Abstract
An automated platform (BeeBlot) was evaluated in parallel with the recommended protocol for the hybridization and detection steps of the Roche Linear Array human papillomavirus (HPV) genotyping test using DNA from 143 cervical specimens. Genotyping profiles showed 100% concordance between the methods, suggesting that automation could complement the Roche Linear Array for enhanced speed and reproducibility.
Infection with high-risk (HR) human papillomavirus (HPV) genotypes is a major causative factor for development of cervical cancer and its precursor lesions (2, 3, 13, 14, 22). There are approximately 40 HPV genotypes known to infect the human anogenital mucosa, which are divided into low-risk and HR types based on their implicated etiology in cervical carcinoma (8, 15). Infections with either HPV risk type may result in abnormal cell growth, though most are transient, asymptomatic, and spontaneously cleared by the immune system. However, persistent infection with HR HPV genotypes is a significant risk factor in the progression of cervical lesions from low grade into high grade and potentially to carcinoma of the cervix (11, 23).
Molecular techniques for HPV detection are widely used, with PCR-based assays providing a sensitive and noninvasive approach for monitoring the presence of active HPV infections (5, 6, 16, 17). Accurate identification of HPV genotypes is important for epidemiological studies, including monitoring persistent HR HPV infections. The Linear Array HPV (LA-HPV) genotyping test (Roche Diagnostics) offers a reliable, sensitive, and standardized approach for detecting HPV DNA in cervical specimens (4, 7, 18, 21). The LA-HPV test is a qualitative in vitro PCR-based test allowing the detection of up to 37 anogenital HPV genotypes, including the major HR types (7, 10). HPV genotyping has important clinical applications: evaluating clearance and reinfection of specific HPV types, monitoring treatment success for high-grade cervical disease, and determining HPV type prevalence in different populations for pre- and postevaluation of prophylactic HPV vaccine impact (1, 17). The LA-HPV test is a highly standardized assay, including reagents, amplification profiles, and hybridization and detection conditions for optimal sensitivity and reproducibility. The test comprises four main processes: DNA extraction, PCR amplification of target sequences, hybridization of PCR products to specific oligonulceotide probes on a nylon strip, and colorimetric detection (4, 7, 9, 18, 21). The recommended protocol for hybridization and detection involves a labor-intensive and time-consuming procedure, which could potentially cause varied reproducibility. With the aim of reducing the labor-intensiveness of the LA-HPV assay, we evaluated the BeeBlot automated platform as an alternative method for the LA-HPV hybridization and detection steps.
Cervical brush specimens (n = 143) were selected from a cohort of 1,679 specimens with different Hybrid Capture 2 results (68 negative and 75 positive specimens were selected) to assess genotyping sensitivity using extracts with low to high HPV viral loads. All specimens were collected in PreservCyt (Cytyc Corporation) between May 2001 and December 2002 from women undergoing ablative treatment for histologically confirmed cervical abnormality at the Royal Women's Hospital, Melbourne, Australia.
DNA was extracted from specimens using the MagNA Pure LC system with a modified procedure, as previously described (18). In brief, a 1-ml aliquot was pelleted, resuspended in 200 μl sterile phosphate-buffered saline, and extracted using the DNA-I protocol into 100 μl. DNA was genotyped using the reverse line-blot LA-HPV test. PCR was performed in a 100-μl volume, using 50 μl LA-HPV master mix (Roche Molecular Systems) and 50 μl DNA template, as previously described (18, 20). Seventy-five microliters of the same denatured PCR product was detected using both protocols, i.e., the air incubator manual method (19) and the BeeBlot automated method, ensuring an accurate comparison.
The BeeBlot (Bee Robotics Ltd., Gwynedd, United Kingdom) is a fully automated platform for the washing and hybridization steps required by strip-based assays, such as the LA-HPV. All reagents were prepared immediately prior to each run. Two reagent priming steps and a preheating (51.5°C) were performed prior to each detection run. A comparison of the incubation and turnaround times for the two methods is summarized in Table 1.
TABLE 1.
Comparison of incubation and turnaround times for the manual and automated detection protocols
| Protocol (no. of tests) | Step | Time (min)a | Incubation temp (°C) |
|---|---|---|---|
| Manual (24) | Hybridization | 30 | 53 |
| Ambient wash | 1 | ||
| Stringent wash | 15 | 53 | |
| Conjugate | 30 | ||
| Ambient wash | 1 | ||
| Ambient wash | 10 | ||
| Ambient wash | 10 | ||
| Ambient wash | 5 | ||
| Citrate | 5 | ||
| Substrate | 10 | ||
| Distilled water | 5 | ||
| Distilled water | 5 | ||
| Distilled water | 0 | ||
| Incubation time | 127 | ||
| Turnaround timeb | 180 (approx) | ||
| BeeBlot (48)c | Preheat | 15d | 51.5 |
| Hybridization | 30 | 51.5 | |
| Ambient wash | 0 | ||
| Conjugate | 15 | ||
| Stringent wash | 12 | 51.5 | |
| Ambient wash aspirate | 0 | ||
| Ambient wash dispense | 5 | ||
| Ambient wash | 5 | ||
| Citrate | 5 | ||
| Substrate | 10 | ||
| Distilled water | 0 | ||
| Distilled water | 0 | ||
| Distilled water | 0 | ||
| Incubation time | 97 | ||
| Turnaround time | 150 (approx) |
Buffer preparation and strip labeling times are similar for both methods.
Turnaround time includes additional hands-on and/or instrument processing times.
Times and incubation temperatures as entered into the BeeBlot instrument.
Strips can be labeled during the preheat incubation.
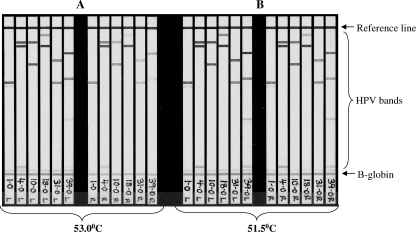
To assess whether positioning within the BeeBlot tray affected hybridization efficiency (including reproducibility of signal intensity), six specimens with multiple HPV genotypes were amplified and then hybridized at 53°C in three positions across the tray (left, center, and right). HPV and β-globin signal intensities decreased from the left side of the tray to the right side at a hybridization temperature of 53°C (Fig. 1A); this was thought to be the result of a 2°C temperature differential identified across the tray, which is within the BeeBlot operational specifications (S. Jones, Bee Robotics, personal communication). To reduce the signal disparity across the tray, subsequent hybridizations (and stringent washing) were performed at 51.5°C, with signal reproducibility markedly improving across the tray (Fig. 1B).
FIG. 1.
BeeBlot detection of LA-HPV strips using hybridization and stringent wash temperatures of 53°C or 51.5°C. The strips shown are those detected in the far left (L) and far right (R) six positions of the BeeBlot tray, as indicated on the strip label.
Following the initial BeeBlot validation, 143 specimens were assessed for a more comprehensive evaluation. Of the 143 DNA extracts, one tested negative for β-globin and HPV by both detection methods and were removed from the analysis. Collectively, specimen adequacy was 99.3% (142/143). Comparing resultant HPV genotyping profiles, a concordance of 100% (142/142) (κ = 1.0) was observed. Levels of background and signal intensities varied marginally between the detection methods, with the manual approach having slightly higher signal intensity levels as well as a minor increase in background. A sample comparison of 15 HPV strips is provided in Fig. 2. Genotyping profiles of the 143 specimens ranged from single HPV infections to multiple HPV infections, with up to seven HPV genotypes detected (Table 2). Approximately one-third of the specimens contained single HPV infections (31.7%), with 29.6% being HPV negative and 38.7% containing multiple genotypes. These findings corroborate the equivalent performances of the manual and automated detection protocols in identifying various quantities of HPV genotypes among clinical specimens.
FIG. 2.
Comparison of manual and BeeBlot LA-HPV detection. Fifteen specimens with various HPV type profiles, detected by both methods, are shown. HPV strips on the left are those detected by the manual method, while those on the right were detected using the BeeBlot.
TABLE 2.
Number of HPV genotypes per specimen detected by manual versus automated detectiona
| No. of HPV types detected | No. (%) of specimens |
|---|---|
| 0b | 42 (29.6) |
| 1 | 45 (31.7) |
| 2 | 22 (15.5) |
| 3 | 11 (7.8) |
| 4 | 9 (6.3) |
| 5 | 6 (4.2) |
| 6 | 4 (2.8) |
| 7 | 3 (2.1) |
One specimen, which was negative for both β-globin and HPV, was excluded.
HPV negativity per the LA-HPV test.
The recently released LA-HPV genotyping test provides a standardized, consistent, and rapid means for identifying HPV genotypes within clinical specimens. This permits the assessment of whether persistence of a specific HPV genotype is the basis of recurrent HPV positivity, thus denoting a substantially increased risk of cervical disease progression (11, 22, 23). Although HPV type persistence can be assessed with the LA-HPV test, there is currently no standardized recommendation for using genotype persistence for patient management. The LA-HPV hybridization and detection steps can be considered labor-intensive and time-consuming, particularly for extensive genotyping studies. Incorporating automation into these steps would greatly facilitate the HPV genotyping test, providing simplicity and improving time and labor efficiency and, most importantly, the accuracy and reproducibility of results.
The BeeBlot, as an automated processing platform for use with the LA-HPV test, was evaluated and validated in this study. This platform can accommodate from 2 to 48 samples (in multiples of 2), with a full run of 48 DNA extracts typically genotyped within a 2 1/2-h period. Signal intensities across the plate (for both HPV and β-globin) were most consistent when hybridization and stringent wash steps were performed at 51.5°C, which is imperative for assay reproducibility. Among 142 valid specimens, the HPV genotyping profiles obtained were identical using either the manual or automated procedure (concordance of 100%). To further improve the consistency and reproducibility of the LA-HPV genotyping test, particularly during interpretation of HPV bands, the use of a scanner or other such automated device to quantify band intensities would be highly beneficial, as recently reported (12).
In conclusion, these findings indicate that the BeeBlot automated platform, as a supplementary tool with the LA-HPV test, has a capacity equal in sensitivity to the current recommended detection protocol for typing single and multiple HPV infections. Laboratories, particularly those involved in large-scale HPV genotyping studies, would find automated platforms, such as the BeeBlot, simpler, less time-consuming, and potentially more reproducible than the recommended manual detection approach. With these findings, the BeeBlot automated hybridization and detection system could quite effectively be utilized for processing LA-HPV strips upon appropriate internal laboratory validation. Other automated hybridization and detection platforms for strip-based assays, such as the ProfitBlot (Tecan Group Ltd.), Genelabs AutoBlot 20/36 systems (Genelabs Diagnostics), and MedTec's AutoBlot 2000/6000 processors (Helvetica Health Care), provide similar advantages, though they also require validation prior to implementation in the laboratory.
Acknowledgments
We thank Roche Molecular Systems for providing the Linear Array HPV genotyping and detection kits, which enabled the completion of this study.
Footnotes
Published ahead of print on 10 September 2008.
REFERENCES
- 1.Bekkers, R. L., W. J. Melchers, J. M. Bakkers, A. G. Hanselaar, W. G. Quint, H. Boonstra, and L. F. Massuger. 2002. The role of genotype-specific human papillomavirus detection in diagnosing residual cervical intraepithelial neoplasia. Int. J. Cancer 102148-151. [DOI] [PubMed] [Google Scholar]
- 2.Bosch, F. X., M. M. Manos, N. Munoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman, K. V. Shah, et al. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J. Natl. Cancer Inst. 87796-802. [DOI] [PubMed] [Google Scholar]
- 3.Bosch, F. X., A. Lorincz, N. Munoz, C. J. Meijer, and K. V. Shah. 2002. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 55244-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castle, P. E., M. Sadorra, F. Garcia, E. B. Holladay, and J. Kornegay. 2006. Pilot study of a commercialized human papillomavirus (HPV) genotyping assay: comparison of HPV risk group to cytology and histology. J. Clin. Microbiol. 443915-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coutlée, F., M. H. Mayrand, D. Provencher, and E. Franco. 1997. The future of HPV testing in clinical laboratories and applied virology research. Clin. Diagn. Virol. 8123-141. [DOI] [PubMed] [Google Scholar]
- 6.Coutlée, F., D. Rouleau, A. Ferenczy, and E. Franco. 2005. The laboratory diagnosis of genital human papillomavirus infections. Can. J. Infect. Dis. Med. Microbiol. 1683-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coutlée, F., D. Rouleau, P. Petignat, G. Ghattas, J. R. Kornegay, P. Schlag, S. Boyle, C. Hankins, S. Vézina, P. Coté, J. Macleod, H. Voyer, P. Forest, S. Walmsley, the Canadian Women's HIV Study Group, and E. Franco. 2006. Enhanced detection and typing of human papillomavirus (HPV) DNA in anogenital samples with PGMY primers and the Linear Array HPV genotyping test. J. Clin. Microbiol. 441998-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Villiers, E. M., C. Fauquet, T. R. Broker, H. U. Bernard, and H. zur Hausen. 2004. Classification of papillomaviruses. Virology 32417-27. [DOI] [PubMed] [Google Scholar]
- 9.Gravitt, P. E., C. L. Peyton, R. J. Apple, and C. M. Wheeler. 1998. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J. Clin. Microbiol. 363020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gravitt, P. E., C. L. Peyton, T. Q. Alessi, C. M. Wheeler, F. Coutlée, A. Hildesheim, M. H. Schiffman, D. R. Scott, and R. J. Apple. 2000. Improved amplification of genital human papillomaviruses. J. Clin. Microbiol. 38357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho, G. Y., R. D. Burk, S. Klein, A. S. Kadish, C. J. Chang, P. Palan, J. Basu, R. Tachezy, R. Lewis, and S. Romney. 1995. Persistent genital human papillomavirus infection as a risk factor for persistent cervical dysplasia. J. Natl. Cancer Inst. 871365-1371. [DOI] [PubMed] [Google Scholar]
- 12.Jeronimo, J., N. Wentzensen, R. Long, M. Schiffman, S. T. Dunn, R. A. Allen, J. L. Walker, M. A. Gold, R. E. Zuna, M. E. Sherman, S. Wacholder, and S. S. Wang. 2008. Evaluation of linear array human papillomavirus genotyping using automatic optical imaging software. J. Clin. Microbiol. 462759-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munoz, N. 2000. Human papillomavirus and cancer: the epidemiological evidence. J. Clin. Virol. 191-5. [DOI] [PubMed] [Google Scholar]
- 14.Munoz, N., F. X. Bosch, S. de Sanjose, L. Tafur, I. Izarzugaza, M. Gili, P. Viladiu, C. Navarro, C. Martos, and N. Ascunce. 1992. The causal link between human papillomavirus and invasive cervical cancer: a population-based case-control study in Colombia and Spain. Int. J. Cancer 52743-749. [DOI] [PubMed] [Google Scholar]
- 15.Munoz, N., F. X. Bosch, S. de Sanjose, R. Herrero, X. Castellsague, K. V. Shah, P. J. Snijders, and C. J. Meijer. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348518-527. [DOI] [PubMed] [Google Scholar]
- 16.Schiffman, M., R. Herrero, A. Hildesheim, M. E. Sherman, M. Bratti, S. Wacholder, M. Alfaro, M. Hutchinson, J. Morales, M. D. Greenberg, and A. T. Lorincz. 2000. HPV DNA testing in cervical cancer screening: results from women in a high-risk province of Costa Rica. JAMA 28387-93. [DOI] [PubMed] [Google Scholar]
- 17.Snijders, P. J., A. J. van den Brule, and C. J. Meijer. 2003. The clinical relevance of human papillomavirus testing: relationship between analytical and clinical sensitivity. J. Pathol. 2011-6. [DOI] [PubMed] [Google Scholar]
- 18.Stevens, M. P., E. Rudland, S. M. Garland, and S. N. Tabrizi. 2006. Assessment of MagNA pure LC extraction system for detection of human papillomavirus (HPV) DNA in PreservCyt samples by the Roche Amplicor and Linear Array HPV tests. J. Clin. Microbiol. 442428-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevens, M. P., S. M. Garland, and S. N. Tabrizi. 2006. Human papillomavirus genotyping using a modified linear array detection protocol. J. Virol. Methods 135124-126. [DOI] [PubMed] [Google Scholar]
- 20.Stevens, M. P., S. M. Garland, E. Rudland, J. Tan, M. A. Quinn, and S. N. Tabrizi. 2007. Comparison of the Digene Hybrid Capture 2 assay and Roche Amplicor and Linear Array human papillomavirus (HPV) tests in detecting high-risk HPV genotypes in specimens from women with previous abnormal Pap smear results. J. Clin. Microbiol. 452130-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Hamont, D., M. A. van Ham, J. M. Bakkers, L. F. Massuger, and W. J. Melchers. 2006. Evaluation of the SPF10-INNO LiPA human papillomavirus (HPV) genotyping test and the Roche Linear Array HPV genotyping test. J. Clin. Microbiol. 443122-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 18912-19. [DOI] [PubMed] [Google Scholar]
- 23.Wallin, K. L., F. Wiklund, T. Angstrom, F. Bergman, U. Stendahl, G. Wadell, G. Hallmans, and J. Dillner. 1999. Type-specific persistence of human papillomavirus DNA before the development of invasive cervical cancer. N. Engl. J. Med. 3411633-1638. [DOI] [PubMed] [Google Scholar]