Abstract
Peripheral blood mononuclear cells are reported to be one of the extrahepatic replication sites contributing to the persistence of hepatitis C virus (HCV) infection. Whole-blood and plasma samples from 61 individuals were compared as sources for the detection of HCV RNA. Forty-four of the individuals were receiving antiviral therapy, while 17 were treatment naïve. The quantitation of HCV RNA was done by a sensitive in-house real-time reverse transcription-PCR. When the viral loads in the two types of samples were compared, a correlation coefficient of 0.858 (P < 0.001) was found, indicating that plasma and whole blood are equally acceptable sources for testing for HCV RNA.
Approximately 18 million people in India are estimated to be infected with hepatitis C virus (HCV) (13). HCV infection is chronic in 75% to 85% of infected individuals (3). Chronic HCV infection can lead to hepatocellular carcinoma after many years. Even though hepatocytes are the major replication site for HCV, a broad range of extrahepatic complications and diseases are associated with chronic HCV infection. These include mixed cryoglobulinemia, non-Hodgkin's lymphoma, cutaneous vasculitis, membranoproliferative glomerulonephritis, neuropathy, lymphoproliferative disorders, porphyria cutanea tarda, lichen planus, and Sjogren's syndrome (1, 7). HCV is reported to replicate in extrahepatic sites like interstitial cells of the kidney, acinar cells of the pancreas, peripheral blood mononuclear cells, and mononuclear cells of lymph nodes (5, 14). Currently, the diagnosis of HCV infection is achieved by the detection of HCV RNA in plasma or serum. Testing for HCV RNA in plasma or serum might give a false-negative result if HCV RNA is present inside peripheral blood mononuclear cells (PBMCs) or as precipitates of immune complexes and cryoglobulin aggregates. Hence, the recovery of RNA from whole blood is likely to help with the diagnosis of HCV infection in individuals with low HCV loads, since both intracellular RNA and plasma RNA are detected. This can also have implications in the monitoring of individuals receiving antiviral therapy (12). Screening for HCV by the use of whole-blood samples will be useful, since all blood components are screened and the time required to separate plasma and serum is saved (leaving the remaining sample usable for serology). This study was done to investigate whether whole blood is more sensitive than plasma for the detection of HCV RNA and to assess the impact of testing of whole blood on the viral load result.
Blood samples were collected from 61 patients who came to the Department of Clinical Virology, Christian Medical College, as referrals from the Departments of Hematology, Gastroenterology, and Nephrology. All patients were recruited after they provided verbal consent, in addition to a general consent that is obtained in our hospital for all investigations as part of our routine patient management. These 61 patients consisted of 44 men and 17 women with a mean age of 42 years (age range, 16 to 66 years). Of these 61 patients, 17 (27.9%) were not receiving treatment and 44 (72.1%) had been receiving pegylated interferon and ribavirin, interferon and ribavirin, pegylated interferon alone, or interferon alone for durations that ranged from 12 to 24 weeks. Whole-blood samples were collected in EDTA tubes, and the contents were mixed by gentle shaking and stored in two aliquots (400 μl each) at −60°C. Plasma was separated from the remaining sample after centrifugation at 1,500 rpm for 10 min and was stored in multiple aliquots at −60°C for testing. Five whole-blood samples were tested in duplicate to assess the reproducibility of the results.
HCV RNA was extracted from 200 μl of whole blood or plasma with a High Pure viral nucleic acid kit (Roche Diagnostics GmbH, Mannheim, Germany), according to the manufacturer's protocol. This extraction protocol was easy to perform and is reproducible. This method can be used for the extraction of viral RNA and DNA from whole-blood and plasma samples. Five microliters of 5 × 10−6 μg brome mosaic virus (BMV) RNA was added to 1 ml of binding buffer, which served as the internal control (IC) to rule out false-negative results. Elution was done with 50 μl of prewarmed (70°C) elution buffer. The eluate was always processed immediately.
The primers used for the in-house real-time reverse transcription (RT)-PCR were HCV TAQ1 (GTC TAG CCA TGG CGT TAG TA), HCV TAQ 2 (GTA CTC ACC GGT TCC GC), BMV TAQ 1 (GTT CAC CGA TAG ACC GCT G), and BMV TAQ 2 (AAG AGC CCG GAA TGT CAA GA); and the probes were HCV TAQPR (6-carboxyfluorescein-CCC TCC CGG GAG AGC CAT AGT G-6-carboxytetramethylrhodamine [TAMRA]) and BMV TAQPR (JOE [6-carboxy-4′,5′-dichloro-2′,7′-dimethoxy-fluorescein]-CCT CAA GCT GAA ATG GCA CGG ATG-TAMRA). The HCV primer and probe sequences were directed against the 5′ noncoding region of the HCV genome. The primer and probe sequences used for this in-house HCV real-time RT-PCR were kindly provided by Richard Tedder (University College London, London, United Kingdom). Amplification was performed with 10 μl of extract in a 25-μl volume containing 12.5 μl of 2× QuantiTect probe RT-PCR buffer containing Taq polymerase; 0.25 μl of reverse transcriptase enzyme mix (Qiagen GmbH, Hilden, Germany); 20 pmol of HCV TAQ 1, HCV TAQ 2, BMV TAQ 1, and BMV TAQ 2; and 10 pmol of HCV TAQPR and BMV TAQPR (Metabion GmbH, Martinsried, Germany). The reaction mixture was amplified by using the following thermal cycling conditions: 50°C for 30 min for the RT step, followed by 95°C for 15 min and amplification for 45 cycles at 95°C for 15 s and 60°C for 60 s. Amplification and detection of HCV RNA and the IC were done with a Rotor-Gene 3000 instrument (Corbett Research, Australia). A standard curve was generated by using the built-in software (Rotor Gene, version 6.0) in the Rotor-Gene 3000 instrument. The HCV RNA viral load in the sample was estimated from the standard curve generated by using five standard concentrations ranging from 10 to 1 × 105 international units (IU)/ml. The in-house standard for HCV real-time RT-PCR was an HCV RNA-positive blood bag with a viral load of 106 IU/ml (obtained courtesy of the Scudder Memorial Blood Bank, Christian Medical College, Vellore, India). This in-house standard was tested in triplicate in parallel with WHO Second HCV International Standard 96/798 to assess the exact viral load. The upper and lower limits of detection of this assay were 106 IU/ml and 39 IU/ml, respectively. The IC served as a control to rule out the presence of PCR inhibitors during the extraction and the amplification steps.
Representative samples (n = 34) from the 61 individuals were genotyped by using the NS5b region sequencing method (6). The distribution of HCV genotypes was as follows: genotype 1, n = 4; genotype 3, n = 25; genotype 4, n = 2; and genotype 6, n = 3. Five of the 34 samples genotyped were from samples previously obtained from these study patients.
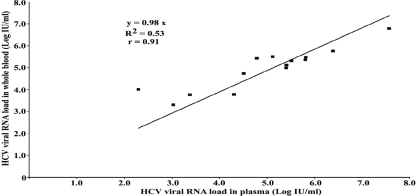
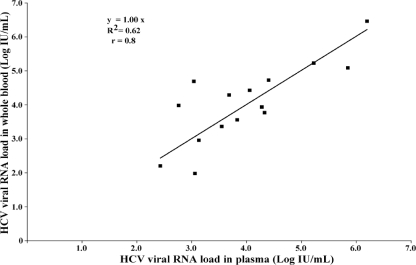
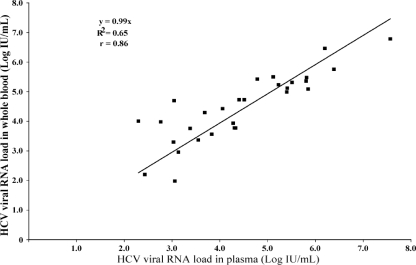
Of the 61 individuals whose whole blood and plasma were tested, 29 (47.5%) were RNA positive and 32 (52.5%) were RNA negative when plasma samples were tested. None of the samples which were negative for HCV RNA showed inhibition of the IC. The coefficient of variation obtained by testing duplicates of five whole-blood samples ranged from 0.1 to 0.5 log unit, thereby showing the good reproducibility of testing of whole blood. Of the 44 patients who were receiving treatment, the plasma samples of 30 (68.2%) were negative and those of 14 (31.8%) were positive for HCV RNA by the in-house HCV real-time RT-PCR assay. The scatter plot in Fig. 1 compares the viral loads between blood and plasma samples from in patients who were receiving therapy. Of the 17 patients who were not receiving therapy, 15 (88.2%) were positive and 2 (11.8%) were negative for HCV RNA. The scatter plot in Fig. 2 depicts the correlation of the viral load between blood and plasma samples from patients who were treatment naïve. The HCV RNA results for whole blood compared with those for plasma for all 29 individuals (15 treatment-naïve individuals and 14 individuals receiving therapy) are shown in Fig. 3.
FIG. 1.
Scatter plot comparing the HCV viral RNA loads in whole-blood and plasma samples from patients receiving antiviral therapy (n = 14).
FIG. 2.
Scatter plot comparing the HCV viral RNA loads in whole-blood and plasma samples from treatment-naïve patients (n = 15).
FIG. 3.
Scatter plot comparing the HCV viral RNA loads in whole-blood and plasma samples from all 29 individuals (15 treatment-naïve patients and 14 patients receiving treatment).
Among 30 whole-blood samples that were positive, 29 plasma samples were positive and 1 plasma sample was negative. For all 31 whole-blood samples that were negative, the matching plasma sample was also negative. All samples that were positive when plasma was used were positive for HCV RNA when whole blood was used. Of the 32 negative plasma samples, 1 sample had a low virus titer (22 IU/ml) in the whole-blood assay. This patient had received interferon for 10 weeks and was lost to subsequent follow-up. The indices of accuracy for whole-blood HCV RNA detection were as follows: the sensitivity was 100.0% (95% confidence interval [CI], 85.4, 100.0), the specificity was 96.9% (95% CI, 82.0, 99.8), the positive predictive value was 96.7% (95% CI, 80.9, 99.8), and the negative predictive value was 100.0% (95% CI, 86.3, 100.0). The kappa coefficient was 0.97 (P < 0.0001). The viral loads obtained by testing plasma and whole blood did not show any significant differences (P = 0.817, t test with paired samples). Comparison of the viral loads showed a significant correlation (Pearson's correlation coefficient, r = 0.858; P < 0.001), indicating that plasma and whole blood are equally acceptable for use for testing for HCV RNA.
Some reports have suggested that HCV replicates in extrahepatic sites, like PBMCs, in HCV-infected individuals. Further reports suggest that the HCV RNA load detected in plasma might not represent the exact viral load of the patient. The low viral load of HCV RNA in plasma samples could be attributed to viruses that adhere to blood cells or HCV RNA found within cryoprecipitates, which might sediment during the centrifugation process. Two earlier studies that used whole-blood and plasma samples have shown that the viral loads in whole blood are higher than those in plasma and that the excess viral load in the whole-blood samples was attributed to the combined viral load of plasma and RNA from the cellular pellet and cell washes following separation of the plasma. In those studies, extraction was done with the cationic surfactant tetradecyltrimethylammonium oxalate (Catrimox-14) and guanidinium isothiocyanate (10, 11). Contrary to these findings reported earlier, our study did not find a significant increase in the HCV viral load in whole blood compared to that in plasma. This finding is in accordance with that of another study in which no significant increase in the viral load in whole-blood samples compared with that in plasma samples was observed. In the earlier study, extraction was done by use of the tetradecyltrimethylammonium oxalate-Trizol method (4).
Watkins-Riedel et al. (12) compared the HCV RNA titers in whole-blood, serum, and plasma specimens obtained from 56 patients who did not respond to initial interferon alpha 2b monotherapy in order to determine whether the specimen type can predict the rate of virologic response to high-dose treatment with interferon and ribavirin (12). Of the 56 patients in that study, serum and plasma obtained from 18 patients tested negative for HCV RNA at the end of treatment, indicating a complete virological response. In contrast, analysis of whole-blood specimens obtained at the same time revealed the presence of viral RNA in 12 of these 18 patients. The testing of whole blood for the detection of HCV RNA was highly predictive of viral relapse (positive predictive value, 100%) and may thus be a useful tool for the monitoring of patient responses to interferon and ribavirin therapy (12). Testing of only serum or plasma specimens could underestimate the true circulating HCV load and could lead to an overestimation of the antiviral response rates (9, 12). Contrary to the findings reported earlier, among the 44 patients receiving antiviral therapy in our study, the plasma sample from only 1 additional patient whose whole blood was positive for HCV RNA (22 IU/ml) was negative for RNA. This patient was lost to follow-up, and hence, the virological response to therapy could not be ascertained.
Amplification of HCV RNA from whole-blood samples is done by different methods. Most of the studies have used RT-PCR for the amplification of HCV RNA from whole blood. Watkins-Riedel et al. (12) compared the LightCycler assay, the Cobas Amplicor assay, and two different in-house RT-PCRs and found that one whole-blood sample gave a positive result by the LightCycler real-time assay but was negative by the other assays evaluated. In the present study, despite the use of a sensitive in-house TaqMan real-time assay, there was no significant difference in the rates of detection of HCV RNA between whole blood and plasma. This may be attributed to the efficacy of the High Pure viral nucleic acid kit for the extraction of RNA from both types of clinical samples.
The detection of HCV RNA in whole blood was previously considered to be difficult due to the RNases present in blood samples and the inhibitory effect of hemoglobin. However, the detection of HCV RNA in whole blood will remove the need to separate plasma. Earlier studies have used tetradecyltrimethylammonium oxalate, a surfactant used for the extraction of HCV RNA from whole-blood samples (2, 9). Newer nucleic acid extraction protocols which can remove these factors from whole blood are now available. In this study, we have used the High Pure viral nucleic acid protocol (Roche Diagnostics GmbH) to extract HCV RNA from whole blood. Our study shows a good correlation between the whole-blood HCV viral load and the plasma HCV viral load in individuals chronically infected with HCV (r = 0.858), as reported earlier (8).
In summary, the rates of detection of HCV RNA in whole blood and plasma compared favorably under the conditions used in this study. The direct detection of viral nucleic acid in whole blood minimizes the need for sample processing and has the potential for widespread use in blood banks and diagnostic laboratories.
Acknowledgments
This study was done with intramural funds from departmental resources.
We gratefully thank Gopalan Sridharan, Department of Clinical Virology, for his generous advice during standardization of the assay and his valuable inputs during preparation of the manuscript.
Footnotes
Published ahead of print on 13 August 2008.
REFERENCES
- 1.Agnello, V., and F. G. De Rosa. 2004. Extrahepatic disease manifestations of HCV infection: some current issues. J. Hepatol. 40341-352. [DOI] [PubMed] [Google Scholar]
- 2.Ali, S. A., B. Kubik, H. Gulle, M. M. Eibl, and A. Steinkasserer. 1998. Rapid isolation of HCV RNA from Catrimox-lysed whole blood using QIAamp spin columns. BioTechniques 25975-978. [PubMed] [Google Scholar]
- 3.Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341556-562. [DOI] [PubMed] [Google Scholar]
- 4.Cook, L., A. M. Ross, G. B. Knight, and V. Agnello. 2000. Use of whole blood specimens for routine clinical quantitation of hepatitis C virus RNA does not increase assay sensitivity. J. Clin. Microbiol. 384326-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong, G. Z., L. Y. Lai, Y. F. Jiang, Y. He, and X. S. Su. 2003. HCV replication in PBMC and its influence on interferon therapy. World J. Gastroenterol. 9291-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris, K. A., and C. G. Teo. 2001. Diversity of hepatitis C virus quasispecies evaluated by denaturing gradient gel electrophoresis. Clin. Diagn. Lab. Immunol. 862-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson, R. J., D. R. Gretch, H. Yamabe, J. Hart, C. E. Bacchi, P. Hartwell, W. G. Couser, L. Corey, M. H. Wener, C. E. Alpers, and R. Willson. 1993. Membranoproliferative glomerulonephritis associated with hepatitis C virus infection. N. Engl. J. Med. 328465-470. [DOI] [PubMed] [Google Scholar]
- 8.Patel, K., J. Albrecht, E. Owens, A. Dev, S. Heaton, S. Pianko, P. J. Pockros, A. Conrad, L. M. Blatt, and J. G. McHutchison. 2005. The clinical utility of using Catrimox-14-treated whole blood in detecting hepatitis C virus RNA. Antivir. Ther. 10535-541. [PubMed] [Google Scholar]
- 9.Schmidt, W. N., P. Wu, D. Brashear, D. Klinzman, M. J. Phillips, D. R. LaBrecque, and J. T. Stapleton. 1998. Effect of interferon therapy on hepatitis C virus RNA in whole blood, plasma, and peripheral blood mononuclear cells. Hepatology 281110-1116. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt, W. N., P. Wu, J. Cederna, F. A. Mitros, D. R. LaBrecque, and J. T. Stapleton. 1997. Surreptitious hepatitis C virus (HCV) infection detected in the majority of patients with cryptogenic chronic hepatitis and negative HCV antibody tests. J. Infect. Dis. 17627-33. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt, W. N., P. Wu, J. Q. Han, M. J. Perino, D. R. LaBrecque, and J. T. Stapleton. 1997. Distribution of hepatitis C virus (HCV) RNA in whole blood and blood cell fractions: plasma HCV RNA analysis underestimates circulating virus load. J. Infect. Dis. 17620-26. [DOI] [PubMed] [Google Scholar]
- 12.Watkins-Riedel, T., P. Ferenci, P. Steindl-Munda, M. Gschwantler, C. Mueller, and M. Woegerbauer. 2004. Early prediction of hepatitis C virus (HCV) infection relapse in nonresponders to primary interferon therapy by means of HCV RNA whole-blood analysis. Clin. Infect. Dis. 391754-1760. [DOI] [PubMed] [Google Scholar]
- 13.WHO. 1997. Hepatitis C: global prevalence. Wkly. Epidemiol. Rec. 72341-344. [PubMed] [Google Scholar]
- 14.Yan, F. M., A. S. Chen, F. Hao, X. P. Zhao, C. H. Gu, L. B. Zhao, D. L. Yang, and L. J. Hao. 2000. Hepatitis C virus may infect extrahepatic tissues in patients with hepatitis C. World J. Gastroenterol. 6805-811. [DOI] [PMC free article] [PubMed] [Google Scholar]