Abstract
The diphtheria epidemic in the Russian Federation in the 1990s made diphtheria a focus of global concern once again. The development of rapid and reproducible typing methods for the molecular characterization of Corynebacterium diphtheriae has become a priority in order to be able to monitor the spread of this important pathogen on a global scale. We report on a comparison of four molecular typing methods (ribotyping, pulsed-field gel electrophoresis [PFGE], random amplification of polymorphic DNA [RAPD], and amplified fragment length polymorphism [AFLP]) for the characterization of C. diphtheriae strains. Initially, 755 isolates originating from 26 countries were analyzed by ribotyping. One strain of each ribotype was then randomly chosen and characterized by PFGE, RAPD, and AFLP. In order to ascertain whether the Eastern European epidemic ribotype could be further discriminated, 10 strains of ribotype D1 (the epidemic ribotype) from different geographical regions were randomly chosen and subjected to analysis by PFGE, RAPD, and AFLP. The results revealed that ribotyping is highly discriminatory and reproducible and is currently the method of choice for typing C. diphtheriae. PFGE and AFLP were less discriminatory than ribotyping and RAPD. An assessment of the transcontinental spread of the organism showed that several genotypes of C. diphtheriae circulated on different continents of the world and that each outbreak was caused by a distinct clone. The ribotypes seen in Europe appeared to be distinct from those seen elsewhere, and certain ribotypes appeared to be unique to particular countries.
Due to the highly effective diphtheria vaccine that became available in the 1940s and 1950s, the incidence of diphtheria declined dramatically in many parts of the world. However, epidemic diphtheria reemerged in Eastern Europe in the 1990s, and the disease spread to all 15 Newly Independent States (NIS) of the former USSR. The disease is endemic in countries such as Turkey, Bangladesh, India, Pakistan, and Vietnam and in Africa and parts of South America (7). The onset of these epidemics and the occurrence of endemicity in some countries highlighted the importance of monitoring the spread of C. diphtheriae from index cases to the community, country, and beyond. It is also necessary to distinguish domestic from imported cases to allow the adequate implementation of local preventive measures. Therefore, the availability of rapid and reproducible typing tools for the molecular characterization of C. diphtheriae became a high priority.
Here we report on a comparison of four molecular typing methods (ribotyping, pulsed-field gel electrophoresis [PFGE], random amplification of polymorphic DNA [RAPD], and amplified fragment length polymorphism [(AFLP]) for characterization of C. diphtheriae. Initially, 755 isolates originating from 26 countries were analyzed by ribotyping. One strain of each ribotype was randomly chosen and characterized by PFGE, RAPD, and AFLP. In order to ascertain whether the Eastern European epidemic ribotype could be further discriminated by other typing methods, 10 strains of ribotype D1 (the epidemic ribotype) from different geographical regions were randomly chosen and were subjected to analysis by PFGE, RAPD, and AFLP. The transcontinental spread of C. diphtheriae was determined by using the extensive set of ribotyping results obtained from this study.
MATERIALS AND METHODS
Bacterial isolates.
Seven hundred fifty-five C. diphtheriae isolates from 26 countries (Armenia, Australia, Belarus, Denmark, Dominican Republic, Estonia, Finland, France, Germany, Italy, Kazakhstan, Kenya, Kyrgyzstan, Latvia, Poland, Romania, Rwanda, Russia, Sweden, Thailand, Turkmenistan, the United Kingdom, Ukraine, Uzbekistan, the United States, and Vietnam) referred to the Streptococcus and Diphtheria Reference Unit at the Centre for Infections, London, United Kingdom, for identification and typing from 1985 to 2000 were chosen for use in the ribotyping studies. The isolates were from patients with diphtheria, pharyngitis, or tonsillitis; asymptomatic carriers; and contacts. The disease status was unknown for the sources of 68% of the isolates. The isolates were from patients aged between 12 months and 83 years (the age was not known for 52% of the patients). Table 1 summarizes the 755 C. diphtheriae strains analyzed by ribotyping. One isolate of each ribotype was randomly chosen for analysis by PFGE, RAPD, and AFLP (Table 2).
TABLE 1.
Summary of the 755 C. diphtheriae isolates analyzed by ribotyping
| Country | No. of isolates | Yr of isolation | Biotype(s) (toxigenicitya) | Ribotype(s) (no. of isolates)b |
|---|---|---|---|---|
| Armeniac | 19 | 1999 | Gravis (18 tox+), mitis (1 tox+) | D1 (11), D4 (7), D7 (1) |
| Australia | 32 | 1992-1995 | Gravis (17 tox−), mitis (1 tox+, 2 tox−) | D9 (18), D15 (1), D46 (1), D47 (1), D48 (2), D49 (1), D50 (1), D51 (3), D52 (1), D56 (2), D57 (1) |
| Belarusc | 79 | 1996-2000 | Gravis (48 tox+, 12 tox−), mitis (16 tox+, 3 tox−) | D1 (13), D4 (26), D5 (1), D6 (2), D7 (7), D10 (25), D15 (1), D19 (2), D30 (1), D40 (1) |
| Denmark | 1 | Mitis (tox+) | D74 (1) | |
| Dominican Republic | 3 | 1995 | Mitis (3 tox+) | D71 (2), D72 (1) |
| Estoniad | 26 | 1993-1995 | Gravis (13 tox+, 11 tox −), mitis (1 tox+, 1 tox −) | D1 (12), D4 (10), D5 (1), D7 (1), D10 (2) |
| Finlandd | 6 | 1993-1995 | Gravis (4 tox+), mitis (1 tox+, 1 tox−) | D1 (4), D10 (2) |
| France | 1 | 1993 | Mitis (tox−) | D46 (1) |
| Germany | 24 | 1993-1994 | Gravis (4 tox+, 9 tox−), mitis (3 tox+, 9 tox−), belfanti (3 tox−) | D1 (3), D4 (1), D11 (8), D13 (1), D14 (3), D15 (3), D16 (1), D17 (1), D18 (1), D26 (2) |
| Italy | 6 | 1994-1996 | Gravis (1 tox+, 4 tox−), mitis (1tox+) | D11 (4), D45 (1), D73 (1) |
| Kazakhstanc | 43 | 1995-1997 | Gravis (39 tox+), mitis (5 tox+) | D1 (7), D4 (32), D7 (4) |
| Kenya | 1 | 1998 | Mitis (1 tox+) | D62 (1) |
| Kyrghyzstanc | 4 | 1993-1995 | Gravis (1 tox+), mitis (1 tox−) | D1 (1), D7 (3) |
| Latviac | 115 | 1999-2000 | Gravis (105 tox+, 1 tox−), mitis (7 tox+, 2 tox−) | D1 (50), D4 (61), D10 (4) |
| Poland | 1 | 1994 | Intermedius (tox+) | D54 (1) |
| Romania | 17 | 1994 | Gravis (2 tox+, 6 tox−), mitis (5 tox+, 1 tox−), belfanti (1 tox−), intermedius (1 tox−) | D11 (6), D20 (1), D21 (6), D22 (1), D23 (1), D24 (1), D25 (1) |
| Ruanda | 2 | 1994 | Mitis (tox−) | D54 (1), D55 (1) |
| Russiac | 218 | 1966, 1993-95 | Gravis (148 tox+, 7 tox−), mitis (63 tox+, 8 tox−) | D1 (81), D2 (1), D3 (2), D4 (56), D5 (1), D6 (3), D7 (56), D8 (1), D9 (3), D10 (11), D11 (2), D12 (1) |
| Swedene | 13 | 1994 | Gravis (2 tox−), mitis (6 tox+, 3tox−), belfanti (2 tox−) | D4 (1), D11 (1), D17 (1), D22 (1), D26 (6), D27 (1), D28 (1), D29 (1) |
| Thailandf | 31 | 1994-1996 | Gravis (1 tox−), mitis (27 tox+, 3 tox+) | D19 (1), D34 (13), D63 (2), D64 (6), D65 (2), D66 (3), D67 (1), D68 (1), D69 (1), D70 (1) |
| Turkmenistanc | 25 | 1995 | Gravis (17 tox+, 4 tox−), mitis (3 tox+, 1 tox−) | D1 (2), D4 (18), D7 (3), D10 (2) |
| United Kingdomg | 22 | 1985-1998 | Gravis (3 tox+), mitis (17 tox+, 2 tox−) | D1 (1), D7 (1) D30 (1), D31 (1), D32 (2), D33 (1), D34 (1), D35 (1), D36 (1), D37 (1), D38 (1), D39 (1), D40 (4), D41 (1), D42 (2), D43 (1), D44 (1) |
| Ukrainec | 19 | 1999 | Gravis (19 tox+) | D1 (19) |
| Uzbekistanc | 11 | 1995 | Gravis (3 tox+, 2 tox−), mitis (6 tox+) | D1 (3), D7 (1), D10 (2), D15 (1), D19 (4) |
| United Statesh | 23 | 1972-82, 1995 | Gravis (8 tox+, 3 tox−), mitis (6 tox−), intermedius (2 tox+, 3 tox−) | D4 (1), D13 (3), D50 (3), D57 (5), D58 (8), D59 (3) |
| Vietnam | 13 | 1995 | Mitis (13 tox+) | D9 (1), D60 (3), D61 (9) |
The numbers of isolate that are toxin positive (tox+) and toxin negative (tox−) are given in parentheses.
International designations for some of the predominant ribotypes are as follows: D1, Sankt Petersburg; D7, Otchakov; D11, Vladimir; D4, Rossija; D10, Cluj.
The majority of the isolates are from the epidemic which began in the Russian Federation in 1990.
Imported cases of diphtheria from the NIS.
Isolates from the Scandinavian outbreak, which occurred in the mid-1980s.
Isolates from the outbreak in Thailand in 1994.
The majority of the isolates are from imported cases of diphtheria from Asia, Africa, and the Far East.
Isolates from the outbreak in Seattle, WA (1971 to 1982).
TABLE 2.
Summary of the 74 strains analyzed by PFGE, RAPD, and AFLP
| Lab no. | Region, country, yr of isolation | Age (yr) | Sexa | Disease | Biotype | Toxb | Ribotype |
|---|---|---|---|---|---|---|---|
| CD93/46 | St. Petersburg, Russia, 1993 | 46 | M | Tonsillitis | Gravis | + | D1 |
| CD95/66 | Omsk region, Russia | Gravis | + | D2 | |||
| CD93/69 | Murmansk, Russia, 1993 | 30 | Diphtheria | Gravis | + | D3 | |
| CD93/78 | Murmansk, Russia, 1993 | 21 | M | Diphtheria | Gravis | + | D4 |
| CD93/266 | St. Petersburg, Russia, 1993 | 4 | F | Diphtheria | Gravis | + | D5 |
| CD93/181 | Moscow, Russia, 1966 | Carrier | Gravis | + | D6 | ||
| CD93/45 | St. Petersburg, Russia, 1993 | 34 | M | Carrier | Mitis | + | D7 |
| CD93/132 | Kaliningrad Oblast, Russia 1993 | 28 | M | Carrier | Mitis | + | D8 |
| CD93/183 | Dagestan, Russia, 1983 | Mitis | − | D9 | |||
| CD93/186 | St. Petersburg, Russia, 1989 | Carrier | Mitis | − | D10 | ||
| CD93/274 | Vladimir, Russia | Carrier | Gravis | − | D11 | ||
| CD93/277 | Moscow, Russia | 19 | Carrier | Gravis | − | D12 | |
| CD94/68 | Erlabrunn, Germany | 5 | Mitis | − | D13 | ||
| CD94/66 | Berlin, Germany | 42 | M | Mitis | + | D14 | |
| CD94/69 | Erlabrunn, Germany | 5 | Gravis | − | D15 | ||
| CD94/72 | Greifswald, Germany | Belfanti | − | D16 | |||
| CD94/76 | Dresden, Germany | Belfanti | − | D17 | |||
| CD94/260 | Germany, 1993 | Bronchus (source) | Belfanti | − | D18 | ||
| CD94/263 | Thailand | 28 | F | Skin lesion (German patient) | Mitis | + | D19 |
| CD94/238 | Romania | Gravis | + | D20 | |||
| CD94/252 | Romania | Gravis | + | D21 | |||
| CD94/249 | Romania | Mitis | − | D22 | |||
| CD94/240 | Romania | Intermedius | − | D24 | |||
| CD94/241 | Romania | Intermedius | − | D25 | |||
| CD94/91 | Sweden (outbreak) | Mitis | + | D26 | |||
| CD94/94 | Sweden | Mitis | + | D27 | |||
| CD94/232 | Sweden | Mitis | − | D28 | |||
| CD94/231 | Sweden | Belfanti | − | D29 | |||
| CD93/32 | United Kingdom | 39 | Immigrant | Gravis | + | D30 | |
| CD94/149 | United Kingdom (Birmingham) | 14 | M | Pharyngitis | Gravis | + | D31 |
| CD85/2 | Swansea, United Kingdom | Tonsillitis | Mitis | + | D32 | ||
| CD85/29 | Swansea, United Kingdom (imported case from Tunisia) | M | Mitis | + | D33 | ||
| CD90/39 | London, United Kingdom | Tonsillitis | Mitis | + | D34 | ||
| CD93/4 | United Kingdom (imported from Australia) | M | Mitis | + | D35 | ||
| CD93/19 | United Kingdom (imported from Bangladesh) | 15 | Diphtheria | Mitis | + | D36 | |
| CD93/117 | United Kingdom | 6 | Mitis | + | D37 | ||
| CD93/121 | United Kingdom | 43 | F | Mitis | + | D38 | |
| CD93/154 | United Kingdom (contact of a Somali) | 14 | F | Mitis | + | D39 | |
| CD98/135 | United Kingdom (imported case from Tanzania) | 19 | Cutaneous | Mitis | + | D40 | |
| CD92/48 | Bristol, United Kingdom | Mitis | + | D41 | |||
| CD94/214 | United Kingdom (imported case from India) | 32 | F | Pharyngitis | Mitis | + | D42 |
| CD94/8 | United Kingdom | 64 | Mitis | − | D43 | ||
| CD94/9 | United Kingdom | 31 | F | Mitis | − | D44 | |
| CD94/16 | Italy (imported case from Peru) | Gravis | + | D45 | |||
| CD93/242 | France | Mitis | − | D46 | |||
| CD93/28 | Perth, Australia | 45 | Mitis | − | D47 | ||
| CD94/38 | Western Australia | 21 | M | Gravis | − | D48 | |
| CD94/39 | Australia | 20 | M | Gravis | − | D49 | |
| CD94/34 | Cairns, Queensland, Australia | 78 | M | Gravis | − | D50 | |
| CD94/35 | New South Wales, Australia | 47 | F | Gravis | − | D51 | |
| CD93/29 | Western Australia | 24 | Gravis | − | D52 | ||
| CD94/62 | Przemysl, Poland | 38 | F | Intermedius | + | D53 | |
| CD94/281 | Rwanda (imported case into Italy) | Wound (source) | Mitis | − | D54 | ||
| CD94/282 | Rwanda (imported case into Italy) | Pharyngeal | Mitis | − | D55 | ||
| CD95/184 | CGH, Western Australia | Gravis | − | D56 | |||
| CD94/126 | Seattle, WA | Intermedius | − | D57 | |||
| CD94/132 | Seattle, WA | Gravis | + | D58 | |||
| CD94/145 | Seattle, WA | Mitis | − | D59 | |||
| CD95/437 | Vietnam | Mitis | + | D60 | |||
| CD95/439 | Vietnam | Mitis | + | D61 | |||
| CD98/138 | Kenya | Cutaneous | Mitis | + | D62 | ||
| CD96/241 | Thailand (patient from Nan Province) | Mitis | + | D63 | |||
| CD96/244 | Thailand | 4 | Mitis | + | D64 | ||
| CD96/264 | Thailand | 11 | Mitis | − | D65 | ||
| CD96/246 | Thailand | 3 | Mitis | + | D66 | ||
| CD96/248 | Thailand | 14 | Mitis | + | D67 | ||
| CD96/250 | Thailand | Mitis | + | D68 | |||
| CD96/260 | Thailand | 8 | Mitis | − | D69 | ||
| CD96/261 | Thailand | 2 | Gravis | − | D70 | ||
| CD95/385 | Dominican Republic | Mitis | + | D71 | |||
| CD95/387 | Dominican Republic | Mitis | + | D72 | |||
| CD95/404 | Italy | Mitis | + | D73 | |||
| CD98/10 | Denmark | Mitis | + | D74 |
M, male; F, female.
Tox, toxigenicity.
A majority of the isolates analyzed were from the NIS of the former USSR. Isolates from Thailand, Sweden, and the United States were outbreak strains; and the isolates from the United Kingdom were mainly from imported cases of diphtheria. All isolates were first identified by standard microbiology techniques at the local microbiology laboratory and sent to the diphtheria reference centers within each country for confirmation of species identity and toxigenicity. The isolates were then transported to the Respiratory and Systemic Infection Laboratory at the Centre for Infections, London, United Kingdom, for additional typing.
The isolates described above were obtained through an extensive collaborative network that was formed through the European Laboratory Working Group on Diphtheria (6).
Biotyping and toxigenicity testing.
Biotyping of all 755 isolates was performed with the API Coryne system, as described previously (8). All isolates were tested for toxin production by the conventional Elek immunoprecipitation test (8). Isolates submitted after 1996 were also tested by the modified Elek test (9).
Ribotyping.
All 755 isolates were ribotyped by using BstEII, as previously described by De Zoysa et al. (4). A cDNA probe derived from the 16S and 23S rRNA of C. diphtheriae type strain NCTC 11397 was used.
PFGE, RAPD, and AFLP.
One isolate of each ribotype (a total of 74 isolates) was randomly chosen and subjected to analysis by PFGE, RAPD, and AFLP. Ten isolates of the most predominant ribotype in Russia (ribotype D1) also randomly chosen from different geographical regions were analyzed by the three methods. All methods were performed as described previously by De Zoysa et al. (2-4). RAPD and AFLP were performed with purified DNA samples.
Data capture for computer analysis of ribotype, PFGE, RAPD, and AFLP profiles.
The ribotype, PFGE, RAPD, and AFLP profiles were scanned (Scanmaker E6; Microtek Lab) and analyzed with the Bionumerics program (version 3.0, Applied Maths, Kortrijk, Belgium). Normalization within each gel was achieved with a bacteriophage λ HindIII marker (Invitrogen) for ribotyping, a bacteriophage λ concatemer (Bio-Rad) for PFGE, and the Gene ruler DNA ladder mix (MBI Fermentas) for RAPD and AFLP. Molecular size markers were placed in every third lane of the gel, and one gel marker lane (selected at random) was used as the normalization standard for the between-gel normalization of all gels in the study. The bands for each profile (ribotype, PFGE, RAPD, and AFLP) were first identified by use of the auto search facility (settings at 10 to 14% minimum profiling and 0.5% minimal area), followed by review and modification after a careful visual comparison of the image provided by the Bionumerics program. The methods were compared by generating a dendrogram by using the Dice similarity coefficient, together with the unweighted pair group method with arithmetic averages clustering method, with the position tolerance set at 1.2% and the optimization set at 0.5%.
Statistical methods.
Two indices were used to provide summaries of the diversity of the different types observed by the typing methods. These were Simpson's index of diversity (D) (λS) and the Hunter and Gaston modified index (λHG) (11, 17). The formulas for these two indices are 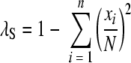 and
and 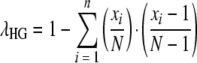 , respectively, where N is the total number of isolates and xi is the number of isolates of the ith type.
, respectively, where N is the total number of isolates and xi is the number of isolates of the ith type.
These indices can be interpreted as the probability that two isolates selected at random are of different types. The isolates used in this study are unlikely to be representative of all clinical C. diphtheriae isolates throughout the world during this time period; thus, these diversity estimates are provided to distinguish whether one typing method provides a greater discrimination between the isolates than another method rather than providing a gauge of the biological diversity in a population. Bootstrap estimates of the 95% confidence intervals (CIs) around the diversity estimates were calculated by using 1,000 resamples taken with replacement.
RESULTS
Biotyping and toxigenicity testing.
A total of 755 isolates were biotyped and tested for toxin production. The number of isolates examined from each country, together with their biotypes and toxigenicity status, are summarized in Table 1. Among the 755 isolates examined, there were 433 (57.3%) toxigenic biotype gravis isolates, 91 (12.05%) nontoxigenic biotype gravis, 176 (23.3%) toxigenic biotype mitis isolates, 42 (5.5%) nontoxigenic biotype mitis isolates, and 6 (0.79%) nontoxigenic biotype belfanti isolates. Seven strains belonged to biotype intermedius (three were toxigenic isolates and four were nontoxigenic isolates). The majority of isolates from the NIS examined were toxigenic and belonged to biotype gravis, and isolates from Australia were mainly nontoxigenic biotype gravis. The isolates from the Far East (Thailand and Vietnam) were predominantly toxigenic biotype mitis.
Ribotyping.
All 755 isolates were typed by ribotyping with BstEII. The technique showed 100% typeability and was highly reproducible. The characteristic patterns of the bands observed for individual strains were found to be independent of such variables as the batch of DNA or the batch of probe used. Each ribotype profile obtained with BstEII comprised 9 to 11 bands, and the ribotype profiles were analyzed by using the Bionumerics computer software program (Applied Maths).
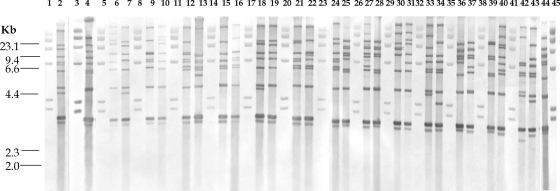
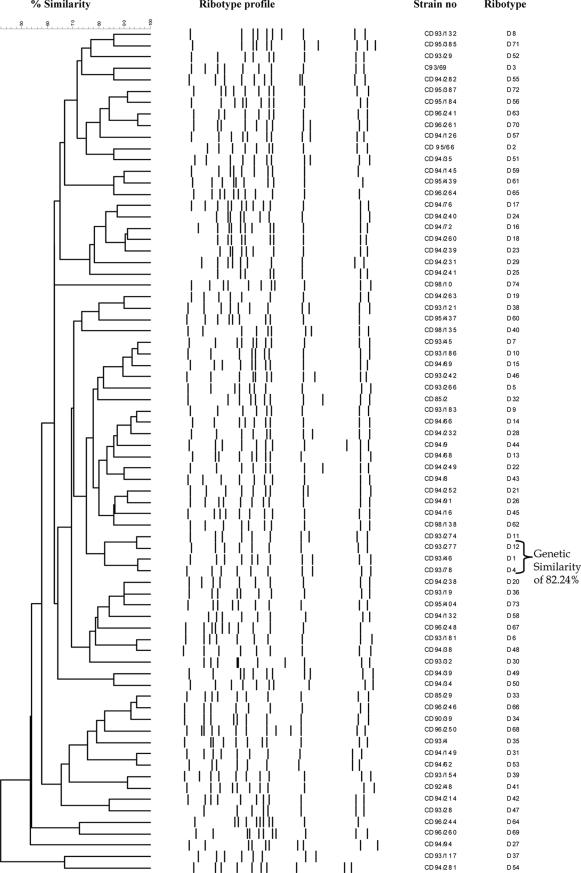
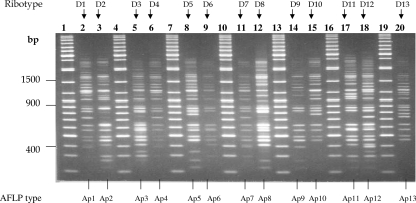
Analysis of the 755 C. diphtheriae ribotype profiles with the Bionumerics program revealed 74 distinct ribotype patterns. The patterns were designated D1 to D74 (Table 2), and Fig. 1 illustrates ribotype profiles D1 to D29. The international designations for some of the predominant ribotypes are listed in Table 1. Figure 2 represents the relationships between the 74 BstEII restriction digestion patterns determined by using the Bionumerics program to generate the dendrogram by use of the criteria described in Materials and Methods. The predominant ribotypes in Russia (ribotypes D1 and D4) appeared to be very closely related (similarity, 95.24%). The profiles of these two ribotypes were also very closely related to the profile of ribotype D12. The profiles of ribotypes D1, D4, and D12 were 82.24% similar.
FIG. 1.
BstEII ribotype profiles for the C. diphtheriae isolates tested. Lanes 1, 3, 5, 8, 11, 14, 17, 20, 23, 26, 29, 32, 35, 38, 41, and 45, bacteriophage lambda HindIII digests used as size standards (sizes are indicated on the left); lane 2, ribotype D1; lane 4, ribotype D2; lane 6, ribotype D3; lane 7, ribotype D4; lane 9, ribotype D5; lane 10, ribotype D6; lane 12, ribotype D7; lane 13, ribotype D8; lane 15, ribotype D9; lane 16, ribotype D10; lane 18, ribotype D11; lane 19, ribotype D12; lane 21, ribotype D13; lane 22, ribotype D14; lane 24, ribotype D15; lane 25, ribotype D16; lane 27, ribotype D17; lane 28, ribotype D18; lane 30, ribotype D19; lane 31, ribotype D20; lane 33, ribotype D21; lane 34, ribotype D22; lane 36, ribotype D23; lane 37, ribotype D24; lane 39, ribotype D25; lane 40, ribotype D26; lane 42, ribotype D27; lane 43, ribotype D28; lane 44, ribotype D29. When the results from different gels are combined, markers from each gel are included.
FIG. 2.
Comparison of the 74 ribotypes obtained by cluster analysis. The dendrogram was generated by using the Dice similarity coefficient, together with the unweighted pair group method with arithmetic averages clustering method, with the position tolerance set at 1.2% and the optimization set at 0.5%.
It must be noted that in a previous study (4) the ribotype profiles of biotype gravis isolates were designated with the prefix G and the profiles of biotype mitis isolates were designated with the prefix M. However, the ribotype nomenclature described in 1995 (4) was later revised and the prefixes G and M were replaced by the prefix D, which is the nomenclature used in this study.
PFGE, RAPD, and AFLP.
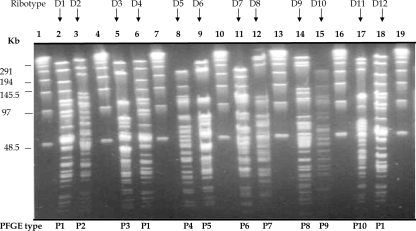
Seventy-four isolates (one strain of each ribotype) were analyzed by PFGE, RAPD, and AFLP. In addition, 10 strains of the most predominant ribotype in Russia (ribotype D1) randomly chosen from different geographical regions were also analyzed by the three methods. All profiles were analyzed by using the Bionumerics program (Applied Maths). PFGE with the restriction endonuclease SfiI produced 72 distinct PFGE profiles, which were designated P1 to P72. The profiles consisted of 15 to 25 DNA fragments ranging from 24 kb to 339.5 kb. Figure 3 illustrates the PFGE profiles of ribotypes D1 to D12.
FIG. 3.
SfiI PFGE profiles for the C. diphtheriae isolates tested. Lanes 1, 4, 7, 10, 13, 16, and 19, bacteriophage lambda concatemer used as a size standard (sizes are indicated on the left); lanes, 2, 3, 5, 6, 8, 9, 11, 12, 14, 15, 17, and 18, PFGE profiles P1 to P10. The ribotype designation for each isolate is indicated above each lane, and the PFGE type is indicated below each lane. When the results from different gels are combined, markers from each gel are included.
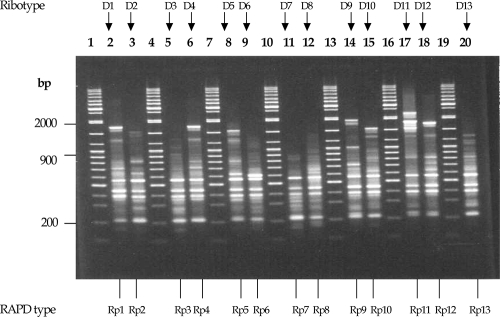
Analysis of the 74 isolates by RAPD revealed 74 distinct profiles, which were designated Rp1 to Rp74. The profiles consisted of 13 to 27 fragments that ranged from 200 bp to 2,072 bp in size. Profiles Rp1 to Rp13 are illustrated in Fig. 4. AFLP revealed 72 profiles among the 74 isolates, and the profiles were designated AP1 to AP72. The AFLP profiles were comprised of 24 to 33 fragments that ranged from 200 bp to 300 bp in size. All three methods showed 100% typeability. PFGE and AFLP did not distinguish between strains of ribotypes D1, D4, and D12 (Fig. 3 and 5), and therefore, the PFGE and AFLP profiles of these three ribotypes clustered together at a similarity level of 100%. The 10 isolates of ribotype D1 from different geographical origins were not distinguished further by these methods. The RAPD profiles of ribotypes D1, D4, and D12 showed a similarity of 91.77%.
FIG. 4.
RAPD profiles for the C. diphtheriae isolates tested. Lanes 1, 4, 7, 10, 13, 16, and 19, 100-bp molecular weight standard (sizes are indicated on the left); lanes, 2, 3, 5, 6, 8, 9, 11, 12, 14, 15, 17, 18, and 20, RAPD profiles Rp1 to Rp13. The RAPD type and ribotype designations for each isolate are indicated below and above each lane, respectively.
FIG. 5.
AFLP profiles for the C. diphtheriae isolates tested. Lanes 1, 4, 7, 10, 13, 16, and 19, 100-bp molecular weight standard (sizes are indicated on the left); the remaining lanes show AFLP profiles Ap1 to Ap13. The AFLP types are given below each lane, and the ribotype designations are given at the top of each lane.
Statistical analysis.
The diversity indices and their 95% confidence intervals calculated for the ribotyping results are shown in Table 3. As expected, the diversity indices were much lower when they were calculated by using the data for all 755 isolates. This is due to the abundance of certain ribotypes (i.e., ribotypes D1 and D4) that have occurred in spatiotemporal clusters. Table 4 shows the diversity of ribotypes and their 95% CIs within disease types (diphtheria, tonsillitis, and carriers). The results show that the ribotypes were less diverse among isolates from cases of diphtheria and were more diverse among isolates from cases of tonsillitis. The relative frequency of particular ribotypes between different countries was also explored by using the diversity indices. The ribotypes observed in very few countries have low diversity indices, while those observed in many countries are likely to have larger diversities. These results are shown in Table 5.
TABLE 3.
Diversity indices and their 95% CIs for the ribotyping methoda
| Sample | λHG | Bootstrap 95% CI for λHG | λS | Bootstrap 95% CI for λS |
|---|---|---|---|---|
| All data | 0.766 | 0.719-0.801 | 0.762 | 0.716-0.797 |
| Unrelated samples | 0.948 | 0.929-0.957 | 0.944 | 0.924-0.952 |
The indices and 95% CIs were calculated by using the results for all 755 strains and the 198 unrelated strains.
TABLE 4.
Diversity of ribotypes within disease types and their 95% CIs for all 755 strains and the 198 unrelated strains
| Disease or status | Data | λHG | Bootstrap 95% CI for λHG | λS | Bootstrap 95% CI for λS |
|---|---|---|---|---|---|
| Diphtheria | All isolates | 0.676 | 0.625-0.708 | 0.671 | 0.620-0.703 |
| 198 unrelated isolates | 0.791 | 0.664-0.835 | 0.763 | 0.641-0.806 | |
| Tonsillitis | All isolates | 0.816 | 0.738-0.853 | 0.800 | 0.723-0.835 |
| 198 unrelated isolates | 0.958 | 0.817-0.950 | 0.898 | 0.766-0.891 | |
| Carrier | All isolates | 0.790 | 0.756-0.812 | 0.784 | 0.751-0.806 |
| 198 unrelated isolates | 0.818 | 0.686-0.859 | 0.794 | 0.666-0.834 |
TABLE 5.
Between-country diversity indices and their 95% CIs for certain predominant and rare ribotypes
| Ribotype | λHG | Bootstrap 95% CI for λHG | λS | Bootstrap 95% CI for λS |
|---|---|---|---|---|
| D1 | 0.766 | 0.719-0.801 | 0.762 | 0.716-0.797 |
| D4 | 0.812 | 0.782-0.830 | 0.808 | 0.779-0.827 |
| D6 | 0.600 | 0.000-0.600 | 0.480 | 0.000-0.480 |
| D9 | 0.325 | 0.091-0.515 | 0.310 | 0.086-0.492 |
| D10 | 0.676 | 0.535-0.762 | 0.662 | 0.524-0.746 |
| D11 | 0.762 | 0.610-0.810 | 0.726 | 0.580-0.771 |
| D13 | 0.500 | 0.000-0.667 | 0.375 | 0.000-0.500 |
| D15 | 0.800 | 0.333-0.867 | 0.667 | 0.278-0.722 |
| D19 | 0.667 | 0.289-0.762 | 0.571 | 0.245-0.653 |
| D26 | 0.429 | 0.000-0.571 | 0.375 | 0.000-0.500 |
| D34 | 0.143 | 0.000-0.363 | 0.133 | 0.000-0.337 |
| D46 | 0.667 | 0.000-0.667 | 0.444 | 0.000-0.444 |
| D50 | 0.500 | 0.000-0.667 | 0.375 | 0.000-0.500 |
| D56 | 0.500 | 0.000-0.667 | 0.444 | 0.000-0.444 |
| D57 | 0.333 | 0.000-0.600 | 0.278 | 0.000-0.500 |
Ribotype D4 appeared to be far more diverse (λHG = 0.812) than ribotype D34 (λHG = 0.143). Ribotype D4 was seen among isolates from 11 countries, and ribotype D34 was seen only among isolates from Thailand and the United Kingdom.
Calculation of the diversity indices and their 95% CIs for PFGE, AFLP, and RAPD showed that the results of RAPD were identical to those of ribotyping. The diversity indices for PFGE and AFLP were much lower than those of ribotyping and very similar to each other, indicating that PFGE and AFLP are less discriminatory than ribotyping.
Transcontinental spread of C. diphtheriae.
The results obtained by the four typing methods (ribotyping, PFGE, RAPD, and AFLP) were used to assess the transcontinental spread and epidemiology of C. diphtheriae. Among the isolates from the Russian epidemic analyzed, 40% were of ribotype D1, which was the predominant ribotype seen in Russia. Other predominant ribotypes in Russia were D4 and D7 (26% were D4 and 25.2% were D7).
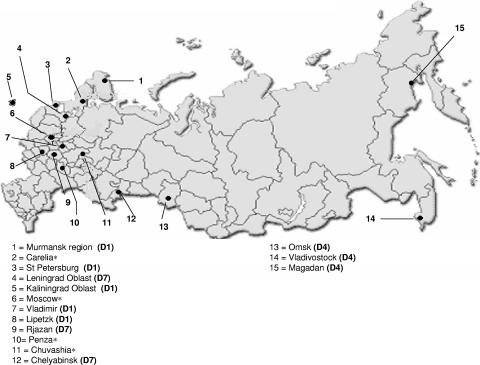
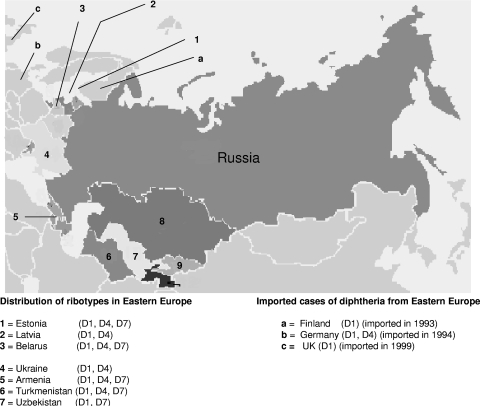
The analysis of 218 C. diphtheriae strains from 15 different regions in Russia showed that certain ribotypes prevailed in certain regions. Figure 6 shows the prevalence of the ribotypes in each region. A total of 568 strains isolated from Eastern Europe between 1993 and 2000 were analyzed. Among these isolates, 195 isolates were of ribotype D1 (189 toxigenic biotype gravis isolates and 6 toxigenic biotype mitis isolates [all 6 biotype mitis isolates were from Latvia]), 215 isolates were of ribotype D4 (196 toxigenic biotype gravis biotype isolates, 17 nontoxigenic biotype gravis isolates, and 2 toxigenic biotype mitis isolates), and 75 isolates were of ribotype D7 (all were toxigenic, and 71 isolates were biotype mitis and 4 were biotype gravis). Figure 7 shows the distribution of the predominant ribotypes in Eastern Europe and cases imported to other neighboring countries. From the results obtained (Fig. 6 and Fig. 7), it is clear that ribotypes D1, D4, and D7 were the epidemic ribotypes in Russia and they had disseminated to all states of the former USSR and to a few neighboring countries. Ribotypes D4 and D7 were documented prior to the Russian epidemic: ribotype D4 was seen for a Swedish isolate, a nontoxigenic biotype mitis isolate that was isolated in 1984, and ribotype D7 was seen for a United Kingdom isolate, a toxigenic biotype mitis isolate that was isolated in 1989.
FIG. 6.
Regions in Russia and the prevalence of ribotypes in each region. *, the number of isolates analyzed from the region was not enough for use for determination of ribotype prevalence.
FIG. 7.
Distribution of predominant ribotypes in Eastern Europe.
Some ribotypes (ribotypes D5, D6, D10, D11, D17, D26, D30, and D40) were seen only among isolates from Eastern and Western Europe. Ribotype D10 was found in Estonia, Finland, Russia, Uzbekistan, Belarus, Latvia, and Turkmenistan. Ribotype D10 appeared to be the fourth most common ribotype in Eastern Europe, and it was seen only among the epidemic isolates from Eastern Europe.
A number of ribotypes were rare and were identified only in particular countries. Quite a few uncommon ribotypes were seen among isolates from the United Kingdom and Russia. Fourteen ribotypes were documented in the United Kingdom; and the majority of these isolates were from patients who had returned from Asia, Australia, Africa, the Middle East, and the Far East. These unusual ribotypes could be endemic strains of C. diphtheriae circulating in individual countries. Ribotypes previously seen among strains responsible for large outbreaks (i.e., Seattle, WA [1971 to 1982], Sweden [1984], Thailand [1994], and Vietnam [1995]) were different from those seen in the Eastern Europe epidemic.
DISCUSSION
Comparison of four molecular typing methods.
Optimal typeability, a high degree of reproducibility, adequate stability, and resolving power characterize a “gold standard” typing technique. We found ribotyping to be highly discriminatory and reproducible. The statistical analysis data calculated for the different typing methods indicate that ribotyping is the most suitable technique and the method of choice for the typing of C. diphtheriae.
In 1997, the method used for the ribotyping if C. diphtheriae was standardized by Regnault et al. (16). In the present study, the majority of the isolates analyzed were collected before 1997, and therefore, the method described by Regnault et al. (16) was not used. The only difference between the two methods is the format of the probe used. Both probes are based on 23S and 16S rRNA, and the probe described by Regnault et al. (16) is commercially available. In 2004, the C. diphtheriae ribotype nomenclature was revised again (ribotypes were named after the geographical origin of the strain) and a ribotype database with international designations was constructed by Grimont et al. (10). The international designations for the predominant ribotypes are presented in Table 1.
The two PCR-based techniques, RAPD and AFLP, proved to be rapid and easier to perform than ribotyping and PFGE. The diversity indices indicate that AFLP and PFGE are less discriminatory than ribotyping and RAPD. However, in a separate study carried out in 2000 (3), we reported that AFLP subdivided certain ribotypes further, but statistical analysis was not performed on the results. RAPD and AFLP are rapid methods which can be used as screening techniques, prior to ribotyping, during outbreak investigations. Use of these methods avoids the need for the undertaking of complex ribotyping analyses with strains which are unrelated.
In conclusion, all four molecular techniques that were used in this study showed 100% typeability. PFGE and AFLP appeared to be less discriminatory than ribotyping, as neither technique could distinguish between strains of ribotypes D1, D4, and D12. These results suggest that isolates of ribotype D1, D4, and D12 may have arisen from a single clonal group. RAPD also supported a clonal relationship between these strains, as they were 91.77% similar by RAPD.
Transcontinental epidemiology.
The results show that the Eastern European epidemic was caused by strains of ribotypes D1, D4, and D7. The cases of diphtheria imported into Finland, Germany, and the United Kingdom may have resulted from the marked increase in travel between Russia and its neighboring countries (5). Our results show that strains of ribotypes D4 and D7 were documented in Russia before the Eastern European epidemic began. A study carried out by Popovic et al. in 1996 (13) with preepidemic and epidemic strains of C. diphtheriae from Russia found that during the preepidemic period, a diverse group of ribotypes was circulating in Russia and ribotype D7 (previously referred to as ribotype M1) was the predominant ribotype among the preepidemic isolates. Popovic et al. (13) also reported that strains of ribotypes D1 and D4 (previously referred to as ribotypes G1 and G4, respectively) were rarely seen in Russia before the epidemic, but since the epidemic, ribotypes D1 and D4 accounted for more than 80% of all ribotypes identified (with ribotype D4 being the predominant one). Those workers found that ribotype D4 was the predominant epidemic ribotype, probably because they focused particularly on one region in Russia (the Vladimir region). In our study, analysis of 218 isolates from 15 regions in Russia revealed that ribotype D1 predominated in some regions and that ribotype D4 or D7 predominated in other regions (Fig. 6). Several other genotypes are also circulating within Russia, and these unique genotypes could be endemic in Russia. In 2002, Skogen et al. (18) analyzed 47 preepidemic C. diphtheriae strains (isolated from 1957 to 1987) from Russia. The authors reported that the earliest reported toxigenic strain of ribotype D4 was identified in Smolensk, Russia, in 1985 and that strains of this ribotype were simultaneously present in several different geographical regions in Russia from 1985 through 1987. Therefore, their findings suggest that the Eastern European epidemic clone was probably an integral part of the endemic reservoir that existed in the former Soviet Union at least 5 years before the epidemic began.
The analysis of sporadic strains from France, Italy, Romania, and Poland and outbreak strains from Thailand, Vietnam, Sweden, and the United States enabled a comparison of isolates from the Eastern European epidemic with those circulating in other parts of the world. It seems that different patterns are seen in different parts of the world and that distinct clones had caused each outbreak. The majority of the cases imported into the United Kingdom from Africa, Asia, and the Middle East were caused by toxigenic biotype mitis isolates, and the ribotypes of these isolates were distinct from those circulating in Europe and the United States and appeared to be characteristic of the ribotypes found in those countries. These strains probably represent endemic strains circulating within those countries.
Endemic strains of toxigenic C. diphtheriae still circulate within certain communities. In 2001, Marston et al. (12) reported on the circulation of toxigenic endemic strains of C. diphtheriae within two communities in the United States and Canada for at least 25 years. The reason why certain strains of C. diphtheriae cause outbreaks and others remain endemic is a subject for speculation. Outbreak strains could be better colonizers and may produce higher levels of toxin than endemic strains. Adherence factors bring the microbe and host cell into close contact to ensure efficient colonization and to deliver the toxin to the specific host cellular target. In 1988, Rappuoli et al. (15) reported that the introduction of a single epidemic strain of C. diphtheriae, which then spread from person to person, resulted in the 1984 to 1985 diphtheria outbreak in Sweden. Those workers also reported that the epidemic strain had an unidentified selective advantage. The genome sequence data for C. diphtheriae strain NCTC 13129 (a representative isolate of ribotype D1 of the Russian epidemic clone) revealed that it possesses a number of putative virulence factors, such as adhesins and fimbria-related proteins (1). Whether toxigenic endemic strains and nontoxigenic C. diphtheriae strains possess these virulence factors or not is still unknown.
The use of molecular methods for studying the epidemiology of C. diphtheriae has identified several new findings that could not have been obtained by conventional epidemiological approaches. The application of molecular typing methods and continuous monitoring of the Eastern European epidemic clone has had a significant public health impact. It was possible to distinguish rapidly between epidemic, endemic, and imported cases, which allowed the implementation of timely and adequate preventive measures when they were needed, and no secondary spread was reported following any of the importations (14). Diphtheria appears to be endemic in some of the countries neighboring Russia, and travel between Russia and its neighboring countries markedly increased in the 1990s, which may have introduced the Russian epidemic clone. Diphtheria may also have been introduced into Russia in the late 1980s with the demobilization of Soviet military forces and their return from the countries neighboring Russia. Few historical and preepidemic isolates were available for evaluation in this study; therefore, knowledge of the extent to which the outbreak strain was introduced or whether the transfer of toxin genes among indigenous strains was more important is limited.
This study has underlined the need for a deeper understanding of the biological properties of C. diphtheriae and their role in diversity and the appearance of epidemic strains. The C. diphtheriae ribotype database, which is curated at the Institut Pasteur, Paris, France, should facilitate the surveillance of clones causing infection and colonizing clones which could acquire tox genes by horizontal gene transfer and cause sporadic cases and outbreaks.
Acknowledgments
We do not have commercial or other associations that might pose conflicts of interest.
Footnotes
Published ahead of print on 10 September 2008.
REFERENCES
- 1.Cerdeño-Tárraga, A. M., A. Efstratiou, L. G. Dover, et al. 2003. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC 13129. Nucleic Acids Res. 316516-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Zoysa, A., and A. Efstratiou. 1999. PCR typing of Corynebacterium diphtheriae by random amplification of polymorphic DNA. J. Med. Microbiol. 48335-340. [DOI] [PubMed] [Google Scholar]
- 3.De Zoysa, A., and A. Efstratiou. 2000. Use of amplified fragment length polymorphisms for typing Corynebacterium diphtheriae. J. Clin. Microbiol. 383843-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Zoysa, A., A. Efstratiou, R. C. George, et al. 1995. Molecular epidemiology of Corynebacterium diphtheriae from northwestern Russia and surrounding countries studied by using ribotyping and pulsed-field gel electrophoresis. J. Clin. Microbiol. 331080-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Zoysa, A., A. Efstratiou, R. C. George, et al. 1993. Diphtheria and travel. Lancet 342446. [PubMed] [Google Scholar]
- 6.Efstratiou, A. 1995. Corynebacterium diphtheriae: molecular epidemiology and characterisation studies on epidemic and sporadic isolates. Microecol. Ther. 2563-71. [Google Scholar]
- 7.Efstratiou, A., and N. T. Begg. 1993. The changing epidemiology of diphtheria. J. Public Health Med. 15203-204. [Google Scholar]
- 8.Efstratiou, A., and P. A. C. Maple. 1994. Manual for the laboratory diagnosis of diphtheria. Report ICP/EPI038 (C). The Expanded Programme on Immunization in the European Region of WHO, Copenhagen, Denmark.
- 9.Engler, K. H., T. Glushkevich, I. K. Mazurova, et al. 1997. A modified Elek test for detection of toxigenic corynebacteria in the diagnostic laboratory. J. Clin. Microbiol. 35495-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimont, P. A., F. Grimont, A. Efstratiou, et al. 2004. International nomenclature for Corynebacterium diphtheriae ribotypes. Res. Microbiol. 155162-166. [DOI] [PubMed] [Google Scholar]
- 11.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 262465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marston, C. K., F. Jamieson, F. Cahoon, et al. 2001. Persistence of a distinct Corynebacterium diphtheriae clonal group within two communities in the United States and Canada where diphtheria is endemic. J. Clin. Microbiol. 391586-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popovic, T., S. Y. Kombarova, M. W. Reeves, et al. 1996. Molecular epidemiology of diphtheria in Russia, 1985-1994. J. Infect. Dis. 1741064-1072. [DOI] [PubMed] [Google Scholar]
- 14.Popovic, T., I. K. Mazurova, A. Efstratiou, et al. 2000. Molecular epidemiology of diphtheria. J. Infect. Dis. 181S168-177. [DOI] [PubMed] [Google Scholar]
- 15.Rappuoli, R., M. Perugini, and E. Falsen. 1988. Molecular epidemiology of the 1984-1986 outbreak of diphtheria in Sweden. N. Engl. J. Med. 31812-14. [DOI] [PubMed] [Google Scholar]
- 16.Regnault, B., F. Grimont, and P. A. D. Grimont. 1997. Universal ribotyping method using a chemically labelled oligonucleotide probe mixture. Res. Microbiol. 148649-659. [DOI] [PubMed] [Google Scholar]
- 17.Simpson, E. H. 1949. Measurement of diversity. Nature 163688. [Google Scholar]
- 18.Skogen, V., V. V. Cherkasova, N. Maksimova, et al. 2002. Molecular characterisation of Corynebacterium diphtheriae isolates, Russia, 1957-1987. Emerg Infect. Dis. 8516-518. [DOI] [PMC free article] [PubMed] [Google Scholar]