Abstract
This study utilized the Bordetella pertussis single-copy PCR target BP3385 as a means of confirming IS481 PCR-positive reactions with cycle threshold (CT) values of >35. IS481 PCRs with CT values of >35 cycles may represent PCR conditions where there is <1 CFU of B. pertussis per PCR.
Pertussis, or whooping cough, is a contagious respiratory disease caused by Bordetella pertussis. Despite the availability of a vaccine, pertussis remains common. Currently, there is no FDA-cleared diagnostic test for Bordetella pertussis, and laboratories have resorted to the use of “home-brewed” tests. A number of targets have been used for real-time PCR assays, yet these targets are often polymorphic and have homology to other Bordetella species. At the Ontario Public Health Laboratories, the multiple-copy target IS481 is used for the diagnosis of B. pertussis. Use of this target leads to the interpretative dilemma where high cycle threshold (CT) values (i.e., >35 cycles) may represent concentrations of DNA target of <1 genome per assay reaction. Cummings et al. (2) suggested BP3385, a gene of unknown function, as a possible diagnostic target because it is well conserved within B. pertussis and has not been observed in any other Bordetella species. The goal of this study was to evaluate the use of this single-copy PCR target as a means of confirming IS481 PCRs with CT values of >35.
Bordetella pertussis ATCC 9797 was used as a control for culture, DNA extraction, and PCR steps. Cultures were maintained on charcoal agar (Oxoid, Cambridge, United Kingdom) (10% [vol/vol] horse blood, Bordetella selective supplement, 20 mg/liter cephalexin) at 37°C in a humidified 5% CO2 incubator for up to 10 days. For real-time PCR optimization, positive control, and initial limit of detection studies, the entire gene sequence of BP3385 (GenBank accession no. EU727200) was cloned and confirmed by Sanger sequence analysis using standard M13 primers. Nasopharyngeal specimens from patients with suspected pertussis were obtained using Dacron-tipped swabs and transported in 500 μl of phosphate-buffered saline. Patient swabs were processed for culture and then subjected to rapid boiling extraction (100°C for 20 min) for real-time PCR amplification using the targets IS481 and IS1001, as described by Kösters et al. (6).
Real-time PCR targeting BP3385 used the primers BP3385_fwd (5′-GGTTTCTTCAGGCCCTAAATGG-3′) and BP3385_rev (5′-TCGTCCTCGACGTGTGTGTAG-3′) and the TaqMan MGB probe 5′-6-carboxyfluorescein-CTCTACCAACGCGCTCT-BNFQ-3′. An internal control (pAcGFP-1; Clontech, Mountain View, CA) was detected using the primers GFP_fwd (5′-AAGCTGACCCTGAAGTTCATCTG-3′) and GFP_rev (5′-AAGTCGTGCTGCTTCATGTGA-3′) and the TaqMan MGB probe 5′-VIC-CCTGAGCTACGGCGTG-BNFQ-3′. PCR mixtures contained 1× TaqMan gene expression master mix (Applied Biosystems), a 0.4 μM concentration of each primer, a 0.2 μM concentration of each probe, 5 fg pAcGFP-1, and 5 μl of template. Real-time PCR was performed using an Applied Biosystems 7900HT instrument (1 cycle of 50°C for 2 min, 1 cycle of 95°C for 15 min, and 45 cycles of 94°C for 15 s and 60°C for 1 min). The specificity of the real-time PCR assay was verified with purified genomic DNAs from 32 different viral, fungal, and bacterial pathogens, including B. parapertussis, B. bronchiseptica, B. holmesii, and B. hinzii.
The limit of detection of BP3385 was determined using Probit regression with a 95% confidence interval, using SPSS15 (SPSS Inc., Chicago, IL). Cell suspensions were 10-fold serially diluted, plated, and grown for 10 days, and the total number of viable cells was determined by direct colony counts. The remaining volume of each suspension was processed for BP3385 and IS481 PCRs.
To test for conservation of the BP3385 gene, sequence analysis was undertaken for 54 culture- and IS481 PCR-positive patient specimens. These specimens had been collected from various locations across the province of Ontario, Canada. Sequencing indicated that BP3385 was 100% conserved throughout the tested population. Furthermore, the BP3385 primers and probe did not cross-react with any other pathogens tested.
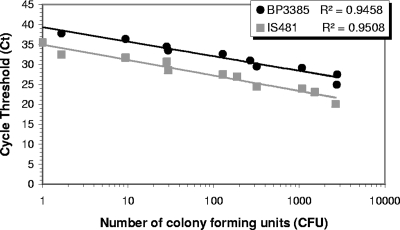
The limit of detection was a CT of 40 ± 0.7, corresponding to 5 ag of cloned BP3385 material, or 1 cell of B. pertussis, as only one copy of the targeted BP3385 gene is present in the genome (http://www.ncbi.nlm.nih.gov/BLAST). A linear correlation between CT value and CFU was observed (Fig. 1), consistent with results from the sensitivity assay using cloned BP3385. A single colony correlates to a BP3385 CT of 40 (1 copy) versus a CT of 35 for IS481 (50 to 200 copies). The detection of IS481 above a CT value of 35 represents <1 CFU, which is consistent with the results of Loeffelholz et al. (8), who determined that IS481 PCR could detect 0.3 to 0.5 CFU/reaction.
FIG. 1.
Analytical sensitivity of BP3385 PCR in comparison to IS481 PCR, using cell suspensions of B. pertussis ATCC 9797 in a real-time PCR assay.
The BP3385 real-time PCR assay was then used to test 3,155 specimens from 2007, which were previously tested for B. pertussis and B. parapertussis by IS481 PCR and culture. This panel included 2,286 negative specimens and 869 pertussis-positive samples, as well as 6 that were culture positive for B. holmesii, 18 that were B. parapertussis positive, and 8 that contained mixtures of B. pertussis and B. parapertussis. Notably, only 90 specimens were culture positive for B. pertussis. A low sensitivity of B. pertussis culture has also been observed in other public health laboratory settings (8). Of the specimens which were negative by IS481 PCR, 99.9% were also negative using the BP3385 target. The two IS481-negative, BP3385-positive specimens were retested against both targets, with the following test results: for the first specimen, the BP3385 CT was 40 and the IS481 CT was 38; and for the second specimen, the BP3385 CT was 41 and the IS481 CT was >45, i.e., there was no detectable amplification. There was less correlation with the 869 B. pertussis-positive specimens, such that only 141 had detectable amplification with BP3385. Of the BP3385-positive specimens, 95% had IS481 CT values of ≤35, largely due to the significant difference in copy number between BP3385 and IS481.
This study indicates that the BP3385 target is specific for B. pertussis. Comparison of BP3385 and IS481 reveals a striking difference in the numbers of positive real-time PCR results for the two targets. Most BP3385-negative, IS481-positive results are associated with IS481 CT values of >35, indicating that there may be <1 CFU of B. pertussis in the PCR samples of these specimens. Such low bacterial loads raise the question of whether there are true clinical cases of pertussis in these specimens, and this dilemma may be the product of several factors. For example, it is possible that an individual who has mounted an effective immune response or has already commenced antimicrobial therapy may have a true clinical case of pertussis yet exhibit a low bacterial burden or no longer carry viable bacteria (1, 7). Similarly, patient age and vaccine status may contribute to the bacterial load of an individual and, consequently, alter the sensitivity of laboratory results (3). Additionally, specimen quality may be affected by transport media and any delay between sampling and testing. Alternatively, the discrepancy may be due to poor sensitivity of the BP3385 assay with specimens containing low levels of B. pertussis. This may include specimens from B. pertussis carriers who might test positive by IS481 real-time PCR yet not meet the World Health Organization's case definition of pertussis. Several studies have shown strong indications that there is indeed a carrier status for B. pertussis (4, 5, 9). Unfortunately, the high sensitivity and lowered specificity of the IS481 real-time PCR assay raise the possibility of “false-positive” or difficult-to-interpret results, especially in instances where there is a low pretest probability. Although amplicon carryover contamination can be a source of false-positive results for PCR-based methods, it is less likely in real-time PCR due to the closed nature of the system. Furthermore, at the Ontario Public Health Laboratories, this is unlikely to be a problem due to the use of uracil-DNA glycosylase in the PCR master mix in addition to the physical separation of the different processes (e.g., extraction on a different floor, PCR reagent preparation and mixing of nucleic acid and clean reagents in different rooms, restrictions on worker flow, and constant quality control for amplicon contamination).
In conclusion, BP3385 is a sensitive and specific target for detection of B. pertussis in clinical samples. Comparison of the BP3385 (single copy) and IS481 (50 to 200 copies) targets indicates that to prevent the detection of <1 CFU or reporting of false-positive results, CT thresholds must be established for multicopy methods. In cases where the IS481 test reveals a CT of >35, reports may require special comments to allow for greater client education. Further studies are required to correlate epidemiological data with IS481 PCR results and to determine the interpretation of CT values of >35 within the context of clinical disease.
Footnotes
Published ahead of print on 10 September 2008.
REFERENCES
- 1.Bamberger, E. S., and I. Srugo. 2008. What is new in pertussis? Eur. J. Pediatr. 167133-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cummings, C. A., M. M. Brinig, P. W. Lepp, S. van de Pas, and D. A. Relman. 2004. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J. Bacteriol. 1861484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dragsted, D. M., B. Dohn, J. Madsen, and J. S. Jensen. 2004. Comparison of culture and PCR for detection of Bordetella pertussis and Bordetella parapertussis under routine laboratory conditions. J. Med. Microbiol. 53749-754. [DOI] [PubMed] [Google Scholar]
- 4.He, Q., H. Arvilommi, M. Viljanen, and J. Mertsola. 1999. Outcomes of Bordetella infections in vaccinated children: effects of bacterial number in the nasopharynx and patient age. Clin. Diagn. Lab. Immunol. 6534-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klement, E., L. Uliel, I. Engel, T. Hasin, M. Yavzori, N. Orr, N. Davidovitz, N. Lahat, I. Srugo, E. Zangvil, and D. Cohen. 2003. An outbreak of pertussis among young Israeli soldiers. Epidemiol. Infect. 1311049-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kösters, K., M. Riffelmann, and C. H. Wirsing von Konig. 2001. Evaluation of a real-time PCR assay for detection of Bordetella pertussis and B. parapertussis in clinical samples. J. Med. Microbiol. 50436-440. [DOI] [PubMed] [Google Scholar]
- 7.Lind-Brandberg, L., C. Welinder-Olsson, T. Lagergard, J. Taranger, B. Trollfors, and G. Zackrisson. 1998. Evaluation of PCR for diagnosis of Bordetella pertussis and Bordetella parapertussis infections. J. Clin. Microbiol. 36679-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loeffelholz, M. J., C. J. Thompson, K. S. Long, and M. J. Gilchrist. 1999. Comparison of PCR, culture, and direct fluorescent-antibody testing for detection of Bordetella pertussis. J. Clin. Microbiol. 372872-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srugo, I., D. Benilevi, R. Madeb, S. Shapiro, T. Shohat, E. Somekh, Y. Rimmar, V. Gershtein, R. Gershtein, E. Marva, and N. Lahat. 2000. Pertussis infection in fully vaccinated children in day-care centers, Israel. Emerg. Infect. Dis. 6526-529. [DOI] [PMC free article] [PubMed] [Google Scholar]