberc. Lung Dis. 9646-653. [PubMed] [Google Scholar]
- 20.Kubica, T., S. Rüsch-Gerdes, and S. Niemann. 2004. The Beijing genotype is emerging among multidrug-resistant Mycobacterium tuberculosis strains from Germany. Int. J. Tuberc. Lung Dis. 81107-1113. [PubMed] [Google Scholar]
- 21.Lavender, C., M. Globan, A. Sievers, H. Billman-Jacobe, and J. Fyfe. 2005. Molecular characterization of isoniazid-resistant Mycobacterium tuberculosis isolates collected in Australia. Antimicrob. Agents Chemother. 494068-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López, B., D. Aguilar, H. Orozco, M. Burger, C. Espitia, V. Ritacco, L. Barrera, K. Kremer, R. Hernandez-Pando, K. Huygen, and D. van Soolingen. 2003. A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin. Exp. Immunol. 13330-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medvedeva, T. V., O. B. Ogarkov, O. M. Nekipelov, I. V. Ushakov, E. S. Koziakova, and R. G. Skvortsova. 2004. MIRU-VNTR genotyping of Mycobacterium tuberculosis strains from East Siberian: Beijing family versus Kilimanjaro family. Mol. Gen. Mikrobiol. Virusol. 433-38. (In Russian.) [PubMed] [Google Scholar]
- 24.Millet, J., C. Miyagi-Shiohira, N. Yamane, C. Sola, and N. Rastogi. 2007. Assessment of mycobacterial interspersed repetitive unit-QUB markers to further discriminate the Beijing genotype in a population-based study of the genetic diversity of Mycobacterium tuberculosis clinical isolates from Okinawa, Ryukyu Islands, Japan. J. Clin. Microbiol. 453606-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mokrousov, I., H. M. Ly, T. Otten, N. N. Lan, B. Vyshnevskyi, S. Hoffner, and O. Narvskaya. 2005. Origin and primary dispersal of the Mycobacterium tuberculosis Beijing genotype: clues from human phylogeography. Genome Res. 151357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mokrousov, I., O. Narvskaya, E. Limeschenko, A. Vyazovaya, T. Otten, and B. Vyshnevskyi. 2004. Analysis of the allelic diversity of the mycobacterial interspersed repetitive units in Mycobacterium tuberculosis strains of the Beijing family: practical implications and evolutionary considerations. J. Clin. Microbiol. 422438-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morcillo, N., B. Di Giulio, C. Chirico, A. Kuriger, A. Dolmann, A. Alito, M. Zumarraga, D. van Soolingen, K. Kremer, and A. Cataldi. 2005. First description of Mycobacterium tuberculosis Beijing genotype in Argentina. Rev. Argent. Microbiol. 3792-95. [PubMed] [Google Scholar]
- 28.Narvskaya, O. 2003. Genome polymorphism of Mycobacterium tuberculosis and its significance in the epidemic process. D.Sc. dissertation, St. Petersburg, Russia.
- 29.Narvskaya, O., I. Mokrousov, T. Otten, and B. Vishnevsky. 2005. Molecular markers: application for studies of Mycobacterium tuberculosis population in Russia, p. 111-125. In M. M. Read (ed.), Trends in DNA fingerprinting research. Nova Science Publishers, New York, NY.
- 30.Narvskaya, O., T. Otten, E. Limeschenko, N. Sapozhnikova, O. Graschenkova, L. Steklova, A. Nikonova, M. L. Filipenko, I. Mokrousov, and B. Vyshnevskiy. 2002. Nosocomial outbreak of multidrug-resistant tuberculosis caused by a strain of Mycobacterium tuberculosis W-Beijing family in St. Petersburg, Russia. Eur. J. Clin. Microbiol. Infect. Dis. 21596-602. [DOI] [PubMed] [Google Scholar]
- 31.Nikolayevskyy, V., K. Gopaul, Y. Balabanova, T. Brown, I. Fedorin, and F. Drobniewski. 2006. Differentiation of tuberculosis strains in a population with mainly Beijing-family strains. Emerg. Infect. Dis. 121406-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun, Y. J., R. Bellamy, A. S. Lee, S. T. Ng, S. Ravindran, S. Y. Wong, C. Locht, P. Supply, and N. I. Paton. 2004. Use of mycobacterial interspersed repetitive unit-variable-number tandem repeat typing to examine genetic diversity of Mycobacterium tuberculosis in Singapore. J. Clin. Microbiol. 421986-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Supply, P., C. Allix, S. Lesjean, M. Cardoso-Oelemann, S. Rusch-Gerdes, E. Willery, E. Savine, P. de Haas, H. van Deutekom, S. Roring, P. Bifani, N. Kurepina, B. Kreiswirth, C. Sola, N. Rastogi, V. Vatin, M. C. Gutierrez, M. Fauville, S. Niemann, R. Skuce, K. Kremer, C. Locht, and D. van Soolingen. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 444498-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 393563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Surikova, O. V., D. S. Voitech, G. Kuzmicheva, S. I. Tatkov, I. V. Mokrousov, O. V. Narvskaya, M. A. Rot, D. van Soolingen, and M. L. Filipenko. 2005. Efficient differentiation of Mycobacterium tuberculosis strains of the W-Beijing family from Russia using highly polymorphic VNTR loci. Eur. J. Epidemiol. 20963-974. [DOI] [PubMed] [Google Scholar]
- 36.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (and other methods) 4.0 beta. Sinauer Associates, Sunderland, MA.
- 37.Toungoussova, O. S., P. Sandven, A. O. Mariandyshev, N. I. Nizovtseva, G. Bjune, and D. A. Caugant. 2002. Spread of drug-resistant Mycobacterium tuberculosis strains of the Beijing genotype in the Archangel Oblast, Russia. J. Clin. Microbiol. 401930-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnik, and P. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Soolingen, D., L. Qian, P. E. W. de Haas, J. T. Douglas, H. Traore, F. Portaels, Z. Quing, D. Enkhasaikan, P. Nymadawa, and J. D. A. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 333234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vishnevskii, B. I., O. V. Narvskaia, S. N. Vasil'eva, N. V. Sapozhnikova, I. V. Mokrousov, and T. F. Otten. 2002. Virulence of Mycobacterium tuberculosis. Probl. Tuberk. 1033-36. (In Russian.) [PubMed] [Google Scholar]
- 41.Yokoyama, E., K. Kishida, M. Uchimura, and S. Ichinohe. 2007. Improved differentiation of Mycobacterium tuberculosis strains, including many Beijing genotype strains, using a new combination of variable number of tandem repeats loci. Infect. Genet. Evol. 7499-508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.