Abstract
We developed two real-time quantitative PCR (qPCR) assays, targeting the 28S rRNA gene, for the diagnosis of zygomycosis caused by the most common, clinically significant Zygomycetes. The amplicons of the first qPCR assay (qPCR-1) from Rhizopus, Mucor, and Rhizomucor species were distinguished through melt curve analysis. The second qPCR assay (qPCR-2) detected Cunninghamella species using a different primer/probe set. For both assays, the analytic sensitivity for the detection of hyphal elements from germinating sporangiospores in bronchoalveolar lavage (BAL) fluid and lung tissue homogenates from rabbits was 1 to 10 sporangiospores/ml. Four unique and clinically applicable models of invasive pulmonary zygomycosis served as surrogates of human infections, facilitating the validation of these assays for potential diagnostic utility. For qPCR-1, 5 of 98 infarcted lung specimens were positive by qPCR and negative by quantitative culture (qCx). None were qCx positive only. Among 23 BAL fluid samples, all were positive by qPCR, while 22 were positive by qCx. qPCR-1 detected Rhizopus and Mucor DNA in 20 (39%) of 51 serial plasma samples as early as day 1 postinoculation. Similar properties were observed for qPCR-2, which showed greater sensitivity than qCx for BAL fluid (100% versus 67%; P = 0.04; n = 15). The assay detected Cunninghamella DNA in 18 (58%) of 31 serial plasma samples as early as day 1 postinoculation. These qPCR assays are sensitive and specific for the detection of Rhizopus, Mucor, Rhizomucor, and Cunninghamella species and can be used for the study and detection of infections caused by these life-threatening pathogens.
The number of cases of zygomycosis have increased over the last six decades, making the diagnosis of these infections imperative (31, 44, 47). Infections with Zygomycetes may advance rapidly, leading to fatality, particularly in patients with underlying illnesses such as diabetes, bone marrow or solid-organ transplantation, and renal failure (44). The modes of transmission are varied for the immunocompromised patient, but the inhalation of sporangiospores with subsequent pulmonary infection is the most common (40, 44). A recent review reported an overall mortality of 76% for patients with pulmonary infections due to Zygomycetes (44). Furthermore, in the immunocompromised host, hematogenous dissemination is frequent and contributes to the high mortality rate, underscoring the urgency of making a rapid and accurate diagnosis of pulmonary zygomycosis (19, 20, 38).
Contributing to the clinical significance of pulmonary infections with Zygomycetes is the limited availability of antifungal agents. Formulations of amphotericin B have long been the primary antifungal agent for the treatment of invasive zygomycosis. The addition of surgical intervention has often been required to contain this highly invasive disease (49). A multifaceted approach to controlling these infections has thus been necessary and has included antifungal therapy, surgical resection, and improving host response (3). Given the importance of an accurate and rapid diagnosis of zygomycosis to guide the timely initiation of amphotericin B and possible surgical intervention, we developed and validated a new diagnostic quantitative PCR (qPCR) assay system for the detection of the four most common medically important genera of Zygomycetes.
Direct examination by wet mount and classical culture techniques are the standard methods for the detection of Zygomycetes in clinical microbiology laboratories. However, the recovery of these organisms from bronchoalveolar lavage (BAL) fluid and tissues can be difficult (40). Fungal elements may not be abundant, and depending on the representative region of tissue sampled, it is possible that organisms causing the infection may not be extracted. Additionally, aggressive tissue grinding may render the fragile, pauciseptate zygomycete nonviable (41). Although the recovery of Zygomycetes in a microaerophilic environment may improve culture yield, these methods are not routinely used in clinical microbiology laboratories (24). Frequently, Zygomycetes are observed upon histopathological analysis of tissues and BAL fluid specimens when culture results are negative. However, even when both culture and histopathological analyses are performed, many suspected infections are not confirmed until postmortem examination. Moreover, discrimination between the histological features of Zygomycetes and those of other filamentous fungi may not be clear (13). Given the increase in the number of these infections in recent years, a molecular approach for the detection of zygomycete molds may increase sensitivity and rapid diagnosis, resulting in earlier, directed therapy.
The most common genera causing pulmonary zygomycosis are Rhizopus species (especially R. oryzae), Mucor species, Rhizomucor pusillus, Cunninghamella bertholletiae, and Absidia species (40, 44). In this report, we describe the development of two qPCR assays for the diagnosis of invasive pulmonary zygomycosis. The first assay (qPCR-1) detects Rhizopus, Mucor, and Rhizomucor species, and the second assay (qPCR-2) detects Cunninghamella species. These assays each utilize a single set of primers and fluorescence resonance energy transfer (FRET) hybridization probes for the detection of species within the genera. Given the paucity of available clinical samples, four rabbit models of the most common forms of invasive pulmonary zygomycosis were developed and studied. Thus, in addition to the assessment of analytical sensitivity and specificity, the assay was validated using BAL fluid, lung tissue homogenates (LTH), and serial plasma samples from rabbits infected with Rhizopus oryzae, Rhizopus microsporus, Mucor circinelloides, and Cunninghamella bertholletiae.
MATERIALS AND METHODS
Both qPCR assays were designed around the 28S rRNA gene in order to optimize the specificity of the assay. This was facilitated by the relative completeness of the sequence data available on the 28S region of the rRNA gene complex.
Primer and probe design (qPCR-1).
An alignment of the 28S rRNA gene of 20 species (22 strains) from five zygomycete genera causing human infections was created using Megalign software (DNASTAR, Inc., Madison, WI). These sequences were chosen from those strains previously used by Voigt et al. to conduct a phylogenetic analysis of Zygomycetes (50). Primers that annealed to sequences within the genera Rhizopus, Mucor, and Rhizomucor but not unrelated fungal genera that might be present in clinical samples such as Penicillium, Aspergillus, or Candida were chosen from this alignment. Primer design was optimized with Primer-3 software (http://frodo.wi.mit.edu/primer3/input.htm). Primers were purchased from Midland Certified Reagent Co. (Midland, TX). The amplicon generated was 180 bp in length. Hybridization FRET probes were chosen using Oligo software (Molecular Biology Insights, Cascade, CO) and synthesized by Operon Biotechnologies, Inc. (Huntsville, AL) (Table 1).
TABLE 1.
Oligonucleotide sequences of primers and probes
| PCR and primer or probe | Oligonucleotide sequence |
|---|---|
| qPCR-1 | |
| Primers | |
| ZYGO(+) sense | 5′-TTC AAA GAG TCA GGT TGT TTG G-3′ |
| ZYGO(−) sense | 5′-CAG TCT GGC TCC AAA CGG TTC-3′ |
| Probes | |
| ZYGO FITC | 5′-GGC GAG AAA CCG ATA GCG AAC-3′ |
| ZYGO RD 640 | 5′-GTA CCG TGA GGG AAA GAT GAA AAG AAC TTT GAA A-3′ |
| qPCR-2 | |
| Primers | |
| CUb(+) sense | 5′-TAG TCA GCC AGG TAA ATA AGT-3′ |
| CU(−) sense | 5′-TCG TCA ATA TTT AGC TTT AGG-3′ |
| Probes | |
| CU FITC | 5′-GCT TGG AAA CGA AGA GTC AGG TTG-3′ |
| CU RD 640 | 5′-TGG GAA TGC AGC CTA AAA TGG GAG TGA-3′ |
Primer and probe design (qPCR-2).
A quantitative, real-time PCR utilizing FRET technology was designed to target the 28S region of the rRNA gene complex. The primers and probes were based on a consensus sequence generated from multiple rRNA alignments of sequences of Cunninghamella species from GenBank utilizing the Sequencher software package (Gene Codes Corp., Ann Arbor, MI). The preliminary sequences of the primers and probes were designed using LightCycler Probe Design software 2.0 (Idaho Technology, Inc., Salt Lake City, UT). The NCBI Basic Local Alignment and Search Tool (BLAST) database search program was then used to determine the ability of the primers to specifically target the rRNA of C. bertholletiae and not to cross-hybridize with any mammalian DNA sequence. Once this was determined, primers and probes were further refined using Oligo primer analysis software, v6.72 (Molecular Biology Insights), to minimize upper, lower, and upper/lower primer duplexes and hairpins to optimize respective interactive melting temperatures (Tms) of the primers and probes (Table 1). The refined primers' sequence specificities were reconfirmed using the NCBI BLAST database search program. The amplicon generated was 148 bp in size.
PCR conditions (qPCR-1 and qPCR-2).
Quantitative real-time PCR was performed with a LightCycler instrument (Roche Applied Sciences, Indianapolis, IN). For each PCR run, a “kit blank” (water processed through the extraction protocol) and a negative master mix control (water) were included. The master mix was set up in a laminar flow hood that was located in a room separate from where DNA extractions were performed. For both assays, 5 μl of extracted specimen was added to 15 μl of master mix. Just prior to adding 5 μl of extracted specimen to the master mix, all extracted specimens were briefly vortexed and then centrifuged for 45 s at 4,500 × g.
The PCR master mix for qPCR-1 consisted of 0.25 μM of each primer, 1× PCR buffer (Invitrogen Corp., Carlsbad, CA), 3 mM MgCl2, 0.025% bovine serum albumin (BSA) (Sigma-Aldrich Corp., St. Louis, MO), 0.025 U Platinum Taq DNA polymerase (Invitrogen Corp.), 0.2 mM PCR Nucleotide MixPlus (dATP, dCTP, dGTP, and 3 dUTP) (Roche Applied Sciences), 0.1 μM each fluorescein isothiocyanate (FITC) and LC Red-640 probes (RD640) (Idaho Technology, Inc.), and 0.002 U/μl HK-uracil N-glycosylase (UNG) (Epicentre, Madison, WI). Uracil was released by incubation at 37°C for 15 min, followed by enzyme inactivation at 95°C for 3 min. Touchdown PCR cycling was performed as follows: 95°C of denaturation for 0 s, followed by annealing in 1°C incremental steps between 68°C and 54°C for 5 s each with a 72°C extension step for 15 s for each cycle. Touchdown cycling was followed by 35 cycles of 95°C for 0 s, 54°C for 5 s, and 72°C for 15 s. Following amplification, melt curve analysis was performed by cooling from 96°C to 40°C for 30 s, followed by a gradual increase in temperature (2°C/s) to 75°C.
For qPCR-2, PCR master mix consisted of 0.5 μM of each of the primers, 1× PCR buffer (Invitrogen Corp.), 4 mM MgCl2, 0.025% BSA (Sigma-Aldrich Corp.), 0.025 U/ml Platinum Taq DNA polymerase (Invitrogen Corp.), 0.2 mM PCR Nucleotide MixPlus (Roche Applied Sciences), and 0.2 μM each FITC and RD640 probes (Idaho Technology, Inc.). As with qPCR-1, HK-UNG was utilized as recommended by the manufacturer to prevent potential amplicon carryover. The cycling conditions were as follows: uracil activation at 37°C for 900 s (slope, 20°C/s) and uracil heat inactivation at 95°C for 180 s (slope 20°C/s) for 1 cycle. Amplification cycles were as follows: denaturation at 95°C for 0 s (slope, 20°C/s), annealing at 57°C for 5 s (slope, 10°C/s), and extension at 72°C for 15 s (slope, 3°C/s) for 50 cycles. A melt cycle (96°C for 0 s [slope, 20°C/s], 40°C for 30 s [slope, 2°C/s], and 75°C for 0 s [slope, 0.2°C/s]) was performed at the end of each run to confirm that the correct amplicon was generated via PCR product melt temperature analysis.
Fungal isolates (qPCR-1 and qPCR-2).
Fungal isolates used to characterize the qPCR assay were previously identified in the Clinical Microbiology Laboratory of the Clinical Center at the NIH or were obtained from the laboratory collection of the Fungus Testing Laboratory of the University of Texas Health Science Center and the Immunocompromised Host Section, Pediatric Oncology Branch, National Cancer Institute, at the NIH. All isolates were identified by colony and microscopic morphology criteria (5, 34).
Analytical sensitivity and quantitation (qPCR-1 and qPCR-2).
The PCR product resulting from the amplification of R. microsporus strain 375, M. circinelloides strain 184, and C. bertholletiae strain 190 were each cloned into vector pCR2.1 using the TA cloning kit (Invitrogen Corp.). Separate clones of these three genera were constructed because sequence differences result in different efficiencies of the amplification curves. The sequence of each clone was verified with either the ABI 3100 capillary sequencer (Applied Biosystems, Inc., Foster City, CA) or confirmed by an outside source (Genewiz, Inc., La Jolla, CA). Plasmid constructs were quantified and diluted in 10-fold dilutions from 1 × 106 to 1 × 100 copies/reaction in Tris-EDTA (pH 8.0) with 33.3 μg/ml glycogen. These dilutions were used to assess assay sensitivity and linearity and were included as quantitation standards with each qPCR run.
Specificity and cross-reactivity studies (qPCR-1 and qPCR-2).
Specificity and cross-reactivity of the assays were assessed using DNA extracted from 9 yeasts, 21 filamentous fungi, and mammalian whole blood from rabbits and humans (Table 2).
TABLE 2.
Specificities of qPCR-1 and qPCR-2 against clinically relevant organisms and potential environmental contaminants
| Genus and species | Amplificationa
|
|
|---|---|---|
| qPCR-1 | qPCR-2 | |
| Rhizopus oryzae | +b | − |
| Rhizopus microsporus | +c | − |
| Mucor circinelloides | +d | − |
| Mucor ramosissimus | +d | − |
| Mucor indicus | + | − |
| Rhizomucor pusillus | +c | − |
| Cunninghamella bertholletiae | −d | +e |
| Cunninghamella echinulata | − | + |
| Absidia corymbifera | − | − |
| Candida albicans | − | − |
| Candida tropicalis | − | − |
| Candida parapsilosis | − | − |
| Candida krusei | − | − |
| Candida glabrata | − | − |
| Saccharomyces cerevisiae | − | − |
| Trichosporon asahii | − | − |
| Trichosporon inkin | − | − |
| Trichosporon inkin | − | − |
| Aspergillus fumigatus | − | − |
| Aspergillus terreus | − | − |
| Aspergillus flavus | − | − |
| Aspergillus niger | − | − |
| Penicillium marnefii | − | − |
| Penicillium notatum | − | − |
| Penicillium purpurogenum | − | − |
| Penicillium chrysogenum | − | − |
| Penicillium citrinum | − | − |
| Fusarium solani | − | − |
| Fusarium boydii | − | − |
| Pseudallescheria boydii | − | − |
| Human (whole blood) | − | − |
| Rabbit (whole blood) | − | − |
Note that one strain was tested unless otherwise specified, and 1 × 106 fg of each DNA sample was tested.
Four strains were tested.
Three strains were tested.
Two strains were tested.
Five strains were tested.
Containment of contamination (qPCR-1 and qPCR-2).
In order to minimize contamination, biological hoods, laboratory rooms, pipettes, and centrifuges, etc., were dedicated either to culture or harvest of hyphal elements or to DNA extraction. Additionally, PCR master mix preparation was conducted in a separate room utilizing a third biological safety hood with dedicated equipment. Thermolabile UNG was incorporated into the master mix to prevent amplicon carryover.
In vitro culture in normal rabbit BAL fluid and LTH (qPCR-1 and qPCR-2).
Fungal sporangiospores were isolated from a fresh growth of R. microsporus strain 375 (obtained from a lung biopsy specimen), M. circinelloides strain 184 (obtained from a blood specimen), or C. bertholletiae strain 190 (obtained from a lung biopsy specimen) grown on potato dextrose agar slants (Remel, Inc., Lenexa, KS). Sporangiospores were harvested with sterile normal saline (NS) with 0.0125% Tween 20 and separated from hyphal elements by filtration through sterile gauze. Following centrifugation at 800 × g for 10 min, sporangiospores were washed and resuspended in phosphate-buffered saline (PBS) (pH 7.2) (Quality Biological, Inc., Gaithersburg, MD). Sporangiospores were counted on a hemocytometer and diluted to achieve the desired inocula. These were confirmed with colony counts on Sabouraud dextrose agar plates. All in vitro cultures were performed in 24-well flat-bottom cell culture plates (Corning, Corning, NY).
Large-scale BALs for these in vitro studies were performed as previously described (45). In brief, upon euthanasia, lungs from normal, uninfected rabbits were excised. The edges of the pulmonary lobes were cut, and 20 ml of PBS at 4°C was injected through each main bronchi three to four times per lobe. The lavage fluid was collected, filtered through sterile gauze, and stored at −30°C until use. When BAL fluid specimens were used as the culture medium, 200 μl of yeast nitrogen broth, 20 μg/ml gentamicin, 20 μg/ml vancomycin, and 800 μl of normal rabbit BAL fluid were added to plate wells. Normal lung tissues were processed as described previously (52). The processed lung tissue was further homogenized utilizing the FastPrep system (Qbiogene, Irvine, CA) using lysing matrix D (LMD) (setting 5 for 30 s three times). Homogenized tissues were stored at −30°C until use. For LTH, the culture medium was the same as for BAL fluid except that the amount of yeast nitrogen broth was decreased to 100 μl.
In vitro reproducibility studies (qPCR-1 and qPCR-2).
The reproducibilities of both the extraction process and the qPCR assay were confirmed through a series of in vitro experiments in the setting of the sample types of clinical importance (rabbit LTH and BAL fluid). For reproducibility studies, R. microsporus and M. circinelloides sporangiospores were inoculated into 800 μl of normal rabbit BAL fluid (n = 10) and LTH (n = 10), resulting in final concentrations of approximately 10 sporangiospores/ml. To determine the reproducibility of qPCR-2, C. bertholletiae sporangiospores were inoculated into either BAL fluid (n = 12) or normal rabbit LTH (n = 10). Cultures were incubated overnight (approximately 20 h) at 37°C to allow sporangiospores to germinate. For M. circinelloides, which has a lower optimum growth temperature, the culture plate was incubated an additional 24 h at room temperature. Additionally, negative controls were evaluated. The presence of hyphae was confirmed microscopically. After incubation, the entire contents of each well were removed and placed into a 1.5-ml screw-cap tube. The well was rinsed with 200 μl sterile PBS to collect any adherent hyphae and added to the collection contents.
In vitro sensitivity studies (qPCR-1 and qPCR-2).
To determine in vitro sensitivity, 100 μl of the sporangiospore suspension at 1 × 103, 1 × 102, and 1 × 101 sporangiospores/ml was added to the medium (BAL fluid or LTH as stated above) in the wells. Each sporangiospore concentration and a set of negative controls were tested in triplicate. Incubation conditions and times were the same as those described above. After incubation, the entire contents of each well were harvested. Each well was rinsed with 200 μl sterile PBS to collect any adherent hyphae and added to the original harvest. The contents of each well's harvest were handled separately. The extraction of DNA was performed as described below.
DNA extraction from fungal isolates (qPCR-1 and qPCR-2).
Fungal sporangiospores harvested from potato dextrose agar slants were inoculated into Sabouraud dextrose broth and incubated at 30°C with gentle shaking for 48 h. Hyphae were harvested from approximately 6 ml of culture by centrifugation at 16,000 × g for 5 min. DNA was extracted from hyphal mats (approximately 100 mg [wet weight]) using the DNeasy Plant Mini kit (Qiagen Inc., Valencia, CA), as described previously by Francesconi et al. (12).
DNA isolation from BAL fluid and LTH (qPCR-1 and qPCR-2).
DNA extraction was performed by the DNeasy Plant Mini kit (Qiagen) and MagNA Pure LC system (Roche Applied Sciences) methods, as described below. Of note, for C. bertholletiae, only the MagNA Pure LC system (Roche Applied Science) was utilized for DNA extraction. To avoid potential contamination, DNA was extracted in an AirClean PCR Workstation (AirClean Systems, Raleigh, NC).
(i) Manual DNA extraction from BAL fluid and LTH.
DNA was extracted from 1 ml of BAL fluid or 200 μl of LTH. Specimens were vortexed and then centrifuged for 10 min at 16,000 × g. Supernatants were discarded, and the pellets were gently resuspended in 100 μl of spheroplast buffer (1.0 M sorbitol, 50 mM monobasic sodium phosphate, 0.1% 2-mercaptoethanol, and 10 mg/ml lyticase) (Sigma-Aldrich Corp.) and 10 μl of lysing enzymes (20 mg/ml Novozyme) (Sigma-Aldrich Corp.). Samples were incubated at 30°C for 30 min in an Eppendorf thermomixer (Eppendorf, Westbury, NY) with shaking at 1,200 gyrations per minute (gpm) for the first 5 min. Following incubation, 400 μl of AP1 buffer (DNeasy Plant kit; Qiagen) was added. Samples were transferred into LMD tubes (Qbiogene/MP Biomedicals, Solon, OH) and processed using the FastPrep instrument (Qbiogene/MP Biomedicals), as described previously (33). This process was performed a total of three times. Samples were centrifuged at 16,000 × g for 60 s and then gently vortexed. Four microliters of 100 mg/ml RNase A and 100 μl of AP1 buffer were then added. The samples were vortexed and incubated for 10 min at 65°C in an Eppendorf thermomixer at 1,200 gpm. The samples were further processed according to the DNeasy Plant Mini kit (Qiagen) protocol with the following modifications: after 200 μl of AE buffer (preheated to 65°C) was applied to the column, and the column and collection tube were placed at 65°C for 5 min before centrifugation to elute DNA (12, 37). LTHs were eluted with 100 μl of AE buffer by the same procedure.
(ii) Automated DNA extraction from BAL fluid and LTH.
DNA was extracted from 1 ml of BAL fluid or 200 μl of LTH as previously described (10). Briefly, samples were centrifuged in FastPrep tubes (without LMD) for 10 min at 16,000 × g, and supernatants were discarded. A 150-μl aliquot of spheroplast buffer and 10 μl of lysing enzymes were added to each specimen. LMD was added to each sample, and samples were briefly vortexed. Samples were incubated at 30°C for 5 min at 1,200 gpm in an Eppendorf thermomixer. Mixing was terminated, and incubation continued for 25 min. Samples were processed using the FastPrep instrument at speed 5 for 30 s and placed on ice for 5 min; this process was performed a total of three times. Samples were centrifuged for 1 min at 1,000 × g, and 130 μl of bacterial lysis buffer (DNA isolation kit III [bacteria and fungi]; Roche Applied Science) and 20 μl proteinase K were then added to each sample. Samples were vortexed for 30 s and then incubated for 10 min at 65°C in an Eppendorf thermomixer at 1,200 gpm. After incubation, specimens were centrifuged for 1 min at 1,000 × g and then gently mixed. Samples were equilibrated to room temperature and transferred into the sample cartridge. Extraction was completed with the MagNA Pure LC instrument (Roche Applied Science), as recommended by the manufacturer.
(iii) Automated DNA extraction from plasma.
Plasma samples (100 μl) were extracted using the MagNA Pure LC system with DNA isolation kit III for bacteria and fungi. A 130-μl aliquot of bacterial lysis buffer and 20 μl of proteinase K were added to the serum. Samples were vortexed and then incubated at 65°C for 10 min in an Eppendorf thermomixer (Eppendorf) at 1,200 rpm. After incubation, samples were centrifuged for 1 min at 1,000 × g and then gently mixed. Samples (250 μl) were equilibrated to room temperature and subsequently added to the sample cartridge prior to being processed by the MagNA Pure LC instrument. A 250-μl aliquot of PBS was also processed as a control. Samples were stored at −20°C until analysis by qPCR.
Control for PCR inhibition (qPCR-1 and qPCR-2).
To determine if PCR inhibition was occurring, DNA isolated from BAL fluid and homogenized lung tissue specimens (5 μl) were each tested in a separate PCR that included an internal amplification control. Linearized plasmid DNA from pBR322 (1 × 106 copies/reaction) was added into a separate master mix used to amplify each sample according to the procedure described previously by Fahle and Fischer (9). The following PCR primer sequences were designed to amplify a region of pBR322, which was not found in eukaryotic organisms: IC-F-5′-ATTGCTAACGCAGTCAGGCACCG-3′ and IC-R-5′-GCGAGCCCGATCTTCCCCAT-3′ (IC-F, internal control forward: IC-R, internal control reverse). FRET hybridization probe sequences were IC-5′-GATATCGTCCATTCCGACAGCATC-FITC-3′ and IC-5′-RD705CCAGTCACTATGGCGTGCTGCTAG-3′ (IC, internal control). PCR master mix consisted of 0.025% BSA (Sigma-Aldrich Corp.), 4 mM MgCl2, 0.025 U Platinum Taq DNA polymerase, 1× PCR buffer, 0.2 mM PCR Nucleotide Mix Plus, 0.002 U/μl HK-UNG, 0.25 μM of each primer, and 0.2 μM of each RD640- and FITC-labeled probe. Following a UNG step as performed for target-specific amplification, the internal control was amplified with 45 cycles of 95°C for 0 s, 54°C for 5 s, and 72°C for 15 s. Melt curve analysis was performed as described above. The PCR crossover point for each specimen was compared to a control composed only of master mix reagents and water. Specimens in which the crossover threshold (CT) was more than two cycles greater than the control were repeated at a 1:10 dilution in PBS.
In vivo studies (qPCR-1 and qPCR-2). (i) Animal model.
Female New Zealand White rabbits (Covance Research Products, Inc., Denver, PA) weighing 2.3 to 3.5 kg at the time of inoculation were used in all experiments. Sixteen rabbits were infected with R. oryzae 176, 17 were infected with M. circinelloides 234, 9 were infected with R. microsporus 230, and 15 were infected with C. bertholletiae 182. All rabbits were individually housed and monitored under humane care and use of standards in facilities accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International and according to the guidelines of the National Research Council for the care and use of laboratory animals and under approval by the Animal Care and Use Committee of the National Cancer Institute (36). Vascular access was established in each rabbit by the surgical placement of a silastic tunneled central venous catheter as described previously (51).
(ii) Organism and inoculation.
Experiments performed with rabbit models of invasive zygomycosis utilized the following isolates: R. oryzae 176, cultured from a patient flank lesion; M. circinelloides 234, cultured from a patient bronchial washing; R. microsporus 230, cultured from a patient lung biopsy; and C. bertholletiae 182, cultured from a patient lung biopsy. Multiple strains were studied in vitro and in vivo in order to test the robustness of the assay. Immunosuppression was established using cytarabine (Cytosar-U; Upjohn Company, Kalamazoo, MI) and methylprednisolone (Abbott Laboratories, North Chicago, IL) in a volume of 250 to 350 μl. Rabbits were inoculated endotracheally with inocula ranging from 1.25 × 108 to 5 × 109 sporangiospores for R. oryzae, M. circinelloides, and R. microsporus and 1 × 109 sporangiospores for C. bertholletiae on day 4 of immunosuppression. The concentrations of the inocula were confirmed by serial dilutions cultured onto 5% Sabouraud glucose agar plates (K-D Medical, Inc., Columbia, MD).
(iii) Fungal cultures.
Lung tissue from each rabbit was sampled by the excision of a standard, representative region of the lung. Each fragment was weighed individually, placed into a sterile bag, homogenized with sterile NS for 30 s per tissue sample, and quantitatively cultured (52). The CFU of the invading organisms were counted and recorded for each lobe, and the CFU/g were calculated. Remaining LTHs were aliquoted and stored at −30°C until DNA extractions were performed.
(iv) Histopathology.
Lesions were excised and fixed in 10% neutral buffered formalin. Paraffin-embedded tissue sections were stained with periodic acid-Schiff and Gomori methenamine silver stains. Tissues were microscopically examined for pulmonary injury and the presence of hyphae.
(v) BAL.
For the in vivo studies, small-scale BALs were performed on each postmortem-resected lung preparation by the instillation and the subsequent withdrawal of 10 ml of sterile NS into the clamped trachea with a sterile 12-ml syringe. This process was repeated for a total infusion of 20 ml of NS. The lavage material was then centrifuged for 10 min at 500 × g. The supernatant was discarded, leaving 2 ml of fluid in which the pellet was then resuspended. A 0.1-ml aliquot of this fluid and a 0.1-ml dilution (10−1) of this fluid were cultured onto Sabouraud glucose agar plates. The remainder was placed at −30°C until DNA extractions were performed.
(vi) Lung tissue.
Lung tissues were excised and processed as previously described (52). Briefly, rabbit lungs were excised, weighed, and inspected for hemorrhagic infarct lesions in each individual lobe.
Lung tissues were sampled by the excision of a representative region of each infarcted lobe. Each fragment was weighed individually, placed into a sterile polyethylene bag, homogenized with sterile saline for 30 s per tissue sample, and quantitatively cultured.
Statistical analysis (qPCR-1 and qPCR-2).
Comparisons between groups were performed by analysis of variance with Bonferroni's correction for multiple comparisons or by the Mann-Whitney U test, as appropriate. All P values were two sided, and a P value of ≤0.05 was considered to be statistically significant. Values were expressed as means ± standard errors of the means. Correlation analysis was determined by Spearman rank correlation coefficient. The sensitivity and specificity of the qPCR assay system and quantitative culture (qCx) were assessed utilizing 2-by-2 contingency table analysis by applying Fisher's exact test. A positive sample was defined by the presence of infarction in lung tissue. The coefficient of variance (CV) was defined as the standard deviation divided by the mean.
RESULTS
In vitro analytical sensitivity and specificity (PCR-1).
The in vitro sensitivity and specificity of qPCR-1 were assessed utilizing the newly formed plasmid constructs from amplicons amplified from genomic DNA of R. oryzae, M. circinelloides, and R. pusillus. The PCR primers were chosen to amplify the 28S rRNA gene from medically relevant zygomycete species. The 5′ primer was 100% homologous to sequences of five genera of Zygomycetes commonly isolated from pulmonary specimens. However, mismatches within the sequence of the 3′ primer between genera were present, permitting the amplification of only Rhizopus, Mucor, and Rhizomucor species under the chosen reaction conditions.
The assay reliably detected as few as 10 copies of each clone and was linear from 1 × 101 to 1 × 106 copies/reaction. Using genomic DNA quantified spectrophotometrically, 10 fg of DNA from R. oryzae, R. microsporus, and M. circinelloides and 10 pg of DNA from R. pusillus were detected.
Multiple strains of several species were studied to verify that any sequence variations between strains would be unlikely to affect the ability of the assay to detect that species. These were R. oryzae (n = 4), R. microsporus (n = 3), M. circinelloides (n = 2), M. indicus (n = 1), M. ramosissimus (n = 2), and R. pusillus (n = 3). Zygomycetes from which DNA was not amplified included C. bertholletiae (n = 2), C. echinulata (n = 1), and Absidia species (n = 1).
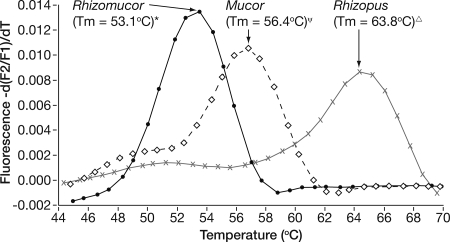
Although DNA from Rhizopus, Mucor, and Rhizomucor species was amplified, these genera were distinguished through melt curve analysis. The FRET hybridization probes bind in regions in which 2 to 4 nucleotide differences exist among the three genera. Amplifying DNA from R. microsporus 375, M. circinelloides 184, and R. pusillus 280 on eight separate runs determined the average peak Tm for each genus. As shown in Fig. 1, these differences result in average Tm values of 63.8, 56.4, and 53.1, respectively. The 95% confidence intervals around these means do not overlap, indicating that an amplified product from any of these three genera was distinguished with peak Tm analysis.
FIG. 1.
Melt curve analysis demonstrating the differentiation of Rhizomucor, Mucor, and Rhizopus species by Tm. The Tm shown is the mean of data from eight replicate reactions. The 95% confidence intervals are indicated as follows: *, 52.9°C to 53.2°C; Ψ, 56.3°C to 56.5°C; ▵, 63.7°C to 63.9°C.
Assay specificity was further characterized with purified genomic DNA from other fungal species that may commonly be isolated from clinical specimens, especially those from the respiratory tract. None of the DNA from these species studied was amplified (Table 2). Additionally, neither human nor rabbit DNA gave a positive amplification signal.
In vitro analytical sensitivity and specificity (qPCR-2).
The sensitivity of qPCR-2 was assessed through multiple sets of serial dilutions of the newly formed plasmid construct containing the C. bertholletiae rRNA amplicon generated by the primers designed in this study. The assay reliably detected between 10 and 100 copies of the new construct per reaction. The fluorescent signal was proportional to the log concentration of the plasmid with a calculated PCR efficiency of 2.00. The assay did not cross-react with any of the clinically relevant organisms tested, nor did the assay cross-react with any of the potential environmental contaminants (Table 2). The assay did detect C. echinulata with equal sensitivity; however, the amplicon of C. echinulata could be distinguished from that of C. bertholletiae by a difference of 3°C in their respective Tm values. This difference in the Tm was due to a single-base mismatch (T to a C) in the region of the FITC probe. This was confirmed by the sequencing of the PCR product.
In vitro amplification from rabbit BAL fluid and lung tissue (qPCR-1).
Both R. microsporus and M. circinelloides were successfully amplified from BAL fluid and LTH.
(i) In vitro reproducibility.
To determine the reproducibility of the qPCR assay for the detection of the Zygomycetes from clinical specimens, sporangiospores from R. microsporus 375 and M. circinelloides 184 were inoculated separately into normal rabbit BAL fluid and LTH (10 replicates each) to achieve a final sporangiospore concentration of approximately 10 sporangiospores/ml. DNA from hyphae was extracted by both the DNeasy and MagNA Pure methods, as described above. The intra-assay CVs for R. microsporus in BAL fluid and lung tissue were 3.3% and 4.2% by the manual method and 6.5% and 5.2% by the automated method, respectively. For M. circinelloides, the intra-assay CVs in BAL fluid and lung tissue were 4.3% and 5.7% by the manual method and 6.1% and 4.6% by the automated method, respectively.
(ii) In vitro sensitivity.
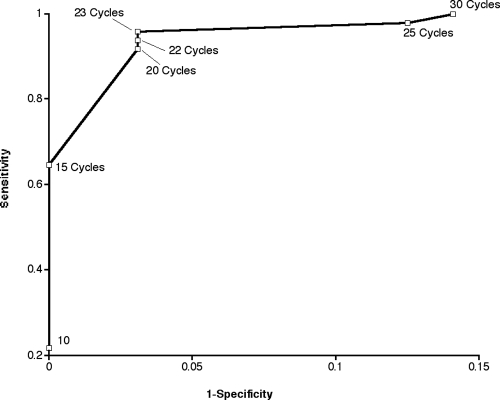
Based upon the receiver-operator curve (ROC) of sensitivity versus specificity for BAL fluid and lung tissue samples, the greatest sensitivity and fewest false positives occurred with a CT cutoff of between 20 and 23 (Fig. 2). Thus, for this assay, positive results were defined as having a CT of ≤22. After the criteria for defining positive and negative results were applied to the in vitro experimental data, one negative control sample was considered to be a false positive. A ROC was not generated for plasma samples, as no plasma sample from an uninfected rabbit generated a false-positive signal. This may be a reflection of the sample type itself. Both BAL fluid and LTH are nonsterile specimens and are thus susceptible to background environmental contamination by zygomycetes. In comparison, plasma samples are sterile specimens that are not susceptible to this environmental background contamination when they are handled aseptically. Thus, given that none of our plasma samples from uninfected rabbits generated a false-positive signal, a designated CT cutoff was unnecessary for plasma samples.
FIG. 2.
ROC demonstrating sensitivity and specificity. The curve was calculated from results of the qPCR-1 assay with in vitro culture experiments. Note that 1 − specificity = false-positive rate.
The absence of a CT cutoff may be a reflection of the sample types themselves, where BAL fluid and lung tissues are nonsterile samples, potentially susceptible to environmental contaminants. Assuming that aseptic techniques are utilized, plasma samples can be considered sterile samples and thus not likely to be susceptible low-level external contaminants when handled properly.
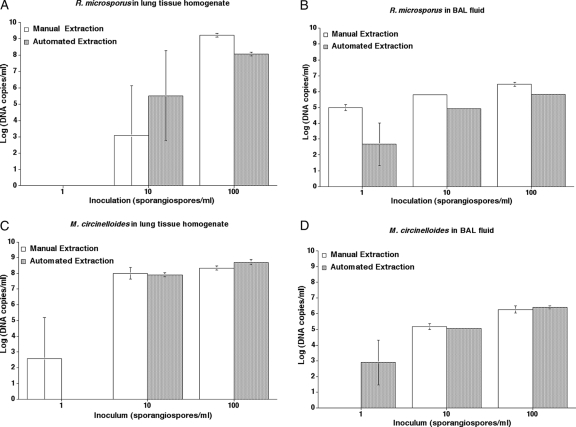
For each experiment, amplification was highly variable at the lowest dilution where 1 sporangiospore was added to each well (Fig. 3). In comparison, the detection of sporangiospores was more consistent with an inoculum of 10 sporangiospores/ml. However, a positive result was obtained for only two of six samples in which 10 R. microsporus sporangiospores were added to lung tissue. At an inoculum of 10 and 100 sporangiospores/ml, the yield of R. microsporus DNA from BAL fluid was better with extraction by the manual method than by the automated method. At the inoculum of 100 Rhizopus sporangiospores/ml, the yields were similar for both methods for LTH samples (Fig. 3A). There was also no significant difference in yield when either extraction method was used for the detection of Mucor from either LTH or BAL fluid (Fig. 3C and D). There was a trend of an increased copy number detected as the inoculum increased. For both Rhizopus and Mucor, higher yields were observed when the sporangiospores were inoculated into LTH than when they were inoculated in BAL fluid.
FIG. 3.
Sensitivity of the qPCR-1 assay to detect zygomycete DNA in lung tissue homogenate and BAL fluid (in vitro). (A) R. microsporus in LTH. (B) R. microsporus in BAL fluid. (C) M. circinelloides in LTH. (D) M. circinelloides in BAL fluid. Data are expressed as means ± standard errors of the means of the log DNA copies/ml.
In vitro amplification from rabbit BAL fluid and lung tissue (qPCR-2).
C. bertholletiae was successfully amplified from BAL fluid and LTH by qPCR-2.
(i) In vitro reproducibility.
The reproducibility of qPCR-2 was determined for DNA extracted from BAL fluid samples (n = 12) and LTH (n = 10) containing germinated hyphae from 10 sporangiospores/ml. Results from DNA extracted from the BAL fluid samples showed a CV of 3.68%, whereas results from DNA extracted from LTH showed a CV of 7.50%.
(ii) In vitro sensitivity.
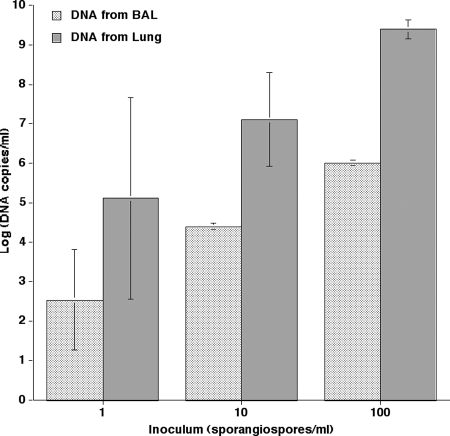
The sensitivity of qPCR-2 was further determined by extracting DNA from 1 × 100, 1 × 101, and 1 × 102 sporangiospores that were allowed to germinate in either BAL fluid or LTH as described above. An inoculum-dependent effect was seen in both spiked BAL fluid (2.54 ± 1.27, 4.39 ± 0.07, and 6.00 ± 0.07, respectively) and spiked LTH (5.11 ± 2.56, 7.10 ± 1.19, and 9.39 ± 0.25, respectively) (Fig. 4).
FIG. 4.
In vitro sensitivity of the qPCR-2 assay for both BAL fluid and LTH inoculated with 1, 10, and 100 sporangiospores. Sporangiospores were allowed to germinate in BAL fluid and LTH (48 h and 24 h, respectively), yielding hyphal structures more closely resembling the organism's early invasive stage.
qPCR-1 inhibition.
For in vitro sensitivity experiments, PCR inhibition was observed for only 1 of 24 BAL fluid specimens extracted by the manual method and 0 of 24 samples extracted by the automated method. Inhibition was more common for extracts of lung tissue. Four of 24 (16%) and 6 of 24 (25%) DNA samples extracted from lung tissues were inhibited when extracted by the manual method and the automated method, respectively. Inhibition was consistently removed when samples were repeated after dilution 1:10 in PBS. Inhibition was also assessed for the samples taken from the rabbit animal model. These samples were extracted with the automated system only. Only 1 of 14 BAL fluid (7%), 4 of 51 (8%) lung tissue, and 0 of the 62 plasma samples processed were initially inhibited. Again, inhibition was consistently removed when samples were repeated after dilution 1:10 in PBS.
qPCR-2 inhibition.
All DNA extractions were performed with the automated system. For in vitro sensitivity and reproducibility experiments, none of the samples extracted showed signs of inhibition in either BAL fluid samples (0/24) or LTH samples (0/24). Samples extracted from the rabbit model showed similar inhibition results of 0/15 and 2/90 of BAL fluid samples and LTH samples, respectively. Again, a 1:10 dilution of the sample in PBS consistently removed any sign of inhibition.
In vivo detection of zygomycete DNA in BAL fluid and lung tissue (qPCR-1).
Rabbit models of pulmonary zygomycosis caused by R. microsporus, R. oryzae, and M. circinelloides were studied for the in vivo detection of genus-specific DNA in BAL fluid, lung tissue, and plasma samples. qPCR and qCx were performed on LTH from lobes demonstrating signs of tissue infarction. Ten rabbits infected with R. oryzae had infarcted lung tissue. Both qCx and qPCR data were available for seven animals for a total of 34 lung and 7 BAL fluid samples. Eleven rabbits infected with M. circinelloides had infarcted lung tissue. Thirty lung tissue and nine BAL fluid samples from 10 of these animals were evaluated. Thirty-four infarcted lung tissue samples and six BAL fluid samples from six rabbits infected with R. microsporus were studied.
There was a significant correlation between qPCR and qCx results for each species for lung tissue specimens (Table 3). Correlation between qPCR and qCx could not be demonstrated for BAL fluid samples. Additionally, there was no statistical difference in sensitivity between the qPCR and qCx for either LTH samples (99% versus 95% [P = 0.12]) or BAL fluid samples (100% versus 96% [P = 1.0]) (Table 4). No samples were negative by qPCR and positive by qCx.
TABLE 3.
Correlation between data for qPCR-1 and qCx for BAL fluid and lung tissue in experimental pulmonary zygomycosis
| Organism | Sample type | Correlation coefficienta | P valued | Total no. of samples | No. of samples positive by qPCR but negative by qCx |
|---|---|---|---|---|---|
| R. oryzae | BAL fluid | 0.61 | 0.17 | 7 | 1 |
| R. oryzae | Lung tissueb | 0.55 | <0.01 | 35 | 2 |
| R. microsporus | BAL fluid | −0.49 | 0.37 | 6 | 0 |
| R. microsporus | Lung tissueb | 0.65 | <0.01 | 34 | 1 |
| M. circinelloides | BAL fluid | −0.26 | 0.47 | 10 | 0 |
| M. circinelloides | Lung tissueb,c | 0.70 | <0.01 | 30 | 2 |
| C. bertholletiae | BAL fluid | 0.63 | 0.01 | 15 | 5 |
| C. bertholletiae | Lung tissueb | 0.68 | <0.01 | 90 | 4 |
Calculated by the Spearman rank correlation method (Instat 3; Graphpad Software).
Only lung tissue samples containing pathological evidence of infarction were processed.
One sample was negative by both qCx and qPCR.
A P value of ≤0.05 is considered to be significant.
TABLE 4.
Sensitivities of qPCR-1, qPCR-2, and culture of BAL fluid and lung tissue in experimental pulmonary zygomycosis
| PCR and culture type | Total no. of samples | No. (%) of samples positive by:
|
P valuea | 95% CIb | |
|---|---|---|---|---|---|
| qPCR | Culture | ||||
| qPCR-1 | |||||
| Lung | 98 | 97 (99) | 92 (95) | 0.12 | 1.00, 1.11 |
| BAL fluid | 22 | 22 (100) | 21 (96) | 1.00 | 0.96, 1.15 |
| qPCR-2 | |||||
| Lung | 90 | 90 (100) | 86 (96) | 0.12 | 1.00, 1.09 |
| BAL fluid | 15 | 15 (100) | 10 (67) | 0.04 | 1.05, 2.15 |
A P value of ≤0.05 is considered to be significant.
CI, confidence interval.
In vivo detection of zygomycete DNA in BAL fluid and lung tissue (qPCR-2).
The log DNA value obtained with BAL fluid (n = 15) utilizing qPCR-2 correlated well with the qCx data obtained with BAL fluid (r = 0.63, P = 0.01) (Table 3). Of note, there were five BAL samples that were positive by qPCR while being culture negative. Consistent with this finding, the qPCR assay was significantly more sensitive than culture methods (100% versus 67% [P < 0.05]) (Table 4). Both methods proved to be 100% specific.
The log DNA value resulting from the qPCR assay of lung tissue (n = 90) also correlated well with qCx data (r = 0.68; P < 0.01) (Table 3). Of the 90 lung tissue samples processed, 4 were negative by culture while being positive by qPCR assay. This resulted in no statistical difference in sensitivity between qPCR and qCx (100% versus 96%, respectively; P = 0.12) (Table 4). Again, the specificity for both methods was 100%. No infected BAL fluid sample or infected LTH sample was found to be negative by PCR but positive by culture.
In vivo detection of zygomycete DNA in plasma (qPCR-1 and qPCR-2).
The presence of species-specific DNA was detectable from serial samples of plasma from all four models of pulmonary zygomycosis. Six (46%) of 13 plasma samples, 8 (35%) of 23 plasma samples, and 6 (40%) of 15 plasma samples were positive for fungal DNA from the pulmonary models of zygomycosis caused by R. oryzae, R. microsporus, and M. circinelloides, respectively. The overall sensitivities of qPCR-1 in plasma for the detection of circulating DNA of R. oryzae, R. microsporus, and M. circinelloides were 10 of 13 (77%) rabbits and 20 of 51 (40%) samples, and those of qPCR-2 were 11 of 14 (79%) rabbits and 18 of 31 (58%) samples.
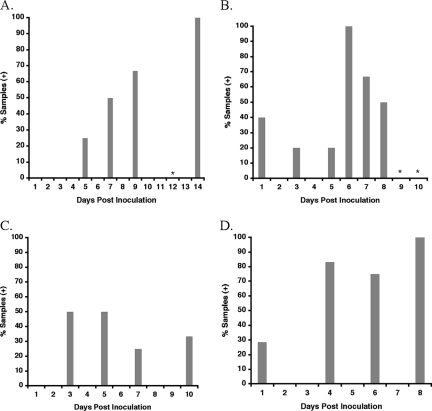
For R. oryzae and R. microsporus, the percentage of plasma samples positive for zygomycete DNA increased after day 1 postinoculation as disease progressed, with the greatest percentage of samples being detected at day 14 (Fig. 5A) and day 6 (Fig. 5B) postinoculation, respectively. The percentage of plasma samples being positive for zygomycete DNA in M. circinelloides infection (Fig. 5C) was relatively lower than the number of positive zygomycete DNA samples from plasma samples from both the R. oryzae and R. microsporus models.
FIG. 5.
Percentage of serial plasma samples positive for circulating DNA of R. oryzae (n = 13 samples) (A), R. microsporus (n = 28 samples) (B), M. circinelloides (n = 15 samples) (C), and C. bertholletiae (n = 29 samples) (D) during the course of infection with experimental invasive pulmonary zygomycosis. *, plasma samples that were qPCR negative.
Of 29 serial plasma samples collected postinoculation from the pulmonary model of C. bertholletiae infection, 18 (62%) were found to be positive by qPCR assay. The overall sensitivity of qPCR-2 was 11 of 14 (79%) animals infected with C. bertholletiae and 18 of 31 (58%) samples. The percentage of plasma samples positive for C. bertholletiae by qPCR at each time point followed a trend similar to that of R. oryzae, whereby as disease progressed, so too did the percentage of plasma samples positive for the pathogen's DNA (Fig. 5D).
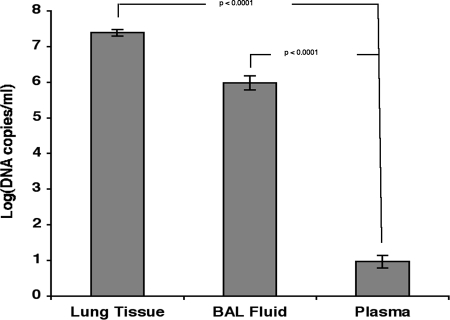
As measured by qPCR-1 and qPCR-2, the mean log DNA copy number in plasma, (0.97 log DNA copies/ml) was significantly lower than that found in either BAL fluid (5.98 log DNA copies/ml; P < 0.0001) or LTH (7.38 log DNA copies/ml; P < 0.0001) (Fig. 6).
FIG. 6.
The mean log DNA copy number in plasma (0.97 log DNA copies/ml) was significantly lower than that found in either BAL fluid (5.98 log DNA copies/ml; P < 0.0001) or lung tissue (7.38 log DNA copies/ml; P < 0.0001).
DISCUSSION
We developed quantitative real-time PCRs that targeted the most common species of Zygomycetes causing human disease in conjunction with several novel animal models of pulmonary zygomycosis. Utilizing the small but important differences in sequences within the probe region among these organisms, we were able to differentiate the genus and species by melt curve analysis. The key to the validation of these assays was the development of novel animal models of pulmonary zygomycosis caused by R. microsporus, R. oryzae, M. circinelloides, and C. bertholletiae. The qPCR assays were shown to be more sensitive than qCx in BAL fluid from animals with invasive pulmonary disease caused by these organisms. Additionally, we were able to detect the DNA of zygomycetes in plasma samples obtained from the in vivo model of invasive pulmonary zygomycosis.
Zygomycosis is an uncommon but frequently lethal infection found in immunocompromised patients. Due to the comparatively limited rate of occurrence, the availability of clinical specimens from emerging fungal pathogens is sparse. For this reason, reproducible and medically relevant animal models are critical to the timely development and validation of new detection systems, such as PCR. The animal models developed and characterized in this study reflect the microbiological, histological, and immunological characterizations of pulmonary zygomycosis in profoundly neutropenic hosts. From these animal models, biological samples such as BAL fluid, lung tissue, and plasma specimens can be obtained, all of which reflect the types of samples submitted to a clinical microbiology laboratory. With these important samples, the validation of new assays can be assessed and allow the potential implementation of new systems for prospective clinical trials and patient care.
In designing primers and probes for qPCR assays for the diagnosis of pulmonary zygomycosis, a strategy of panfungal amplification or genus-specific amplification was possible. Panfungal amplification assays have been developed to target multiple yeast and mold species from clinical specimens. Most of these assays have utilized either the small-subunit (18S) rRNA gene sequences (7, 21, 22, 30) or internally transcribed spacer (ITS) regions (ITS1 and ITS2) (17, 26, 28, 39) as targets because of the conserved nature and high copy number of these regions. As the name implies, panfungal primers are designed to amplify DNA from multiple fungal genera. Detection to the genus or species level is then accomplished by various methods such as sequencing (26, 30), hybridization with specific probes (7, 17, 39), melt curve analysis with genus-specific real-time hybridization probes (22), or performing a second nested PCR with specific primers (21). The nested PCR and probe-based assays have been developed largely to detect Candida and Aspergillus species. Little work has been done with these systems for the detection of Zygomycetes. Although these panfungal PCRs are more rapid than conventional culture methods, the addition of sequencing and hybridization steps results in a delay of 1 or 2 days before results to the species level are available.
The development of a single PCR for the diagnosis of pulmonary zygomycosis is challenging. Amplification of sequences that are highly conserved may result in the detection of other nonpathogenic, environmental organisms such as Penicillium spp. While the detection of a zygomycete from sterile body sites represents a clinically significant infection with limited therapeutic options, the differentiation of genera within the Zygomycetes is important because some genera, such as Rhizopus and Cunninghamella, may be more pathogenic than other genera (40).
Different regions of the rRNA operon have most frequently been the targets for the detection of Zygomycetes with previously reported PCRs for zygomycosis. Several reports previously described the utilization of universal fungal primers from the 18S or ITS rRNA gene regions for PCR amplification followed by sequencing (23, 46) or hybridization of the product to specific probes (7, 32, 43). Bialek et al. previously developed a seminested PCR targeting the 18S rRNA gene of Zygomycetes, followed by sequencing of any amplified product (1). This assay was reported to have a sufficient sensitivity for detection in paraffin-embedded tissues (1). Nagao et al. previously described a multiplex PCR method that could also amplify DNA from paraffin-embedded tissue specimens as well as three samples of blood from a single patient (35). Primers were designed to yield different-sized PCR products from the ITS region that correlate to four species within the genus Rhizopus. Another assay developed previously by Machouart et al. used a mix of four genus-specific sense primers and one degenerate antisense primer from the 18S rRNA gene region to detect four major genera within the Mucorales (29). Digestion of the amplified product with restriction endonucleases was then used to delineate some isolates to the species level. A more recent study by Hata and colleagues reported a real-time PCR that utilized a conserved region of the zygomycete cytochrome b gene (15).
Our goal was to develop a very rapid, highly sensitive, quantitative real-time PCR for the detection of the most common, clinically significant Zygomycetes. qPCR-1 assay was designed to detect Rhizopus spp. and Mucor spp. because these two genera comprise approximately 65% of Zygomycetes isolated at our institution and elsewhere (44).
To increase the specificity of the PCR, we used a touchdown PCR strategy for primer annealing; i.e., for the first 14 cycles, the annealing temperature was lowered from the stringent temperature of 68°C to 54°C to provide a more permissive temperature (16). Unless this procedure was used, there were some other fungal species for which a minimal PCR signal was detected. Increasing specificity in this manner diminished the analytic sensitivity for the detection of other genera such as Rhizomucor species. Although the 5′ primer will anneal to all five of the most common clinically significant Zygomycetes, the lower analytical sensitivity for Rhizomucor may be explained by four base pair mismatches between the 3′ primer and R. pusillus sequences. Mismatches present in the 3′ primer also prevented the amplification of Cunninghamella spp., Absidia spp., and other fungal species using the above-described PCR conditions. As we recognized the importance of other Zygomycetes as a cause of infection in immunocompromised patients (4, 23, 40, 44), a quantitative real-time PCR for the detection of C. bertholletiae was also developed.
An advantage of our qPCR-1 assay design is that three genera within the class Zygomycetes can be detected in a single reaction, and these can be distinguished with melt curve analysis in real time without resorting to a multiplex format. This ability to distinguish among the most common pathogens causing zygomycosis without the need for further procedures, such as sequencing, results in a faster diagnosis time. In a clinical laboratory, it is likely that the sequencing of a PCR product would add a day to the turnaround time, while our qPCR assay can be completed in approximately 2 h once the DNA has been extracted from the original sample. Furthermore, approaches that rely on less specific primers are much more likely to amplify environmental Zygomycetes or other fungal species, and this would not be apparent until sequencing or hybridization is complete (1, 23, 32, 43).
The automated extraction method reported here was previously shown to have a similar sensitivity for the extraction of DNA from Aspergillus conidia spiked into BAL fluid and blood compared to that of the manual QiaAmp tissue kit (Qiagen) (27). DNA from a broad range of fungal isolates, including R. oryzae, was also successfully amplified following automated extraction. Thus, we anticipated that the automated system would have an adequate sensitivity for the extraction of Rhizopus spp. and Mucor spp. from BAL fluid and LTH. The automated and manual DNA extraction methods showed similar reproducibilities and extraction sensitivities. For some extractions of BAL fluid, yields were better with the manual method; however, this trend was not observed with lung tissue extractions. Although the initial steps generating spheroplasts and mechanically disrupting the cells are performed manually, the majority of the processing with the MagNA Pure system is automated, and for a batch of 24 specimens, the time savings for sample preparation is approximately 1.25 h compared to the manual method. However, an additional 1.3 h of time is saved during the automated steps of the process (11). Thus, we chose to process the in vivo lung tissue and BAL fluid samples by the automated system. Many laboratories that perform molecular diagnostic tests on clinical specimens have adopted an automated extraction method to improve productivity and decrease cross-contamination (8, 14, 48). Our results support the incorporation of the MagNA Pure automated system into the processing of lung tissue and BAL fluid samples prior to qPCR for Zygomycetes.
Some inhibition was present when either the automated or manual extraction methods were used. Inhibition was much more common with LTH than with BAL fluid specimens. Since BAL fluid is mostly saline, it would be expected that more inhibitors of PCR would be present in tissue specimens.
The PCR data for our animal models suggest that qPCR may have greater sensitivity for the detection of Rhizopus spp. and Mucor spp. from clinical samples than culture. Five of 98 lung tissue specimens were positive by qPCR and negative by culture, but no specimens were positive by culture and negative by qPCR. These five samples tended to be low positive, with a mean of 4.3 log copies/ml (range, 3.4 to 5.8 log copies/ml). There was a significant correlation between qPCR and qCx results. The log copies/ml detected by PCR tended to be 2 to 3 logs greater than the log CFU/ml. This could represent both the number of copies of the 28S rRNA gene per CFU and increased sensitivity of qPCR over culture. Results from other investigators also suggest that PCR has better sensitivity than conventional methods for the detection of Zygomycetes. The detection of Zygomycetes by PCR from frozen tissue was more sensitive than Calcofluor White direct staining (46). Case reports have utilized PCR methods to detect Zygomycetes when cultures of sputum were negative (23) and from blood specimens (23, 35, 43). Prospective studies comparing PCR to culture, histology, and direct staining are lacking in the literature but are necessary to determine the diagnostic sensitivity of PCR for the detection of Zygomycetes.
Initially, we attempted to develop a single qPCR assay that would encompass all the major organisms that cause zygomycosis. However, due to the highly conserved nature of this region, we were unable to develop a pair of primers that allowed the amplification of all the major organisms causing zygomycosis without one or both of the primers also hybridizing to mammalian DNA and, more specifically, human DNA. This would have resulted in an assay with poor amplification efficiency and/or poor specificity. Due to the lethality of C. bertholletiae, we sought to develop a qPCR assay specifically targeting this pathogen.
Once the primers and probes were designed for the qPCR-2 assay, we sought to characterize the performance of the assay. This qPCR assay reliably detected between 10 and 100 copies of the amplicon construct. The assay was also highly specific for Cunninghamella spp., reflected by the lack of a FRET signal when screened against DNA from 8 yeasts, 17 filamentous fungi, and 2 sources of mammalian DNA (rabbit and human whole blood). Notably, both C. bertholletiae and the less commonly encountered C. echinulata are detected by qPCR-2 but distinguishable by melt curve analysis. For Cunninghamella elegans, a rare cause of zygomycosis (25), a BLAST search demonstrated 100% homology, with both primer and probe sets, suggesting that this organism may be detectable. Hence, while C. bertholletiae was used to develop this qPCR assay as the most commonly medically important member of the genus, C. echinulata and C. elegans may also be detectable. Specificity becomes critical in avoiding misdiagnosis, which impacts not only the initiation of therapy but also the selection of the appropriate antifungal agent or agents.
The qPCR-2 data correlated well with qCx data for both the BAL fluid and lung tissue samples. The correlation of the qPCR assay with qCx of BAL fluid would have been greater; however, the observed r value is reflective of the greater sensitivity of the qPCR assay over standard culture techniques. Five (33%) of the 15 BAL fluid samples were negative by culture. Of the five BAL samples that were negative by culture, all were positive for the organism by qPCR assay. Similarly, four lung tissue samples were negative by culture but positive by qPCR. These data suggest that conventional homogenization and culture techniques may not be optimal for open lung biopsies in patients. Furthermore, qPCR on BAL fluid may be useful for detecting deep pulmonary infection without a lung biopsy being performed.
A circulating biomarker for zygomycosis could greatly facilitate the diagnosis of this life-threatening infection. In a clinical setting, obtaining deep-tissue samples or BAL fluid from a patient with suspected zygomycosis often may not be feasible. Antigen tests that screen for serological biomarkers, such as galactomannan and (1→3)-β-d-glucan, have proven effective in the diagnosing and monitoring of other invasive fungal infections such as aspergillosis and candidemia. However, levels of galactomannan and (1→3)-β-d-glucan are very low in isolates of Zygomycetes (G.-D. J. Kurukularatne, S. Kemmerly, D. Reed, et al., presented at the Focus on Fungal Infections, Las Vegas, NV, 8 to 10 March 2006), and therefore, these assays do not detect infections caused by this family of organisms. In investigating fungal DNA as a potential biomarker, we found that we were able to detect DNA in the plasma obtained from all four models of pulmonary zygomycosis very early during the infectious event, as early as day 1 postinoculation. Although the sensitivity of the assays applied to plasma is less than those for BAL fluid and lung tissue, the frequency of positive results may be helpful in ruling in a diagnosis, but not excluding a diagnosis, of pulmonary zygomycosis. Further studies of diabetic models as well as of patients with zygomycosis are warranted.
The relative amounts of zygomycete-specific DNA recovered from plasma samples were found to be lower than those of DNA found in either lung tissue samples or BAL fluid samples obtained from the animal model. DNA from both lung tissue samples and BAL fluid samples were extracted from organisms recovered from infected tissue or lavaged from infected lung. However, DNA from plasma was non-cell associated (naked DNA). Therefore, levels recovered from plasma might be expected to be lower than levels of DNA extracted from recovered organisms in either lung tissue or BAL fluid samples collected from infected animals. Of note, DNA levels from both BAL fluid samples and tissue samples were end-point samples (samples collected postmortem). Plasma samples were serially collected from day 1 postinoculation through day 8 postinoculation. Further studies are ongoing to fully understand the kinetics of circulating fungal DNA in the host.
Our approach to utilizing this qPCR assay targeting these particular pathogens should include a strategy for identifying the susceptible population of patients, thus increasing the positive predictive value of the assay's results. A dual approach focusing on both a particular patient population that fulfills specific clinical radiological criteria and selection of the appropriate clinical samples should result in increasing the positive predictive value of this assay (6). Attention would be focused on patients who were immunocompromised and had focal pneumonia of unknown cause, thereby increasing the Bayesian prior probability of invasive fungal infection. Based on results obtained from the in vivo experiments used in validating the assays developed in our laboratory, patient samples would include BAL fluid, lung tissue, and plasma. Future studies are currently being designed to validate the potential value of these qPCR assays in a clinical setting to improve the diagnostic specificity and therapeutic monitoring of disease progression or resolution.
The quantitative capacity of qPCR-1 and qPCR-2 may allow the initial detection and potentially subsequent therapeutic monitoring of patients receiving antifungal therapy. The quantitative monitoring would be most practically implemented in the serial sampling of serum. This approach would be more practical then serial BAL. Further studies using experimental models are warranted to further understand the relationship between the therapeutic response and serum PCR signal as a quantitative biomarker for the organism burden in pulmonary zygomycosis.
Although there have been several studies that have applied PCR technology to studying zygomycetes (2, 18, 23, 42), to our knowledge, this is the first report of the systematic application of a quantitative PCR assay to the detection of pulmonary circulating zygomycete-specific DNA. The assays were shown to be both specific and sensitive in their utilization to detect and quantify infection in experimental animal models. The finding of zygomycete-specific DNA in plasma from the model of experimental zygomycete pneumonia suggests that the detection of this molecular biomarker may be a useful and practical clinical tool for the diagnosis of pulmonary zygomycosis. Investigation of the qPCR assay's potential application in a clinical microbiology laboratory setting is warranted in the hopes of improving the early and specific diagnosis and accurate treatment of zygomycosis.
Acknowledgments
We are grateful to Yvonne Shea for assistance with collecting fungal isolates and data from the Clinical Microbiology Laboratory of the Warren G. Magnuson Clinical Center of the National Institutes of Health.
Footnotes
Published ahead of print on 24 September 2008.
REFERENCES
- 1.Bialek, R., F. Konrad, J. Kern, C. Aepinus, L. Cecenas, G. M. Gonzalez, G. Just-Nubling, B. Willinger, E. Presterl, C. Lass-Florl, and V. Rickerts. 2005. PCR based identification and discrimination of agents of mucormycosis and aspergillosis in paraffin wax embedded tissue. J. Clin. Pathol. 581180-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakrabarti, A., A. Ghosh, G. S. Prasad, J. K. David, S. Gupta, A. Das, V. Sakhuja, N. K. Panda, S. K. Singh, S. Das, and T. Chakrabarti. 2003. Apophysomyces elegans: an emerging zygomycete in India. J. Clin. Microbiol. 41783-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chayakulkeeree, M., M. A. Ghannoum, and J. R. Perfect. 2006. Zygomycosis: the re-emerging fungal infection. Eur. J. Clin. Microbiol. Infect. Dis. 25215-229. [DOI] [PubMed] [Google Scholar]
- 4.Darrisaw, L., G. Hanson, D. H. Vesole, and S. C. Kehl. 2000. Cunninghamella infection post bone marrow transplant: case report and review of the literature. Bone Marrow Transplant. 251213-1216. [DOI] [PubMed] [Google Scholar]
- 5.De Hoog, G. S., J. Guarro, J. Gené, and M. J. Figueras. 2000. Atlas of clinical fungi, 2nd ed. ASM Press, Washington, DC.
- 6.De Pauw, B., T. J. Walsh, J. P. Donnelly, D. A. Stevens, J. E. Edwards, T. Calandra, P. G. Pappas, J. Maertens, O. Lortholary, C. A. Kauffman, D. W. Denning, T. F. Patterson, G. Maschmeyer, J. Bille, W. E. Dismukes, R. Herbrecht, W. W. Hope, C. C. Kibbler, B. J. Kullberg, K. A. Marr, P. Munoz, F. C. Odds, J. R. Perfect, A. Restrepo, M. Ruhnke, B. H. Segal, J. D. Sobel, T. C. Sorrell, C. Viscoli, J. R. Wingard, T. Zaoutis, and J. E. Bennett. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 461813-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Einsele, H., H. Hebart, G. Roller, J. Loffler, I. Rothenhofer, C. A. Muller, R. A. Bowden, J. van Burik, D. Engelhard, L. Kanz, and U. Schumacher. 1997. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 351353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espy, M. J., P. N. Rys, A. D. Wold, J. R. Uhl, L. M. Sloan, G. D. Jenkins, D. M. Ilstrup, F. R. Cockerill III, R. Patel, J. E. Rosenblatt, and T. F. Smith. 2001. Detection of herpes simplex virus DNA in genital and dermal specimens by LightCycler PCR after extraction using the IsoQuick, MagNA Pure, and BioRobot 9604 methods. J. Clin. Microbiol. 392233-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fahle, G. A., and S. H. Fischer. 2000. Comparison of six commercial DNA extraction kits for recovery of cytomegalovirus DNA from spiked human specimens. J. Clin. Microbiol. 383860-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francesconi, A., M. Kasai, S. M. Harrington, M. G. Beveridge, R. Petraitiene, V. Petraitis, R. L. Schaufele, and T. J. Walsh. 2007. Automated and manual methods of DNA extraction for Aspergillus fumigatus and Rhizopus oryzae analyzed by quantitative real-time PCR. J. Clin. Microbiol. 461978-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francesconi, A., M. Kasai, S. M. Harrington, M. G. Beveridge, R. Petraitiene, V. Petraitis, R. L. Schaufele, and T. J. Walsh. 2008. Automated and manual methods of DNA extraction for Aspergillus fumigatus and Rhizopus oryzae analyzed by quantitative real-time PCR. J. Clin. Microbiol. 461978-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francesconi, A., M. Kasai, R. Petraitiene, V. Petraitis, A. M. Kelaher, R. Schaufele, W. W. Hope, Y. R. Shea, J. Bacher, and T. J. Walsh. 2006. Characterization and comparison of galactomannan enzyme immunoassay and quantitative real-time PCR assay for detection of Aspergillus fumigatus in bronchoalveolar lavage fluid from experimental invasive pulmonary aspergillosis. J. Clin. Microbiol. 442475-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frater, J. L., G. S. Hall, and G. W. Procop. 2001. Histologic features of zygomycosis: emphasis on perineural invasion and fungal morphology. Arch. Pathol. Lab. Med. 125375-378. [DOI] [PubMed] [Google Scholar]
- 14.Germer, J. J., M. M. Lins, M. E. Jensen, W. S. Harmsen, D. M. Ilstrup, P. S. Mitchell, F. R. Cockerill III, and R. Patel. 2003. Evaluation of the MagNA pure LC instrument for extraction of hepatitis C virus RNA for the COBAS AMPLICOR Hepatitis C Virus Test, version 2.0. J. Clin. Microbiol. 413503-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hata, D. J., S. P. Buckwalter, B. S. Pritt, G. D. Roberts, and N. L. Wengenack. 2008. Real-time PCR method for detection of zygomycetes. J. Clin. Microbiol. 462353-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hecker, K. H., and K. H. Roux. 1996. High and low annealing temperatures increase both specificity and yield in touchdown and stepdown PCR. BioTechniques 20478-485. [DOI] [PubMed] [Google Scholar]
- 17.Hendolin, P. H., L. Paulin, P. Koukila-Kahkola, V. J. Anttila, H. Malmberg, M. Richardson, and J. Ylikoski. 2000. Panfungal PCR and multiplex liquid hybridization for detection of fungi in tissue specimens. J. Clin. Microbiol. 384186-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hopfer, R. L., P. Walden, S. Setterquist, and W. E. Highsmith. 1993. Detection and differentiation of fungi in clinical specimens using polymerase chain reaction (PCR) amplification and restriction enzyme analysis. J. Med. Vet. Mycol. 3165-75. [DOI] [PubMed] [Google Scholar]
- 19.Horger, M., H. Hebart, H. Schimmel, M. Vogel, H. Brodoefel, K. Oechsle, U. Hahn, M. Mittelbronn, W. Bethge, and C. D. Claussen. 2006. Disseminated mucormycosis in haematological patients: CT and MRI findings with pathological correlation. Br. J. Radiol. 79e88-95. [DOI] [PubMed] [Google Scholar]
- 20.Husain, S., B. D. Alexander, P. Munoz, R. K. Avery, S. Houston, T. Pruett, R. Jacobs, E. A. Dominguez, J. G. Tollemar, K. Baumgarten, C. M. Yu, M. M. Wagener, P. Linden, S. Kusne, and N. Singh. 2003. Opportunistic mycelial fungal infections in organ transplant recipients: emerging importance of non-Aspergillus mycelial fungi. Clin. Infect. Dis. 37221-229. [DOI] [PubMed] [Google Scholar]
- 21.Jaeger, E. E., N. M. Carroll, S. Choudhury, A. A. Dunlop, H. M. Towler, M. M. Matheson, P. Adamson, N. Okhravi, and S. Lightman. 2000. Rapid detection and identification of Candida, Aspergillus, and Fusarium species in ocular samples using nested PCR. J. Clin. Microbiol. 382902-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordanides, N. E., E. K. Allan, L. A. McLintock, M. Copland, M. Devaney, K. Stewart, A. N. Parker, P. R. Johnson, T. L. Holyoake, and B. L. Jones. 2005. A prospective study of real-time panfungal PCR for the early diagnosis of invasive fungal infection in haemato-oncology patients. Bone Marrow Transplant. 35389-395. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi, M., K. Togitani, H. Machida, Y. Uemura, Y. Ohtsuki, and H. Taguchi. 2004. Molecular polymerase chain reaction diagnosis of pulmonary mucormycosis caused by Cunninghamella bertholletiae. Respirology 9397-401. [DOI] [PubMed] [Google Scholar]
- 24.Kontoyiannis, D. P., G. Chamilos, S. A. Hassan, R. E. Lewis, N. D. Albert, and J. J. Tarrand. 2007. Increased culture recovery of zygomycetes under physiologic temperature conditions. Am. J. Clin. Pathol. 127208-212. [DOI] [PubMed] [Google Scholar]
- 25.Kwon-Chung, K. J., R. C. Young, and M. Orlando. 1975. Pulmonary mucormycosis caused by Cunninghamella elegans in a patient with chronic myelogenous leukemia. Am. J. Clin. Pathol. 64544-548. [DOI] [PubMed] [Google Scholar]
- 26.Lau, A., S. Chen, T. Sorrell, D. Carter, R. Malik, P. Martin, and C. Halliday. 2007. Development and clinical application of a panfungal PCR assay to detect and identify fungal DNA in tissue specimens. J. Clin. Microbiol. 45380-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loeffler, J., K. Schmidt, H. Hebart, U. Schumacher, and H. Einsele. 2002. Automated extraction of genomic DNA from medically important yeast species and filamentous fungi by using the MagNA Pure LC system. J. Clin. Microbiol. 402240-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo, G., and T. G. Mitchell. 2002. Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J. Clin. Microbiol. 402860-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Machouart, M., J. Larché, K. Burton, J. Collomb, P. Maurer, A. Cintrat, M. F. Biava, S. Greciano, A. F. Kuijpers, N. Contet-Audonneau, G. S. de Hoog, A. Gérard, and B. Fortier. 2006. Genetic identification of the main opportunistic Mucorales by PCR-restriction fragment length polymorphism. J. Clin. Microbiol. 44805-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makimura, K., S. Y. Murayama, and H. Yamaguchi. 1994. Detection of a wide range of medically important fungi by the polymerase chain reaction. J. Med. Microbiol. 40358-364. [DOI] [PubMed] [Google Scholar]
- 31.Marr, K. A., R. A. Carter, F. Crippa, A. Wald, and L. Corey. 2002. Epidemiology and outcome of mould infections in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 34909-917. [DOI] [PubMed] [Google Scholar]
- 32.Monecke, S., K. Hochauf, B. Gottschlich, and R. Ehricht. 2006. A case of peritonitis caused by Rhizopus microsporus. Mycoses 49139-142. [DOI] [PubMed] [Google Scholar]
- 33.Muller, F. M., K. E. Werner, M. Kasai, A. Francesconi, S. J. Chanock, and T. J. Walsh. 1998. Rapid extraction of genomic DNA from medically important yeasts and filamentous fungi by high-speed cell disruption. J. Clin. Microbiol. 361625-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray, P., E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller. 2007. Manual of clinical microbiology, 9th ed., vol. 2. ASM Press, Washington, DC.
- 35.Nagao, K., T. Ota, A. Tanikawa, Y. Takae, T. Mori, S. Udagawa, and T. Nishikawa. 2005. Genetic identification and detection of human pathogenic Rhizopus species, a major mucormycosis agent, by multiplex PCR based on internal transcribed spacer region of rRNA gene. J. Dermatol. Sci. 3923-31. [DOI] [PubMed] [Google Scholar]
- 36.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC.
- 37.O'Sullivan, C. E., M. Kasai, A. Francesconi, V. Petraitis, R. Petraitiene, A. M. Kelaher, A. A. Sarafandi, and T. J. Walsh. 2003. Development and validation of a quantitative real-time PCR assay using fluorescence resonance energy transfer technology for detection of Aspergillus fumigatus in experimental invasive pulmonary aspergillosis. J. Clin. Microbiol. 415676-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pagano, L., P. Ricci, A. Tonso, A. Nosari, L. Cudillo, M. Montillo, A. Cenacchi, L. Pacilli, F. Fabbiano, A. Del Favero, et al. 1997. Mucormycosis in patients with haematological malignancies: a retrospective clinical study of 37 cases. Br. J. Haematol. 99331-336. [DOI] [PubMed] [Google Scholar]
- 39.Playford, E. G., F. Kong, Y. Sun, H. Wang, C. Halliday, and T. C. Sorrell. 2006. Simultaneous detection and identification of Candida, Aspergillus, and Cryptococcus species by reverse line blot hybridization. J. Clin. Microbiol. 44876-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ribes, J. A., C. L. Vanover-Sams, and D. J. Baker. 2000. Zygomycetes in human disease. Clin. Microbiol. Rev. 13236-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richardson, M. D., and P. Koukila-Kahkola. 2007. Rhizopus, Rhizomucor, Absidia, and other agents of systemic and subcutaneous zygomycoses, p. 1839-1856. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed., vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 42.Rickerts, V., G. Just-Nubling, F. Konrad, J. Kern, E. Lambrecht, A. Bohme, V. Jacobi, and R. Bialek. 2006. Diagnosis of invasive aspergillosis and mucormycosis in immunocompromised patients by seminested PCR assay of tissue samples. Eur. J. Clin. Microbiol. Infect. Dis. 258-13. [DOI] [PubMed] [Google Scholar]
- 43.Rickerts, V., J. Loeffler, A. Bohme, H. Einsele, and G. Just-Nubling. 2001. Diagnosis of disseminated zygomycosis using a polymerase chain reaction assay. Eur. J. Clin. Microbiol. Infect. Dis. 20744-745. [DOI] [PubMed] [Google Scholar]
- 44.Roden, M. M., T. E. Zaoutis, W. L. Buchanan, T. A. Knudsen, T. A. Sarkisova, R. L. Schaufele, M. Sein, T. Sein, C. C. Chiou, J. H. Chu, D. P. Kontoyiannis, and T. J. Walsh. 2005. Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin. Infect. Dis. 41634-653. [DOI] [PubMed] [Google Scholar]
- 45.Roilides, E., C. A. Lyman, T. Sein, C. Gonzalez, and T. J. Walsh. 2000. Antifungal activity of splenic, liver and pulmonary macrophages against Candida albicans and effects of macrophage colony-stimulating factor. Med. Mycol. 38161-168. [PubMed] [Google Scholar]
- 46.Schwarz, P., S. Bretagne, J. C. Gantier, D. Garcia-Hermoso, O. Lortholary, F. Dromer, and E. Dannaoui. 2006. Molecular identification of zygomycetes from culture and experimentally infected tissues. J. Clin. Microbiol. 44340-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siwek, G. T., K. J. Dodgson, M. de Magalhaes-Silverman, L. A. Bartelt, S. B. Kilborn, P. L. Hoth, D. J. Diekema, and M. A. Pfaller. 2004. Invasive zygomycosis in hematopoietic stem cell transplant recipients receiving voriconazole prophylaxis. Clin. Infect. Dis. 39584-587. [DOI] [PubMed] [Google Scholar]
- 48.Smith, T. F., M. J. Espy, J. Mandrekar, M. F. Jones, F. R. Cockerill, and R. Patel. 2007. Quantitative real-time polymerase chain reaction for evaluating DNAemia due to cytomegalovirus, Epstein-Barr virus, and BK virus in solid-organ transplant recipients. Clin. Infect. Dis. 451056-1061. [DOI] [PubMed] [Google Scholar]
- 49.Tedder, M., J. A. Spratt, M. P. Anstadt, S. S. Hegde, S. D. Tedder, and J. E. Lowe. 1994. Pulmonary mucormycosis: results of medical and surgical therapy. Ann. Thorac. Surg. 571044-1050. [DOI] [PubMed] [Google Scholar]
- 50.Voigt, K., E. Cigelnik, and K. O'Donnell. 1999. Phylogeny and PCR identification of clinically important Zygomycetes based on nuclear ribosomal-DNA sequence data. J. Clin. Microbiol. 373957-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walsh, T. J., J. Bacher, and P. A. Pizzo. 1988. Chronic silastic central venous catheterization for induction, maintenance and support of persistent granulocytopenia in rabbits. Lab. Anim. Sci. 38467-471. [PubMed] [Google Scholar]
- 52.Walsh, T. J., C. McEntee, and D. M. Dixon. 1987. Tissue homogenization with sterile reinforced polyethylene bags for quantitative culture of Candida albicans. J. Clin. Microbiol. 25931-932. [DOI] [PMC free article] [PubMed] [Google Scholar]