Abstract
The involvement of two R2R3-MYB genes from Pinus taeda L., PtMYB1 and PtMYB8, in phenylpropanoid metabolism and secondary cell wall biogenesis was investigated in planta. These pine MYBs were constitutively overexpressed (OE) in Picea glauca (Moench) Voss, used as a heterologous conifer expression system. Morphological, histological, chemical (lignin and soluble phenols), and transcriptional analyses, i.e. microarray and reverse transcription quantitative PCR (RT-qPCR) were used for extensive phenotyping of MYB-overexpressing spruce plantlets. Upon germination of somatic embryos, root growth was reduced in both transgenics. Enhanced lignin deposition was also a common feature but ectopic secondary cell wall deposition was more strongly associated with PtMYB8-OE. Microarray and RT-qPCR data showed that overexpression of each MYB led to an overlapping up-regulation of many genes encoding phenylpropanoid enzymes involved in lignin monomer synthesis, while misregulation of several cell wall-related genes and other MYB transcription factors was specifically associated with PtMYB8-OE. Together, the results suggest that MYB1 and MYB8 may be part of a conserved transcriptional network involved in secondary cell wall deposition in conifers.
Introduction
The plant cell wall is a composite assembly of complex polymers that confer structure and provide protection to the cell and to the whole plant. The major polymers of the primary cell wall include cellulose, hemicelluloses, and pectins (Cosgrove, 1999). The secondary cell wall that is formed mostly in vascular cells and fibres also contains a large proportion of lignin. In trees, lignin synthesis is of major importance because of the production of wood which is typically made up of 20–30% lignin on a dry weight basis. Therefore, the formation of secondary xylem (i.e. wood) entails the partitioning of a significant proportion of fixed carbon resources into the synthesis of lignin-building blocks through the phenylpropanoid pathway (Amthor, 2003; Boerjan et al., 2003). Although the enzymes and genes of phenylpropanoid and monolignol biosynthetic pathways have been extensively studied (Humphreys and Chapple, 2002; Boerjan et al., 2003), the mechanisms regulating their expression are still a matter of debate. In addition, molecular mechanisms regulating metabolic flux through the phenylpropanoid biosynthetic pathway are very complex and, to date, remain unclear.
Recent experimental evidence indicates that monolignol synthesis genes are under transcriptional control (Rogers and Campbell, 2004). Among the different classes of transcription factors (TFs) directly or indirectly implicated in lignification, R2R3-MYBs are strong candidates for the regulation of phenylpropanoid enzymes and monolignol biosynthesis (Rogers and Campbell, 2004; Groover and Robischon, 2006). The R2R3-MYBs form one of the largest families of TFs in plants. Numerous studies characterizing their molecular function have assigned diverse roles to these TFs, including cell fate or organ identity (Rogers and Campbell, 2004; Paux et al., 2005; Chen et al., 2006; Groover and Robischon, 2006). R2R3-MYBs also govern many aspects of secondary metabolism in Arabidopsis (Vom Endt et al., 2002) as well as in angiosperm and gymnosperm trees (Rogers and Campbell, 2004; Groover and Robischon, 2006).
The involvement of R2R3-MYBs in the transcriptional control of phenylpropanoid and flavonoid metabolic pathways has been well documented in plant model systems. Different R2R3-MYBs have been shown to bind the promoters of most genes in these pathways (Rogers and Campbell, 2004); they may bind either alone (Patzlaff et al., 2003a, b; Gomez-Maldonado et al., 2004; Goicoechea et al., 2005), or in heteroduplex with bHLH proteins (Goff et al., 1992; Debeaujon et al., 2003) or with WD-40 proteins (Koes et al., 2005; Morita et al., 2006). In woody species, however, only a few functional studies have addressed this question for MYBs. For example, the poplar gene PttMYB21a was proposed to be a negative regulator of CCoAOMT expressed in vascular tissues (Karpinska et al., 2004). On the other hand, the genes PtMYB1 and PtMYB4 from loblolly pine (Patzlaff et al., 2003a, b; Gomez-Maldonado et al., 2004) and EgMYB2 from eucalyptus (Goicoechea et al., 2005) appear to be transcriptional activators of genes encoding lignin synthesis enzymes. These three MYBs are preferentially expressed in developing xylem tissues, bind AC elements, and activate transcription from lignin biosynthetic gene promoters in transient assays in yeast or plant cells (Patzlaff et al., 2003a, b; Gomez-Maldonado et al., 2004; Goicoechea et al., 2005). Overexpression of PtMYB4 resulted in ectopic lignification in tobacco (Patzlaff et al., 2003b) and in Arabidopsis (Newman et al., 2004). Overexpression of EgMYB2 in tobacco plants leads to altered lignin structure, to thicker secondary cell walls, and to the up-regulation of lignin-related genes (Goicoechea et al., 2005). Although data from these studies and others implicated MYBs in the lignification of tree woody tissues, knowledge is still limited about the number and the respective roles of MYB TFs during wood formation.
This report represents overexpression experiments carried out with two Pinus taeda R2R3-MYB genes, namely PtMYB1 and PtMYB8, aiming to develop new insights into the involvement of MYBs in gymnosperm trees. PtMYB1 has been hypothesized to regulate lignin biosynthesis in differentiating xylem because of its ability to bind AC elements and to activate transcription from the phenylalanine ammonia-lyase (PAL) promoter (Patzlaff et al., 2003a). This hypothesis remained to be tested in planta. PtMYB8 was investigated because its closest homologue in spruce, PgMYB8, had transcript accumulation that was strongly preferential to secondary xylem and that increased during compression wood formation (Bedon et al., 2007). Overexpression was preferred to knockout/knockdown analysis since it is less affected by functional redundancy, which can be especially acute for TFs from large gene families such as MYBs (Schwechheimer et al., 1998; Zhang, 2003). This problem has been recently underlined for T-DNA knockout lines of AtMYB46 where no phentotypes were observed (Zhong et al., 2007). In addition, conifers possess a very large genome with a larger proportion of multigenic families compared with angiosperms (Kinlaw and Neale, 1997), which may add another level of complexity for the design of knockout/knockdown experiments. For this report, the use of spruce [Picea glauca (Moench) Voss, a member of the Pinaceae] for transformation provided an expression system that is taxonomically much closer to pine than Arabidopsis and tobacco employed in previous reports (Patzlaff et al., 2003a, b). The extent of ectopic secondary cell-wall deposition that is reported in transgenic spruce plantlets overexpressing these MYBs as well as their microarray, and reverse transcription quantitative PCR (RT-qPCR) transcript profiles helped to assess and compare the putative involvement of PtMYB1 and PtMYB8 in lignification and secondary cell wall biogenesis.
Materials and methods
Vector construction and spruce transformation
PtMYB1 (AY356372; Patzlaff et al., 2003a) and PtMYB8 (DQ399057; Bedon et al., 2007) cDNA from Pinus taeda L. (Loblolly pine) were used to conduct gain-of-function experiments in Picea glauca (Moench) Voss (white spruce). Gain of function was obtained by inserting the full-length cDNA in front of the maize ubiquitin promoter (Christensen et al., 1992), followed at the 3′ end by the 35S terminator. This constitutive expression vector was obtained by modifying the pRT106 (Topfer et al., 1993) plant expression vector. For Agrobacterium tumefaciens-mediated stable transformation, cassettes were digested by HindIII and cloned into pCAMBIA1305.2 (www.cambia.org). The resulting plasmid was then transferred into the A. tumefaciens strain C58 pMP90 (Koncz and Schell, 1986).
The white spruce embryogenic line Pg653 was used in the present study, and was initiated and maintained as described by Klimaszewska et al. (2001). Genetic transformations were carried out also as described in Klimaszewska et al. (2004). Once co-cultivated, explants were decontaminated from A. tumefaciens with cefotaxim and transferred onto fresh medium containing cefotaxim and kanamycin. Kanamycin-resistant embryonal tissues were screened for positive X-gluc staining (Klimaszewska et al., 2004) and assayed for PtMYB1 and PtMYB8 mRNAs accumulation by RT-qPCR (see below). Transgenic lines (each representing an independent transformation event) exhibiting a range of PtMYB mRNAs levels were selected for somatic embryo maturation and plantlet production.
Transgenic lines and culture conditions used for somatic plantlet production
For in vitro monitoring, somatic embryos were produced from selected transgenic lines overexpressing PtMYB1 (lines 4, 12, 14, 24, and 26) and PtMYB8 (lines 1–13) as well as wild-type and pCAMBIA control lines; 50–100 embryos per transgenic line were germinated according to Klimaszewska et al. (2004) and the experiment was repeated twice or three times. When allowed by the phenotype, 10- to 14-week-old transgenic plantlets were transplanted in a mix of moss, vermiculite, and turface (ratio 4:2:1, v/v/v) and grown in a mist environment for 15 d before being transferred to the greenhouse under a photoperiod of 16 h light at 24 °C, and 8 h dark at 20 °C.
For the microarray experiments, somatic embryos were germinated for 3 weeks on MLVG medium supplemented with 58 mM sucrose (Klimaszewska et al., 2004). Two transgenic lines per transgene (lines 4 and 14 for PtMYB1, and lines 1 and 2 for PtMYB8), as well as four control lines (Pg653), were used in a randomized complete block design, with four biological replicates per line and 50 whole plantlets per replicate.
Plantlet growth quantification, tissue sampling, and histology
Image capturing was used for plantlet growth quantification. The length of hypocotyl and root was determined by using ImageJ 1.32 Software (W Rasband, National Institute of Health, USA; http://rsb.info.nih.gov/ij/). Elongations were calculated from five replicates, with 6–10 plantlets per replicate, and the experiment was repeated twice. Afterwards, tissues of in vitro plantlets were sampled as follows: roots and hypocotyls were rapidly separated on the medium by using a scalpel and were immediately fixed as described below when used for histology, or frozen into liquid nitrogen and stored at –80 °C for further molecular analysis.
For histology, the tissue samples (three plantlets per transgenic line per replicate, with five replicates) were fixed for 24 h under low vacuum in 2% (v/v) paraformaldehyde, 3% (v/v) glutaraldehyde, 0.1 M cacodylate buffer (pH 7.2) supplemented with 1 mM CaCl2, and 1% (w/v) sucrose. Samples were dehydrated in graded ethanol series and toluene, before infiltration with paraffin for 7 d. Thin sections (5 μm) were prepared by using a microtome. Paraffin-free sections were transferred to water and, after incubation in the mordant 2% (w/v) ZnCl2, were stained in Sharman's safranin O–orange G–tannic acid (Sharman, 1943). All sections were observed under a Zeiss Axioskop microscope (Jena, Germany) fitted with a digital camera.
RNA extraction, cDNA synthesis, and real-time RT-qPCR analysis
RNA extraction, cDNA synthesis and real-time RT-qPCR analyses were performed as described in Bedon et al. (2007) with minor modifications. Briefly, total RNA samples were obtained by using a Mixer Mill MM300 engine (Retsch, Germany) to grind tissue (from spruce and pine) in 1.5 ml Ependorf microtubes, and were extracted following the procedure of Chang et al. (1993). Total RNA samples were treated with amplification grade DNase I (Invitrogen, Carlsbad, CA, USA) and purified using RNeasy columns (Qiagen). cDNAs were synthesized from 1 μm of purified RNA using SuperScript II (Invitrogen) according to the manufacturer's instructions. For RT-qPCR quantification of each target RNA, a 5-fold dilution of cDNA mixture was used as template. Thermocycling, conducted using an Opticon2 DNA Engine (MJ Research Inc.), was initiated by a 2 min incubation at 95 °C followed by 35 cycles (95 °C for 10 s; 55 °C for 40 s; 72 °C for 6 s) with a single fluorescent reading taken at the end of each cycle. A standard dilution series covering five orders of magnitude were prepared for the target and reference genes from 1 ng μl−1 PCR amplicon of each cDNA to produce solutions covering 10−1 to 10−6 ng μl−1. All mRNA levels were calculated from threshold cycle values, normalized with respect to the transcript level of ELONGATION FACTOR 1-alpha (EF1a with 100% identity to Picea abies AJ132534) and expressed as a ratio relative to the control. For endogenous spruce MYB genes, absolute quantity of transcripts was calculated using standard curves as described by Rutledge and Côté (2003). Technical duplicates were performed for each sample amplification to assess variation in RT-qPCR data; a mean variation in the threshold cycle of 0.31±0.10 and 0.36±0.07 (corresponding to 23.6% and 28.3% of the fold change, respectively) was obtained between technical duplicates for sequences related to cell wall and secondary metabolism, and for MYB sequences, respectively. The means of these duplicates were used to calculate fold change ratios for each biological replicate, the mean ratio being obtained from three or four biological replicates. The specific primer pairs of target and reference genes, designed using Primer3 software (Rozen and Skaletsky, 2000), are given in Table S1 (see Supplementary data available at JXB online).
Microarray experiment
All the information regarding microarray manufacture and quality control are detailed in Appendix S1 (see Supplementary data available at JXB online). The microarray experiment was designed in total compliance with MIAME guidelines. Briefly, total RNA was extracted as described above. For each sample, 1.0 μg of total RNA was used for indirect RNA amplification performed with the Superscript™ Indirect RNA Amplification System kit (Invitrogen) according to the manufacturer's instructions. Dye coupling was performed on 5 μg of amplified RNA with either Alexa Fluor® 555 or Alexa Fluor® 647 reactive dyes (Invitrogen).
Pre-hybridization and hybridization protocols are described in Appendix S1 (see Supplementary data available at JXB online). Eight hybridizations (including four dye swaps) were carried out for each of the PtMYB1 and PtMYB8 versus wild-type (WT) comparisons. Array images were scanned using ScanArray Express (Perkin Elmer) and image analysis was performed with Quantarray software (Packard BioSciences, version 3.0, 2001).
Statistical analysis of microarray data
A complete list of microarray data from PtMYB1-OE and PtMYB8-OE in spruce is presented in Table S2 (see Supplementary data available at JXB online). Data were analysed in R (Ihaka and Gentleman, 1996), mainly using the BioConductor suite of packages (Gentleman et al., 2004). Spot intensities were analysed with the LIMMA package from Bioconductor (Smyth, 2005). Data normalization was performed using the composite method based on Lowess curves (Yang et al., 2002). Normalized data were then statistically analysed using the linear model and empirical Bayes analyses in LIMMA. The results were corrected using the Benjamini and Hochberg (1995) method of false discovery rate (1%). In this study, the focus was placed on differentially expressed genes that gave a P value <0.01 (from the LIMMA), and met a log2 ratio threshold of 0.8 (1.75-fold change) between WT and PtMYB-OE transgenics. For each transgenic construct, data from the two different lines were analysed separately and only the differentially expressed genes identified in both lines for a given transgene were considered (i) to reduce the impact of positional effects of the transgene, and (ii) to ensure that the differential expression was not the effect of only one transgenic line. Therefore, genes showing differential expression for only one transgenic line were not retained.
Soluble phenolic metabolite analysis
Spruce hypocotyl tissue from 7-week-old plantlets (∼20 mg) was suspended in 1.5 ml of methanol:water:HCl (48.5:48.5:1, v/v/v), pulverized in a cell disrupter (FastPrep) at high power for 20 s, and then extracted for 4 h at 50 °C in a heating block. The homogenate was centrifuged for 10 min at 18900 g, and the supernatant equally divided into two 600 μl aliquots. Subsequently, 1 ml of distilled water was added to methanolic extracts followed by an equal volume of ethyl-ether. After mixing thoroughly, the ether phase was retained. This extraction was performed twice and the ether phases were pooled. The ether phase was concentrated to dryness in a speedvac and resuspended in 100 μl methanol before injection in a Summit HPLC system (Dionex) fitted with a 0.2×150 mm Pursuit column (Waters; 5 μm particle size), an autosample, and a photodiode array detector. The elution at a rate of 1 ml min−1 was performed at 45 °C using a linear gradient from 100% eluant A (CH3COOH, 5%) to 80% eluant B [CH3COOH (20%):CH3CN, 75:25, v/v] over 60 min, followed by a 10 min wash with 100% B, and 10 min reacclimatizing with 100% A.
Acetyl bromide (AcBr) lignin determination
Samples were first produced to generate a standard curve for lignin determinations via the AcBr method as follows. Oven-dried 2-year-old spruce stems (Pg653) were ground in a Wiley mill to pass through a 40-mesh screen, and then soxhlet-extracted with acetone for 24 h. Holocellulose was purified from the extractive free wood by reacting 200 mg of ground wood with 1 ml of NaClO2 solution (400 mg 80% sodium chlorite, 4 ml distilled water, 0.4 ml glacial acetic acid) in a 25 ml round-bottom flask maintained at 90 °C in an oil bath. An additional millilitre of NaClO2 solution was added every 30 min and the samples removed to a cold water bath after 2 h. Samples were then filtered through a coarse crucible and dried overnight. Holocellulose composition was determined gravimetrically and characterized for residual lignin content by Klason lignin determination. For lignin isolation, ground, extract-free 2-year-old spruce wood was ball milled as per Stewart et al. (2006) and lignin was isolated according to Björkmann (1956). Isolated holocellulose and lignin samples were then recombined in varying ratios, ranging from 100:0 to 0:100, and validated by Klason analysis, to produce a standard curve for the AcBr technique.
Seven-week-old hypocotyls from transgenic and wild-type spruce were ground to a powder in a mortar and pestle with liquid nitrogen; they were subsequently pre-extracted overnight in hot acetone to remove free phenolics prior to lignin determination using the AcBr method. Acetone-extracted samples, as well as the standard curve mixtures (∼5 mg), were accurately weighed into Teflon-capped glass vials, in which 0.5 ml of AcBr solution was added [25% (v/v) AcBr in glacial acetic acid]. Samples were incubated in a heating block at 50 °C for 2 h with occasional mixing. After 2 h, the solution was transferred to a 10 ml volumetric flask with ∼5 ml of glacial acetic acid and 2 ml of 2 M NaOH. Then, 0.35 ml of 0.5 M hydroxylamine was added. The volume was made up to exactly 10 ml with glacial acetic acid and the solution thoroughly mixed. The hypocotyls and standard curve solution were then read at 280 nm and lignin content interpreted.
Results
PtMYB1 and PtMYB8 display overlapping transcript profiles preferential to differentiating secondary xylem
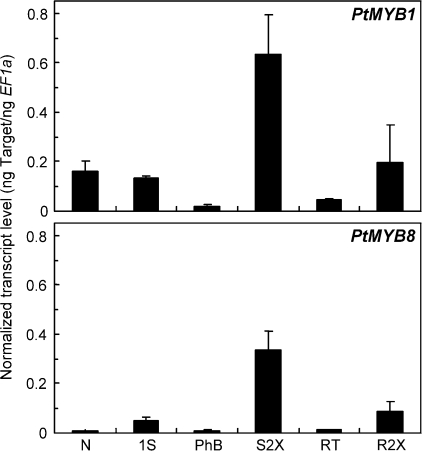
Transcript abundance of PtMYB1 and PtMYB8 was surveyed in several organs or tissues from 2-year-old Pinus taeda trees by using real time RT-qPCR analysis. As shown in Fig. 1, PtMYB1 and PtMYB8 transcripts preferentially accumulated in S2X and R2X, congruent with a role in xylem differentiation. The results also showed that PtMYB1 transcript levels in those tissues are higher than the ones of PtMYB8. Interestingly, the expression profiles of these two MYB genes were similar to those reported for their closest homologues from spruce, PgMYB1 and PgMYB8 (Bedon et al., 2007). For these pine and spruce pairs of MYB sequences, pair-wise optimal alignments gave amino acid identities of 99.1% and 98.3% for the DBD and of 87.1% and 93.1% for the complete coding sequence (Bedon et al., 2007).
Fig. 1.
Transcript profiles of PtMYB1 and PtMYB8 in pine. Total RNA was isolated from various tissues of 2-year-old Pinus taeda trees. Transcript levels were determined by RT-qPCR from three biological replicates and were normalized relative to EF1-alpha expression level. Error bars represent standard deviation. N, Needle; 1S, primary shoot, corresponding to elongating shoot (no apical bud) with no sign of secondary growth; PhB, phloem-bark; S2X, shoot secondary xylem; RT, root tip; R2X, root secondary xylem.
PtMYB1- and PtMYB8-OE in spruce leads to ectopic secondary cell wall deposition
Transgenic spruces overexpressing (OE) PtMYB1 and PtMYB8 were generated to gain insight into their role as potential regulators of lignification. Five PtMYB1-OE lines and 13 PtMYB8-OE lines with strong transgene expression (determined by RT-qPCR analysis of transgene RNA levels, not shown) were selected for in vitro growth monitoring and phenotypic analyses.
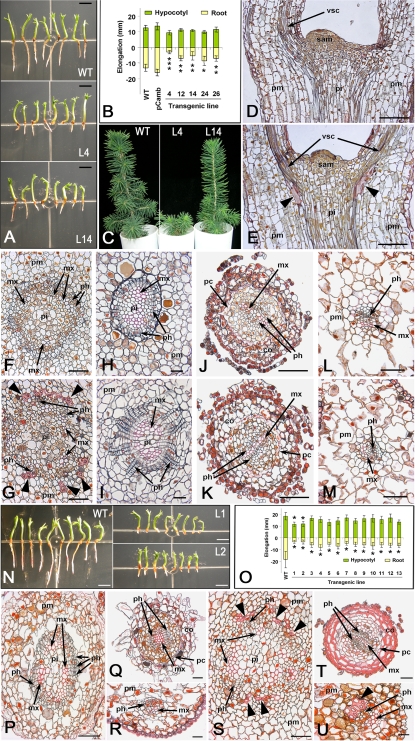
The PtMYB1 transgenic spruce plantlets produced roots that were 30–80% shorter (mean decrease of 40%) depending on the transgenic line compared with WT and pCAMBIA controls (Fig. 2A, B). PtMYB1-OE plantlets survived subsequent transfer to soil but their growth was moderately to severely delayed, particularly for line 4 (Fig. 2C). Histological observations of PtMYB1 transgenic plantlets provided evidence of ectopic secondary cell wall deposition in a few cells displaying the pink colour that is diagnostic of lignified cell walls following Sharman's staining (Sharman, 1943). Significant cell wall thickening characteristic of sclerenchyma-like elements was observed among the parenchyma cells, located close to the shoot apical meristem (Fig. 2E) and peripheral to vascular tissues (Fig. 2G); this phenomenon was never observed in WT control plantlets (Fig. 2D, F). The hypocotyls of PtMYB1 transgenic plantlets also had altered vascular radial patterning as the phloem zone was expanded and contained longer files of phloem cells (Fig. 2I) compared with the WT control (Fig. 2H). No unusual secondary cell wall deposition was noticed in the root or needles of PtMYB1 overexpressors (Fig. 2K, M) compared with the WT control (Fig. 2J, L).
Fig. 2.
Phenotypes induced by PtMYB1 and PtMYB8 overexpression in spruce. (A–M) PtMYB1-OE phenotypes: (A) 7-week-old in vitro plantlets in wild type (WT) and two independent PtMYB1-OE transgenic lines (L4, L14); (B) root and hypocotyl growth in 7-week-old plantlets produced from five independent PtMYB1-OE lines [significant differences in elongation between PtMYB1-OE lines and control one are indicated by asterisk according to Student's t-test at a level of 0.05 (*), 0.01 (**), or 0.001 (***)]; (C) plantlet morphology in wild type (WT) and two independent transgenic lines (L4, L14) of PtMYB1-OE transgenic spruce after transfer to soil; (D–M) 5 μm sections in hypocotyl, root, and needle of WT (D, F, H, J, L) and PtMYB1-OE (E, G, I, K, M) in vitro plantlets. Longitudinal (D, E) and cross- (F, G) sections in hypocotyl of 16-d-old in vitro plantlets. Sclerenchyma-like elements around the vascular cylinder are shown (black arrowheads) by pink staining characteristic of lignified cell walls (Sharman, 1943). Cross-sections in hypocotyl (H, I), root (J, K), and needle (L, M) are of 7-week-old plantlets. Supernumerary phloem cells can be observed in PtMYB1-OE section (I). No ectopic lignification was observed in root and needle of PtMYB1-OE transgenic spruce (K, M) compared with the control (J, L). (N–U) PtMYB8-OE phenotypes: (N) 7-week-old in vitro plantlets in WT and PtMYB8-OE independent transgenic lines (L1, L2); (O) root and hypocotyl growth in 7-week-old plantlets from 13 independent transgenic lines; no plantlet survived transfer to soil. Significant differences in elongation between PtMYB8-OE lines and the control one are indicated by an asterisk according to Student's t-test at a level of 0.01 (*). (P–U) Sections (5 μm) of paraffin-embedded hypocotyl (P, S), root (Q, T), and needle (R, U) in wild-type (P–R) and PtMYB8-OE (S–U) in vitro plantlets. Sclerenchyma-like elements around the vascular cylinder can be seen in the PtMYB8-OE transgenic (black arrowheads) in hypocotyl (S) and needle (U). Lignified cell wall staining (pink) was observed in parenchyma (S) and cortical (T) cells only in the transgenics. All histological sections were stained in safranin O–orange G–tannic acid after mordanting in 2% ZnCl2 (Sharman, 1943). Scale bars correspond to 5 mm (A, N), 50 μm (D, E, L, M, P, Q, S, T), 25 μm (F, G, R, U), 20 μm (H, I), and 100 μm (J, K). mx, metaxylem; ph, phloem; pm, parenchyma; pi, pith; co, cortex; pc, pericycle; sam, shoot apical meristem; vsc, vascular cells.
In PtMYB8 transgenic plantlets, the root growth was strongly reduced compared with the untransformed control (Fig. 2N), with a mean decrease in length of 68% (Fig. 2O). Two PtMYB8 transgenic lines also developed shorter hypocotyls (L1 and L2, Fig. 2N, O). In vitro development of PtMYB8-OE plantlets was poor, and none of them survived the subsequent transfer to soil. Histological analyses using Sharman's staining provided evidence of extensive ectopic secondary cell wall thickening (pink staining) in PtMYB8 transgenics after 7 weeks in germination. Sclerenchyma-like elements were observed among the parenchyma cells, located in the periphery of vascular tissues (Fig. 2S). The pink staining also revealed cell wall thickening in root cortical cells (Fig. 2T), as well as the presence of sclerenchyma-like elements in the needle (Fig. 2U). In WT control plantlets, the staining was restricted to vascular elements in the hypocotyl, root, and needle (Fig. 2P–R).
Lignin and phenolics in PtMYB1 and PtMYB8 overexpressing spruce
Secondary cell walls are normally composed of a large proportion of lignin (Campbell and Sederoff, 1996; Amthor, 2003). In light of the observed altered secondary cell wall deposition in PtMYB1- and PtMYB8-OE transgenic spruces (Fig. 2), lignin content and phenolic compound profiles were quantified. As a consequence of the small sample size, lignin determinations used the AcBr technique on 7-week-old hypocotyls. Both the PtMYB1 and PtMYB8 overexpressors had significant increases in lignin content, which ranged from 10.1% to 11.2% increase in PtMYB1-OE, and 5.3% to 12.1% in PtMYB8-OE, compared with controls (Fig. 3A). The amount of soluble phenolic compounds that could be recovered by methanol extraction from PtMYB1 and PtMYB8 transgenic and wild-type plantlet tissues was also examined. Visual inspection of the HPLC chromatograms clearly indicates that there is an overall decrease in the amount of low molecular weight phenolics in both transgenics compared with WT (Fig. 3B).
Fig. 3.
Lignin content and free phenolic profiles resulting from PtMYB1 and PtMYB8 overexpression in spruce. (A) Lignin content (± SD) of 7-week-old hypocotyls in wild type (WT) and transgenic spruce overexpressing PtMYB1 (L4, L14), and PtMYB8 (L1, L2). Lignin content was quantified by the AcBr method, expressed as a percentage of dry weight, acetone-extracted whole hypocotyls, and was calculated from three biological replicates per line and 10 plantlets per replicate. Asterisks indicate that transgenic means were significantly different from wild type, according to Student's t-test at P ≤0.05 (*) and P ≤0.01 (**). (B) HPLC profiles of low molecular weight phenolic compounds in 7-week-old wild-type (WT), and PtMYB1 and PtMYB8 overexpressing spruce (L4 and L1, respectively). IS, 3,4,5-Trimethoxycinnamic acid as internal standard.
PtMYB1- and PtMYB8-OE have overlapping impact on spruce transcriptome
As described above, similarities were observed in the PtMYB1 and PtMYB8 overexpression phenotypes, including ectopic secondary cell wall deposition. However, the phenotypes conferred by the two transgenes were clearly distinct, implying that the functions of these two genes may not be completely redundant. Transcript profiling was carried out with custom 9K cDNA microarrays in order to further characterize similarities and differences in the effects of the two MYB genes. Two independent transgenic lines per construct were used for comparison with wild-type background. The focus was placed on statistically significant genes that gave a 1.75-fold difference or greater (log2 ratio of 0.8) between the transgenic and controls (P value <0.01; false discovery rate, 1%). Each transgenic line was analysed separately: for PtMYB1, the number of up- and down-regulated sequences were, respectively, 52 and 35 for line 4, and 37 and 16 for line 14; for PtMYB8, the number of up- and down-regulated sequences were, respectively, 107 and 187 for line 1, and 89 and 159 for line 2. However, only those genes that had differential transcript levels in both lines for each MYB transgenic were considered, so as to reduce the potential influence of position effects of the transgene. Based on these criteria, the overexpression of PtMYB1 had a moderate impact on the spruce transcriptome with a total of 34 up-regulated and 12 down-regulated genes. A stronger effect on spruce transcriptome was observed in PtMYB8-OE, with a total of 79 up-regulated and 138 down-regulated genes. Functional annotations obtained from the blastx procedure against Uniref100 and a search against PGI5.0 and PFAM databases indicate that a large proportion of differentially expressed genes had a predicted function that could be linked to cell wall biogenesis, secondary metabolism (phenylpropanoid, flavonoid, and terpenoid) and related pathways (shikimate and S-adenosyl methionine), as well as stress and detoxification (Table 1; see Table S2 in Supplementary data available at JXB online). Many genes with unknown function were also misregulated in PtMYB1 (19%) and PtMYB8 (31%) overexpressors (see Table S2 in Supplementary data available at JXB online). Overlapping and distinct responses in both transgenics are described below.
Table 1.
Selected genes differentially expressed in PtMYB1- and PtMYB8-OE transgenic spruce
 |
Overlapping response in terms of the pattern of differential expression was observed when comparing PtMYB1 and PtMYB8 transgenics (Table 1, genes listed in bold). All the up-regulated genes that overlapped in these transgenics were also preferentially expressed in differentiating secondary xylem (Table 1). They included: four genes that were associated with the phenylpropanoid pathway, i.e. phenylalanine ammonia-lyase (PAL), 4 coumarate coA ligase (4CL), hydroxycinnamoyl transferase (HCT), and caffeic acid O-methyl transferase (COMT) (Table 1); two pinoresinol-lariciresinol reductase (PLR1 and PLR2) genes, that are involved in lignan synthesis primarily derived from the monolignols, E-coniferyl, and E-p-coumaryl alcohols (Kwon et al., 2001); and transcripts encoding 3-deoxy-D-arabino-heptulosonate 7-phosphate synthetase (DAHP), an important enzyme of the shikimate pathway, which provides precursors for phenylpropanoid metabolism leading to monolignols and flavonoids (Herrmann, 1995; Amthor, 2003). Both transgenics also displayed a strong up-regulation for an uclacyanin, a plant-specific blue copper protein that might participate in redox processes occurring during the primary defence response and/or lignin formation in plants (Nersissian et al., 1998). Conversely, the overlapping transcripts that had reduced expression in both transgenics were preferential to phloem-bark or needle tissue or did not shown any tissue preference (Table 1). Reduced transcript levels were observed in both transgenics for a xyloglucan endo-transglycosylase (XTH) that could be associated with cell wall biogenesis and reassembly, as well as for a canonical class III peroxidase (MN5232900). Three other genes, that could be associated with stress and detoxification, such as glyoxalase, LEA EMB7 (MN5251934), and r4g30 protein coding gene, were strongly down-regulated, particularly in PtMYB8-OE. A beta-primeverosidase (MN5233446), which could be linked to geraniol hydrolysis, was also strongly down-regulated in both transgenics, as well as two genes of unknown function. Only a single transcript, a carbonic anhydrase, was strongly up-regulated in PtMYB1 transgenics and down-regulated in PtMYB8 transgenics. Carbonic anhydrases have been shown to facilitate the transport of inorganic carbon and to catalyse carboxylation–decarboxylation reactions essential to photosynthesis and/or respiration in plants (Shiraiwa and Miyachi, 1985).
Distinct transcriptional responses were also observed between the transgenics. Transcripts distinctly misregulated in either PtMYB1 or PtMYB8-OE were generally preferential to secondary xylem when up-regulated, whereas they were assigned to needles, phloem-bark, or did not show preference to any particular tissue when down-regulated (Table 1). Large parts of the sequences which were only differentially expressed in PtMYB8-OE were associated with cell-wall organization and biogenesis (Table 1). Indeed, several genes related to cellulose (CesA), hemicellulose (CLS, GT2), and pectin (GT8) metabolism were up-regulated, together with an epimerase (NSI) and an arabinogalactan protein (AGP1). In parallel, several genes related to cell wall expansion and reassembly were down-regulated in PtMYB8-OE, such as GH17, GH28, UDPGT, XTH, and PAE, in addition to needle preferential transcripts encoding a CesA and a CSL-like. Upstream of the phenylpropanoid pathway, additional transcripts representing genes from the shikimate pathway were either up-regulated (chorismate mutase) or down-regulated (prephenate dehydratase). Other down-regulated sequences were similar to phenylpropanoid pathway genes, like coumaroyl CoA O-methyltransferase-like (CCoAOMT-like) and cinnamoyl CoA reductase-like (CCR-like) genes, as well as an N-hydroxycinnamoyl benzoyltransferase (HCBT), and were preferential to needle and/or phloem/bark tissues (Table 1). Genes linked to S-adenosyl-L-methionine, a potential methyl group donor for methoxylation reactions in the monolignol biosynthetic pathway (Amthor, 2003), were also misregulated in PtMYB8-OE (Table 1). Transcripts corresponding to enzymes involved in the early steps of the flavonoid pathway, such as chalcone synthase (CHS), chalcone isomerase (CHI), dihydroflavonol 4-reductase (DFR), and flavonol synthase (FS), also showed clear down-regulation in both PtMYB8 transgenic lines (Table 1). A clear decrease in transcript abundance was observed for several peroxidase genes, some of which could be associated with lignin polymerization (MN5235950; Koutaniemi et al., 2005) or linked to other oxidative processes (MN5233396). Genes associated with stress and detoxification were generally down-regulated, as shown for glutathione S-transferase, as well as those related to the terpenoid pathway. In PtMYB1-OE, additional up-regulated sequences were related to the phenylpropanoid pathway such as trans-cinnamate 4-hydroxylase (C4H), and to the cell wall as an alpha-expansin which was up-regulated 2-fold (Table 1). In turn, a peroxidase, as well as several genes involved in the terpenoid pathway, and stress response and detoxification were down-regulated.
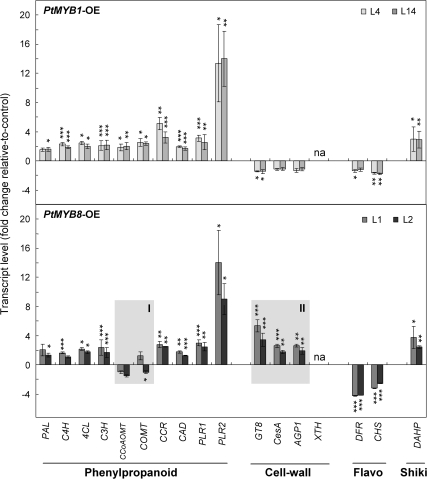
Targeted RT-qPCR analysis of phenylpropanoid metabolism, cell-wall, and R2R3-MYB-related genes
RT-qPCR analyses were performed on all the genes involved in the phenylpropanoid pathway to complement microarray transcript profiles, and were extended to a subset of genes that were related to cell wall biogenesis and to shikimate and flavonoid pathways in order to validate the results from microarray analysis (Fig. 4; see Fig. S1 in Supplementary data available at JXB online). Compared with controls, the significant up-regulation of genes involved in phenylpropanoid metabolism was confirmed by Student's t-test (Fig. 4). In both transgenics, transcript abundance was increased by ∼2-fold for pathway genes going from C4H to CAD, except for COMT and CCoAOMT in PtMYB8-OE for which transcript abundance was similar to control (Fig. 4, shaded box I). In both transgenics, PAL gave smaller and less consistent increases. In addition, the strong up-regulation of the two PLR transcripts (around 3-fold for PLR1, and >10-fold for PLR2) linked to phenylpropanoid metabolism was confirmed in both transgenics compared with the control (Fig. 4). The down-regulation of flavonoid- (DFR and CHS) and up-regulation of shikimate (DAHP)-related genes was also confirmed in both transgenics, as shown in Fig. 4. The differential effects on cell wall-related genes in PtMYB1-OE compared with PtMYB8-OE were also validated (Fig. 4, shaded box II). Compared with control and PtMYB1-OE, GT8, CesA, and AGP1 transcripts were all increased in PtMYB8-OE, while XTH was strongly down-regulated in both transgenics.
Fig. 4.
Targeted RT-qPCR analysis in PtMYB1 and PtMYB8 transgenic plantlets: validation of microarray and expression data from genes related to secondary metabolism and cell wall assembly. Phenylpropanoid-related genes are phenylalanine ammonia-lyase (PAL), cinnamic acid 4-hydroxylase (C4H), 4-coumarate-CoA ligase (4CL), coumarate 3-hydroxylase (C3H), caffeate O-methyltransferase (COMT), caffeoyl-CoA 3-O-methyltransferase (CCoAOMT), cinnamoyl-CoA reductase (CCR), cinnamyl-alcohol dehydrogenase (CAD), and pinoresinol-lariciresinol reductase 1 and 2 (PLR1 and PLR2); cell wall-related genes are cellulose synthase (CesA), glycosyltransferase family 8 (GT8), arabinogalactan (AGP1), and xyloglucan endotransglycosylase/hydrolase (XTH). Flavonoid related genes (Flavo) are chalcone synthase (CHS), dihydroflavonol 4-reductase (DFR); the shikimate-related gene (Shiki) is 3-deoxy-D-arabino-heptulosonate 7-phosphate synthetase (DAHP). For both transgenics, transcript accumulation was assessed by RT-qPCR on 3-week-old plantlets produced from two independent transgenic lines for each gene construct (L4 and L14 for PtMYB1; L1 and L2 for PtMYB8). Transcript levels were determined from four biological replicates (25 plantlets per replicate) and were normalized relative to EF1-alpha expression level. Transcript levels were then expressed relative to control plants, and the significance of differential transcript accumulation was evaluated with Student's t-test (two-sample, unpaired, one-sided) at P ≤0.05 (*), P ≤0.01 (**), and P ≤0.001 (***). na, Not amplified in MYB transgenics.
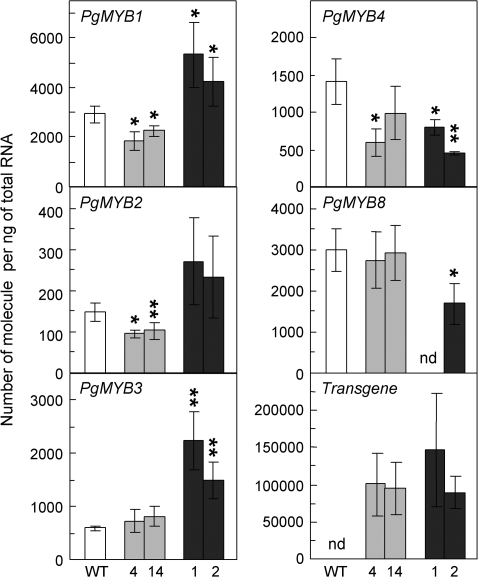
The RNA levels of PgMYB1 and PgMYB8, previously identified as putative spruce orthologues of PtMYB1 and PtMYB8 (Bedon et al., 2007), were examined together with those of the transgenes. In addition, transcript levels of three other spruce MYBs, PgMYB2, PgMYB3, and PgMYB4, that were shown to be preferentially expressed in differentiating xylem from 33-year-old white spruce trees (Bedon et al., 2007), were also evaluated. Transgene transcript levels in PtMYB1 and PtMYB8 overexpressors were quite similar when normalized relative to EF1-alpha, representing a 33-fold and a 40-fold increase in transcript abundance compared with their endogenous counterparts PgMYB1 and PgMYB8, respectively (Fig. 5). PtMYB1 overexpression led to a small but significant decrease (P ≤0.05) in steady-state RNA of its putative homologue PgMYB1 (Fig. 5) and of PgMYB2 and PgMYB4, but PgMYB3 and PgMYB8 were unaffected. In PtMYB8-OE plantlets, transcript levels of PgMYB1 and PgMYB3 were significantly up-regulated compared with the control according to Student's t-test at P ≤0.05 and P ≤0.01, respectively (Fig. 5), whereas transcript abundance was reduced for PgMYB4 (P ≤0.01), as well as for PgMYB8 (P ≤0.05).
Fig. 5.
Transcript accumulation of endogenous spruce MYBs and pine transgenes in wild-type and PtMYB1 and PtMYB8 transgenic plantlets. For both transgenics, transcript accumulation was assessed by RT-qPCR on 3-week-old plantlets produced from two independent transgenic lines (L4 and L14 for PtMYB1, grey columns; L1 and L2 for PtMYB8, black columns). Transcript levels were normalized relative to EF1a expression level and were calculated using standard curves as described by Rutledge and Côté (2003). Bars represent means (±SD) from three biological replicates (15 plantlets per replicate). nd, Not detected. The significance of differential transcript accumulation (up or down) between control and transgenic plantlets was evaluated with Student's t-test (two-sample, unpaired, one-sided) at P ≤0.05 (*), or P ≤0.01 (*).
Discussion
Previous studies have inferred that lignin biosynthesis and deposition in wood are regulated by R2R3-MYBs (see reviews: Rogers and Campbell, 2004; Groover and Robischon, 2006; Demura and Fukuda, 2007). In gymnosperm trees, however, only Pinus taeda PtMYB4 has been shown to induce ectopic lignification when heterologously expressed in tobacco and Arabidopsis (Patzlaff et al., 2003b; Newman et al., 2004). Based on transcriptional activation assays in yeast, PtMYB1 was also proposed to act as a transcriptional regulator of lignification (Patzlaff et al., 2003a) but this hypothesis remained to be tested in planta. The present gain-of-function experiments using a conifer expression system provide experimental evidence that is highly consistent with the involvement of PtMYB1 as well as PtMYB8 in lignification. Furthermore, PtMYB8-OE additionally led to the misregulation of cell wall-related transcripts concomitantly with the ectopic deposition of secondary cell wall in different cell types. The putative effects of these MYBs on phenylpropanoid metabolism and related pathways, as well as secondary cell wall biogenesis, in conifer trees are interpreted in more detail below.
PtMYB1 and PtMYB8 impact upon phenylpropanoid and shikimate pathways
The transcript profiles of phenylpropanoid-related genes clearly overlapped in PtMYB1- and PtMYB8-OE spruce. The overexpression of each MYB led to a clear up-regulation of most of the genes evaluated in the phenylpropanoid pathway (from C4H to CAD), except that PtMYB8-OE had no effect on the transcript levels of COMT and CCoAOMT (Table 1; Fig. 4, shaded box I). It appears that different MYB genes may act differentially on genes within the lignin biosynthetic pathway in spruce, as it was shown in other plant systems (Patzlaff et al., 2003b; Goicoechea et al., 2005). Other authors have also proposed that more than one MYB may act on the same target gene to regulate its expression (Moyano et al., 1996; Tamagnone et al., 1998; Borevitz et al., 2000) and thus provide multiple levels of control for lignin biosynthesis in plants (Tamagnone et al., 1998; Borevitz et al., 2000). The present results provide evidence that several lignin biosynthetic genes can be recruited in spruce as a result of PtMYB1 or PtMYB8 overexpression, congruent with the increased lignin deposition (Fig. 3A) and the decrease of low molecular weight phenolic compounds (Fig. 3B) observed in both transgenics.
Despite these observations, restricting the impact of PtMYB1 and PtMYB8 to the sole phenylpropanoid biosynthetic pathway may lead to a biased interpretation or to a restricted view of the putative metabolic pathways affected by R2R3-MYB overexpression in spruce. Microarray profiling was useful to assess the variations of transcripts of upstream, and/or downstream biosynthetic pathways linked to phenylpropanoid metabolism, and also provided a potential overview of metabolic routing in PtMYB1 and PtMYB8 overexpressors (Table 1). The clear up-regulation of DAHP observed in both PtMYB1-OE and PtMYB8-OE (Table 1; Fig. 4) may indicate a routing of metabolic flow into the upstream shikimate pathway, which provides precursors required for lignin biosynthesis (Herrmann, 1995; Amthor, 2003). This situation is similar to that observed in petunia where the R2R3-MYB gene ODO1 was shown to be a key regulator of benzenoid compounds responsible for floral scent through the activation of genes in the upstream shikimate pathway (Verdonk et al., 2005). It would thus appear that PtMYB1 and PtMYB8 are potential regulators of phenylpropanoid metabolism in conifers, probably by directing metabolic flux into shikimate and monolignol pathways. However, the different transcriptional responses observed for COMT and CCoAOMT (Fig. 4, shaded box I) may indicate that the two MYBs have different impacts in terms of methoxylation in the monolignol biosynthetic pathway. Different responses were also observed for transcripts linked to SAM/SAH metabolism (Table 1), which forms the methyl group donor for lignin monomers (Amthor, 2003).
Comparative analysis of MYB overexpressors also suggested an effect on lignan biosynthesis as indicated by a strong up-regulation of two putative pinoresinol/lariciresinol reductase (PLR) coding sequences (Fig. 4). Lignans constitute an abundant class of phenylpropanoid compounds which have been supposed to act in defence against pathogens (Gang et al., 1999; Schmitt and Petersen, 2002) but also as important components of heartwood formation (Swan et al., 1969). The general routing for lignan biosynthesis has been strongly suggested to be the result of the conversion from coniferyl alcohol to secoisolariciresinol, and then to matairesinol (Susuki and Umezawa, 2007). This routing starts with stereoselective coupling of coniferyl alcohol monomers to form pinoresinol, the first substrate of the PLR enzyme (Davin et al., 1997). A possible explanation for the accumulation of PLR transcripts may involve a feed-forward response to the up-regulated monolignol pathway in MYB-OE spruces. If such was the case, it would be an indirect effect of MYB overexpression. Another hypothesis would be that PLR genes are in fact direct targets for PtMYB1 and/or PtMYB8. The >10-fold up-regulation of PLR2 in both types of MYB overexpressors is not inconsistent with this possibility (Fig. 4). As far as is known, no MYB has been associated with transcriptional control of genes coding for lignan synthesis enzyme. A gel shift experiment with PtMYB1/PgMYB1 and PtMYB8/PgMYB8 recombinant proteins and PLR1 and PLR2 promoter regions should be further considered to test this hypothesis.
PtMYB8 as a putative actor in secondary cell wall biosynthesis
The involvement of R2R3-MYB TFs in the regulation of secondary cell wall biosynthesis has only been recently reported for AtMYB26 (Yang et al., 2007) and AtMYB46 (Zhong et al., 2007) in the model plant Arabidopsis thaliana. Overexpression of the Eucalyptus EgMYB2 in tobacco has been associated with secondary cell wall thickening but not to the regulation of genes associated with secondary cell wall biogenesis, other than those involved in monolignol biosynthesis (Goicoechea et al., 2005). The data presented here show that PtMYB8 clearly impacted secondary cell wall biogenesis as reflected by the distribution of ectopic schlerenchyma-like elements, and by the deposition of secondary cell walls in parenchyma, and cortical cells (Fig. 2). Congruent with these observations, overexpression of PtMYB8 led to the misregulation of several genes associated with biosynthetic pathways of cellulose and hemicellulose (Table 1; Fig. 4, shaded box II). In this context, PtMYB8/PgMYB8 may be considered as putative regulators of secondary cell wall biosynthesis, in agreement with their expression being restricted to secondary xylem in pine and spruce (Bedon et al., 2007; Fig. 1). Consistent with such a role, PgMYB8, the closest homologue of PtMYB8 in spruce, displayed increased transcript accumulation upon the induction of compression wood (Bedon et al., 2007). Here, PtMYB8-OE led to the up-regulation of AGP1 arabinogalactan transcripts (Table 1; Fig. 4) whose up-regulation has been associated with compression wood in spruce (Bedon et al., 2007) and pine (Zhang et al., 2000) and tension wood in poplar (Lafarguette et al., 2004). In conifers, compression wood is produced in bent or leaning trees, and is characterized by an enhanced deposition of secondary cell wall compounds (Timell, 1986), by increased lignin content, and an enrichment in H lignin units in spruce (Lange et al., 1995) and pine (Yeh et al., 2006).
Constitutive overexpression of PtMYB8 in spruce was also associated with strong decreases in transcript levels of flavonoid, terpenoid, and benzenoid biosynthesis enzymes (Table 1; Fig. 4). Most of these down-regulated sequences were identified as needle or phloem-bark preferential (Table 1), a tissue where PtMYB8 transcripts normally accumulated at very low level (Fig. 1). Due to the highly conserved DNA binding domain observed among conifer MYBs (Bedon et al., 2007), ectopic overexpressing PtMYB8 in such tissues may have led to the indirect effects linked to non-specific binding to gene promoters caused by MYB overexpression. Such a dosage-linked effect would influence most strongly the expression of genes with similar cis regulatory motifs, as suggested in Arabidopsis (Jin et al., 2000). MYB overexpression may also repress promoter activity by competing with cognate TFs (squelching) and thus sequestrating components of the transcriptional machinery away from cis regulatory DNA elements (Gill and Ptashne, 1988).
A parallel may be drawn between the present study and the transcriptional network regulating secondary cell wall biosynthesis recently proposed by Zhong et al. (2006, 2007). In this model, the A. thaliana NAC domain protein SND1 activates a transcriptional cascade involving R2R3-MYB and KNOX TFs, which themselves are strong candidates for the regulation of genes encoding enzymes required for secondary cell wall deposition. These authors further suggested that the control of secondary cell wall biosynthesis by SND1-like genes may be a common mechanism in plants (Zhong et al., 2006). The results presented here could indeed indicate that secondary cell wall deposition in conifer trees is controlled by a transcriptional network similar to the SND1 cascade, in which PtMYB8/PgMYB8 and PtMYB1/PgMYB1 may represent putative members. Most of the spruce MYB sequences which are misregulated following PtMYB8 overexpression (Fig. 5) are close homologues of the MYB partners described in the SND1 cascade in Arabidopsis (Zhong et al., 2007). These spruce sequences were shown to be highly similar to the Arabidopsis R2R3-MYB genes AtMYB20 (At1g66230) and AtMYB85 (At4g22680) for Pg/PtMYB1, AtMYB103 (At1g63910) for Pg/PtMYB2, and AtMYB46 (At5g12870) for Pg/PtMYB4 and Pg/PtMYB8 (Bedon et al., 2007). These later sequences (AtMYB46, Pg/PtMYB4 and -8) are also highly similar to EgMYB2 from Eucalyptus, shown to control secondary cell wall thickening (Goicoechea et al., 2005). All these spruce MYBs are also the most clearly preferential to differentiating secondary xylem tissue (Bedon et al., 2007) supporting their putative involvement in secondary cell wall deposition. In addition, both of the xylem preferential CesA coding sequences from spruce (MN5175268 and MN5193335), which were up-regulated in PtMYB8-OE (Fig. 4; Table 1), were also most similar to AtCesA7 and AtCesA8 associated with SND1 transcriptional cascade in Arabidopsis (Zhong et al., 2007).
In conclusion, Pg/PtMYB1 and Pg/PtMYB8 appear to be potentially important players in conifers in the regulation of secondary cell wall biosynthesis, including lignin deposition. Their potential roles were examined in comparison with the transcriptional cascade controlling the formation of xylem secondary cell wall in the model plant Arabidopsis thaliana. Together these two systems may provide a comparative framework to assess further hierarchy between TFs but also—and perhaps most interestingly—gain insights into the evolution of regulatory processes of wood formation and related biosynthetic pathways. If Pg/PtMYB1 and/or Pg/PtMYB8 are part of a transcriptional network that is conserved between gymnosperm and angiosperm species, it would be of great interest to identify upstream actors such as NAC-type master genes similar to SND1 (Zhong et al., 2007), VND, and NTS (Demura and Fukuda, 2007; Mitsuda et al., 2007), which could be pivotal TFs in secondary cell wall biogenesis in trees.
Supplementary data
The following supplementary material is available at JXB online.
Table S1. Primer sequences used for RT-qPCR.
Table S2. Complete list of expression data relative to PtMYB1 and PtMYB8 overexpression in spruce.
Fig. S1. RT-qPCR analysis of transcript accumulation in wild type, PtMYB1 and PtMYB8 transgenic plantlets: validation of microarray and expression data from genes related to secondary metabolism and cell wall assembly.
Appendix S1. Procedure used for microarray manufacturing.
Supplementary Material
Acknowledgments
We thank Dr RR Sederoff (North Carolina Sate University, Raleigh, NC) for kindly providing clones of Pinus taeda, F Morency for assistance in tissue culture, and P-O Nadeau for RNA extractions and quality control. Thanks are extended to Dr B Boyle for critical comments on the manuscript. This work was supported by Genome Québec and Genome Canada to JM and AS for the ARBOREA project.
References
- Amthor J. Efficiency of lignin biosynthesis: a quantitative analysis. Annals of Botany. 2003;91:673–695. doi: 10.1093/aob/mcg073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedon F, Grima-Pettenati J, Mackay J. Conifer R2R3-MYB transcription factors: sequence analysis and gene expression in wood-forming tissues of white spruce (Picea glauca) BMC Plant Biology. 2007;7:17. doi: 10.1186/1471-2229-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- Björkmann A. Studies on finely divided wood. 1. Extraction of lignin with neutral solvents. Suen Papperstidn. 1956;59:477. [Google Scholar]
- Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annual Reviews in Plant Biology. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. The Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MM, Sederoff RR. Variation in lignin content and composition: mechanisms of control and implications for the genetic improvement of plants. Plant Physiology. 1996;110:3–13. doi: 10.1104/pp.110.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 1993;11:113–116. [Google Scholar]
- Chen Y, Yang X, He K, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Molecular Biology. 2006;60:107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- Christensen AH, Sharrock RA, Quail PH. Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Molecular Biology. 1992;18:675–689. doi: 10.1007/BF00020010. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Enzymes and other agents that enhance cell wall extensibility. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:391–417. doi: 10.1146/annurev.arplant.50.1.391. [DOI] [PubMed] [Google Scholar]
- Davin LB, Wang H-B, Crowell AL, Bedgar DL, Martin DM, Sarkanen S, Lewis NG. Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science. 1997;275:362–366. doi: 10.1126/science.275.5298.362. [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L. Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. The Plant Cell. 2003;15:2514–2531. doi: 10.1105/tpc.014043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demura T, Fukuda H. Transcriptional regulation in wood formation. Trends in Plant Science. 2007;12:64–70. doi: 10.1016/j.tplants.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Gang DR, Kasahara H, Xia Z-Q, Van der Mijnsbrugge K, Bauw G, Boerjan W, Van Montagu M, Davin LB, Lewis NG. Evolution of plant defense mechanisms: relationships of phenylcoumaran benzylic ether reductases to pinoresinolariciresinol and isoflavone reductases. The Journal of Biological Chemistry. 1999;274:7516–7527. doi: 10.1074/jbc.274.11.7516. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G, Ptashne M. Negative effect of transcriptional activator GAL4. Nature. 1988;324:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- Goff SA, Cone KC, Chandler VL. Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes & Development. 1992;6:864–875. doi: 10.1101/gad.6.5.864. [DOI] [PubMed] [Google Scholar]
- Goicoechea M, Lacombe E, Legay S, et al. EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. The Plant Journal. 2005;43:553–567. doi: 10.1111/j.1365-313X.2005.02480.x. [DOI] [PubMed] [Google Scholar]
- Gomez-Maldonado J, Avila C, Torre F, Canas R, Canovas FM, Campbell MM. Functional interactions between a glutamine synthetase promoter and MYB proteins. The Plant Journal. 2004;39:513–526. doi: 10.1111/j.1365-313X.2004.02153.x. [DOI] [PubMed] [Google Scholar]
- Groover A, Robischon M. Developmental mechanisms regulating secondary growth in woody plants. Current Opinion in Plant Biology. 2006;9:55–58. doi: 10.1016/j.pbi.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Herrmann KM. The shikimate pathway: early steps in the biosynthesis of aromatic compounds. The Plant Cell. 1995;7:907–919. doi: 10.1105/tpc.7.7.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys JM, Chapple C. Rewriting the lignin roadmap. Current Opinion in Plant Biology. 2002;5:224–229. doi: 10.1016/s1369-5266(02)00257-1. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. Journal of Computational and Graphical Statistics. 1996;5:299–314. [Google Scholar]
- Jin HL, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO Journal. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinska B, Karlsson M, Srivastava M, Stenberg A, Schrader J, Sterky F, Bhalerao R, Wingsle G. MYB transcription factors are differentially expressed and regulated during secondary vascular development in hybrid aspen. Plant Molecular Biology. 2004;56:255–270. doi: 10.1007/s11103-004-3354-5. [DOI] [PubMed] [Google Scholar]
- Kinlaw CS, Neale DB. Complex gene families in pine genomes. Trends in Plant Science. 1997;2:356–359. [Google Scholar]
- Klimaszewska K, Lachance D, Pelletier G, Lelu MA, Séguin A. Regeneration of transgenic Picea glauca, P. mariana, and P. abies after cocultivation of embryogenic tissue with Agrobacterium tumefaciens. In vitro Cellular & Developmental Biology – Plant. 2001;37:748–755. [Google Scholar]
- Klimaszewska K, Rutledge RG, Séguin A. Genetic transformation of conifers utilizing somatic embryogenesis. In: Peña L, editor. Methods in molecular biology. Totowa, NJ: Humana Press; 2004. pp. 151–164. [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Molecular and General Genetics. 1986;204:383–396. [Google Scholar]
- Koutaniemi S, Toikka MM, Kärkönen A, Mustonen M, Lundell T, Simola LK, Kilpeläinen IA, Teeri TH. Characterization of basic p-coumaroyl and coniferyl alcohol oxidizing peroxidases from lignin-forming Picea abies suspension culture. Plant Molecular Biology. 2005;58:141–157. doi: 10.1007/s11103-005-5345-6. [DOI] [PubMed] [Google Scholar]
- Kwon M, Davin LB, Lewis NG. In situ hybridization and immunolocalization of lignan reductases in woody tissues: implications for heartwood formation and other forms of vascular tissue preservation. Phytochemistry. 2001;57:899–914. doi: 10.1016/s0031-9422(01)00108-x. [DOI] [PubMed] [Google Scholar]
- Lafarguette F, Leplé JC, Déjardin A, Laurans F, Costa G, Lesage-Descauses MC, Pilate G. Poplar genes encoding fasciclin-like arabinogalactan proteins are highly expressed in tension wood. New Phytologist. 2004;164:107–121. doi: 10.1111/j.1469-8137.2004.01175.x. [DOI] [PubMed] [Google Scholar]
- Lange BM, Lapierre C, Sandermann H., Jr Elicitor-induced spruce stress lignin (structural similarity to early developmental lignins) Plant Physiology. 1995;108:1277–1287. doi: 10.1104/pp.108.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagia M. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. The Plant Cell. 2007;19:270–280. doi: 10.1105/tpc.106.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S. Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant and Cell Physiology. 2006;47:457–470. doi: 10.1093/pcp/pcj012. [DOI] [PubMed] [Google Scholar]
- Moyano E, Martinez-Garcia JF, Martin C. Apparent redundancy in myb gene function provides gearing for the control of flavonoid biosynthesis in Antirrhinum flowers. The Plant Cell. 1996;8:1519–1532. doi: 10.1105/tpc.8.9.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nersissian AM, Immoos C, Hill MG, Hart PJ, Williams G, Herrmann RG, Valentine JS. Uclacyanins, stellacyanins, and plantacyanins are distinct subfamilies of phytocyanins: plant-specific mononuclear blue copper proteins. Protein Science. 1998;7:1915–1929. doi: 10.1002/pro.5560070907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman LJ, Perazza DE, Juda L, Campbell MM. Involvement of the R2R3-MYB, At MYB61, in the ectopic lignication and dark-photomorphogenic components of the det3 mutant phenotphype. The Plant Journal. 2004;37:239–250. doi: 10.1046/j.1365-313x.2003.01953.x. [DOI] [PubMed] [Google Scholar]
- Patzlaff A, McInnis S, Courtenay A, et al. Characterisation of a pine MYB that regulates lignification. The Plant Journal. 2003b;36:743–754. doi: 10.1046/j.1365-313x.2003.01916.x. [DOI] [PubMed] [Google Scholar]
- Patzlaff A, Newman LJ, Dubos C, Whetten RW, Smith C, McInnis S, Bevan MW, Sederoff RR, Campbell MM. Characterisation of PtMYB1, an R2R3-MYB from pine xylem. Plant Molecular Biology. 2003a;53:597–608. doi: 10.1023/B:PLAN.0000019066.07933.d6. [DOI] [PubMed] [Google Scholar]
- Paux E, Carocha V, Marques C, Mendes de Sousa A, Borralho N, Sivadon P, Grima-Pettenati J. Transcript profiling of Eucalyptus xylem genes during tension wood formation. New Phytologist. 2005;167:89–100. doi: 10.1111/j.1469-8137.2005.01396.x. [DOI] [PubMed] [Google Scholar]
- Rogers LA, Campbell MM. The genetic control of lignin deposition during plant growth and development. New Phytologist. 2004;164:17–30. doi: 10.1111/j.1469-8137.2004.01143.x. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods in Molecular Biology. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Rutledge RG, Côté C. Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Research. 2003;31 doi: 10.1093/nar/gng093. e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J, Petersen M. Influence of methyl jasmonate and coniferyl alcohol on pinoresinol and matairesinol accumulationin a Forsythia × intermedia suspension culture. Plant Cell Reports. 2002;20:885–889. [Google Scholar]
- Schwechheimer C, Zourelidou M, Bevan MW. Plant transcription factor studies. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:127–150. doi: 10.1146/annurev.arplant.49.1.127. [DOI] [PubMed] [Google Scholar]
- Sharman BC. Tannic acid and iron alum with safranin and Orange G in studies of the shoot apex. Stain Technology. 1943;18:105–111. [Google Scholar]
- Shiraiwa Y, Miyachi S. Role of carbonic anhydrase in photosynthesis of blue-green alga (Cyanobacterium) Anabaena variabilis ATCC 29413. Plant and Cell Physiology. 1985;26:109–116. [Google Scholar]
- Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and computational biology solutions using R and bioconductor. New York, NY: Springer-Verlag; 2005. pp. 397–420. [Google Scholar]
- Stewart JJ, Kadla JF, Mansfield SD. The influence of lignin chemistry and ultrastructure on the pulping efficiency of clonal aspen (Populus tremuloides Michx.) Holzforschung. 2006;60:111–122. [Google Scholar]
- Susuki S, Umezawa T. Biosynthesis of lignans and norlignans. Journal of Wood Science. 2007;53:273–284. [Google Scholar]
- Swan EP, Jiang KS, Gardner AF. The lignans of Thuja plicata and the sapwood-heartwood transformation. Phytochemistry. 1969;8:345–351. [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C. The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. The Plant Cell. 1998;10:135–154. doi: 10.1105/tpc.10.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timell TE. Compression wood in gymnosperms. Berlin: Springer Verlag; 1986. [Google Scholar]
- Topfer R, Maas C, Horicke-Grandpierre C, Schell J, Steinbiss HH. Expression vectors for high-level gene expression in dicotyledonous and monocotyledonous plants. Methods in Enzymology. 1993;217:67–78. doi: 10.1016/0076-6879(93)17056-b. [DOI] [PubMed] [Google Scholar]
- Verdonk JC, Haring MA, van Tunen AJ, Schuurink RC. ODORANT1 regulates fragrance biosynthesis in Petunia flowers. The Plant Cell. 2005;17:1612–1624. doi: 10.1105/tpc.104.028837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vom Endt D, Kijn JW, Memelink J. Transcription factors controlling plant secondary metabolism: what regulates the regulators? Phytochemistry. 2002;61:107–114. doi: 10.1016/s0031-9422(02)00185-1. [DOI] [PubMed] [Google Scholar]
- Yang C, Xu Z, Song J, Conner C, Vizcay Barrena Gm, Wilson ZA. Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. The Plant Cell. 2007;19:534–548. doi: 10.1105/tpc.106.046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Research. 2002;30 doi: 10.1093/nar/30.4.e15. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh TF, Braun JL, Goldfarb B, Chang HM, Kadla JF. Morphological and chemical variations between juvenile wood, mature wood, and compression wood of loblolly pine (Pinus taeda L.) Holzforschung. 2006;60:1–8. [Google Scholar]
- Zhang JZ. Overexpression analysis of plant transcription factors. Current Opinion in Plant Biology. 2003;6:430–440. doi: 10.1016/s1369-5266(03)00081-5. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sederoff RR, Allona I. Differential expression of genes encoding cell wall proteins in vascular tissues from vertical and bent loblolly pine trees. Tree Physiology. 2000;20:457–466. doi: 10.1093/treephys/20.7.457. [DOI] [PubMed] [Google Scholar]
- Zhong R, Demura T, Ye Z-H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. The Plant Cell. 2006;18:3158–3170. doi: 10.1105/tpc.106.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye Z-H. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. The Plant Cell. 2007;19:2776–2792. doi: 10.1105/tpc.107.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.