Abstract
The chloroplast protein CP12 has been shown to regulate the activity of two Calvin cycle enzymes, phosphoribulokinase (PRK) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), by the reversible formation of a multiprotein complex. In Arabidopsis there are three CP12 genes, CP12-1, CP12-2, and CP12-3, and expression analysis suggested that the function of these proteins may not be restricted to the Calvin cycle. Reverse transcription-PCR analysis was used here to investigate further the expression patterns of the three CP12 Arabidopsis genes together with the genes encoding plastid GAPDH (GAPA-1 and GAPB), PRK (PRK), and plastid NAD-dependent GAPDH (GAPCp1 and GAPCp2) during development, in response to changes in light, temperature, and anaerobic conditions. Expression of the CP12-2 gene was similar to that of the Calvin cycle enzymes PRK and GAPDH. However, this was not the case for CP12-1 and -3 which were both expressed in roots. Analysis of transgenic Arabidopsis lines expressing CP12::GUS fusion constructs revealed that the CP12 genes display different spatiotemporal expression patterns. The CP12-1 gene was expressed in root tips whilst CP12-3::GUS expression was evident throughout the root tissue. The most unexpected finding was that all three CP12 genes were expressed in floral tissues; CP12-1 and CP12-2 expression was detected in the sepals and the style of the flower, while in contrast CP12-3::GUS expression was restricted to the stigma and anthers. Taken together, the data suggest that the redox-sensitive CP12 proteins may have a wider role in non-photosynthetic plastids, throughout the plant life cycle.
Keywords: Calvin cycle, non-photosynthetic plastids, redox, thioredoxin
Introduction
CP12 was originally identified as a small, redox-sensitive, chloroplast protein that interacts with two enzymes of the Calvin cycle, phosphoribulokinase (PRK) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), in cyanobacteria, a green alga, and higher plants, forming a high molecular weight complex (PRK/GAPDH/CP12) (Wedel et al., 1997; Wedel and Soll, 1998; Scheibe et al., 2002; Graciet et al., 2004). Evidence demonstrating the presence of this complex in red algae and in diatoms has recently been presented (Boggetto et al., 2007; Oesterhelt et al., 2007). The function proposed for the PRK/GAPDH/CP12 complex was as an additional mechanism, over and above that of thioredoxin, for the light/dark regulation of the activity of the component enzymes in response to changes in the NADPH/NADP ratio (Wedel et al., 1997). More recently it has been shown that changes in the status of the stromal PRK/GAPDH/CP12 complex, mediated by thioredoxin f, allow the activity of the Calvin cycle to be modulated in response to rapidly changing light levels that occur in the natural environment (Howard et al., 2008).
CP12-like proteins have been found exclusively in photosynthetic organisms, and all of the CP12 proteins so far identified contain a highly conserved region, proposed to form an α-helix. In green algae and plants, this region is flanked by two pairs of cysteine residues at the N- and C-terminal ends of the protein (Wedel et al., 1997; Wedel and Soll, 1998; Graciet et al., 2003; Trost et al., 2006). Interestingly the C-terminal region of the CP12 protein shares homology with the C-terminal extension of the Gap B subunit of chloroplastic GAPDH, known to be involved in thioredoxin-mediated redox activation of this enzyme (Baalmann et al., 1996; Pohlmeyer et al., 1996; Sparla et al., 2002; Marri et al., 2008). There is evidence that under oxidizing conditions the two cysteine pairs of the CP12 protein each form an intramolecular disulphide bridge which is essential for reconstitution of the PRK/GAPDH/CP12 complex in vitro (Graciet et al., 2003; Marri et al., 2005b; Sparla et al., 2007). Further in vitro analysis has shown that the oxidized form of CP12 interacts first with GAPDH and then this binary complex interacts with PRK, forming the PRK/GAPDH/CP12 complex (Lebreton et al., 2006; Marri et al., 2008). The recently elucidated redox properties of the CP12 protein match well with that of the GAPDH A2B2 complex and that of PRK, providing evidence that this scenario is feasible in vivo (Marri et al., 2008). The structure of oxidized and reduced CP12 was investigated by circular dichroism (CD) and nuclear magnetic resonance (NMR), and revealed a lack of structure, which led to the suggestion that this protein may belong to a group of proteins called intrinsically unstructured proteins or IUPs (Graciet et al., 2003; Gardebien et al., 2006). Interestingly it has been shown that a number of IUPs have the ability to interact with a number of different molecules in the cell (Tompa, 2002; Dyson and Wright, 2005). This raises the possibility that the CP12 protein may also be able to interact with proteins in the chloroplast other than PRK and GAPDH.
Bioinformatic analysis of genome sequence and expressed sequence tag (EST) data has revealed that in higher plants CP12 is encoded by a small multigene family. In Arabidopsis there are three CP12 gene sequences: CP12-1 (At2g47400), CP12-2 (At3g62410), and CP12-3 (At1g76560). Amino acid sequence comparisons reveal that two of the proteins encoded by the CP12-1 and CP12-2 genes are highly similar and share 98% identity. However, CP12-3 encodes a protein with <50% identity with CP12-1 and CP12-2, and no function for this protein has yet been elucidated. Previously it has been shown that expression of Calvin cycle genes is highly co-ordinated in response to light, development, and tissue specificity (Raines et al., 1991; Yang et al., 1993; Sun et al., 2003), but analysis of the CP12 genes had suggested that the expression of the CP12-1 gene was not strictly in keeping with that of the Calvin cycle genes (Pohlmeyer et al., 1996; Marri et al., 2005a; Trost et al., 2006). These results are interesting and may suggest that the CP12-1 protein has a role in the metabolism of plastids in non-green tissues. Further support for this suggestion comes from analysis of CP12-transgenic antisense tobacco and Arabidopsis plants which displayed a range of developmental phenotypes including altered leaf morphology, stunted growth, reduced fertility, fused cotyledons, and loss of apical dominance (Raines and Paul, 2006; Singh, 2007). This work clearly showed that the CP12 proteins are essential for normal growth and development and that it is likely that these proteins have additional functions apart from that of regulation of PRK and GAPDH in the Calvin cycle. To gain further knowledge of the potential function of the Arabidopsis CP12 genes, reverse transcription-PCR (RT-PCR) expression analysis is presented of not only the CP12-1 and CP12-2 genes, but also CP12-3 for which there are no data. To look for co-ordinate expression with genes encoding CP12-interacting proteins, the Calvin cycle genes encoding GAPDH (GAPA and GAPB) and PRK (PRK), and the genes encoding a novel NAD-dependent plastid GAPDH (GAPCp1 and GAPCp2) have also been included in this study (Backhausen et al., 1998; Petersen et al., 2003). The detailed analysis of the spatiotemporal expression patterns of the CP12 gene family in both vegetative and floral tissues is also presented for the first time.
Materials and methods
Plant growth
Arabidopsis thaliana (Col-0) plants were grown in an environmentally controlled room at 22 °C with a 14 h light/10 h dark cycle. For dark treatments, 4-week-old plants were grown under a normal light–dark regime, transferred to the dark for 48 h, and then illuminated for 1, 4, and 24 h in the light. To study further the effect of dark treatments, plants were grown for 3 weeks under a light–dark regime and then transferred to darkness for 7 d. For cold stress treatments, plants were grown normally under 14 h light/10 h dark conditions in a controlled environment growth cabinet at 22 °C. Plants were then transferred to 5 °C for 24 h and 48 h, and transferred back to normal growth temperature for 24 h. For anaerobic induction, Arabidopsis seeds were germinated on Petri dishes containing MS (1/2) medium and allowed to grow for 14 d in 14 h light/10 h dark cycles at 25 °C; seedlings were submerged in situ into a tank containing sterilized water.
RNA extraction and RT-PCR analysis
Total RNA from leaves, flowers, roots, and stems was isolated using TRI REAGENT (Sigma) according to the manufacturer's instructions. Isolation of total RNA from Arabidopsis seeds was essentially as described (Penfield et al., 2005).
RNA transcripts were converted to cDNA using the superscript first-strand synthesis system. The cDNA synthesis was performed by using oligo(dT)s. All the primers were designed using the program Primer 3 Input (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The cDNA was used as a template for RT-PCR with gene-specific primer sets as follows: CP12-1 forward, 5′-CGACCTGTGGCTCGTGATTT-3′ and reverse 5′-CTGCCGTGTACCTTCTCCAT-3′; CP12-2 forward 5′-ACTACGGCACTCTTGGTCT-3′ and reverse 5′-CGACACTCATCAGCTTCACGAT-3′; CP12-3 forward 5′-AGAGTGTTGCTTCCGTCACA-3′ and reverse 5′-CGGTGTAAGTGAACACGATGA-3′; GapA-1 (At3g26550) forward 5′-AACCGAAACCCGTCTCTTCT-3′ and reverse 5′-CTTTGAGACCTGCAAACGA-3′; GapB (At1g42970) forward 5′-CCAACTGTTTGGCCCTTTT-3′ and reverse 5′-GGCTGTAACCCCACTCGTTA-3′; PRK (At1g32060) forward 5′-ACACTCATCAGCGACACGAC-3′ and reverse 5′-AGGAATCCACGAGATGGTTG-3′; GapCp-1 (At1g79530) forward 5′-CTCCCACTTCCGAATCAA-3′ and reverse 5′- GCTGTACTGGAACAGAACAAAAATA-3′; and GapCp-2 (At1g16300) forward 5′-TCTGTTTCTGGTAGCTTAGCAAT-3′ and reverse 5′-ATATATAGAAAAGGTTTCGACTAGCTG-3. A Perkin Elmer Gene Amp system 2400 and a Perkin Elmer Gene Amp PCR system 9700 were used for PCR and RT-PCR analysis. PCR programs included a hot start at 94 °C for 5 min followed by 24 or 45 cycles of 1 min at 94 °C plus 1 min at 45 °C (CP12-1, GapA-1, GapB, PRK, and EF) or 47 °C (GapCp-1 and GapCp-2), 45 cycles of 1 min at 45 °C for CP12-3, and finally 72 °C for 1 min.
Arabidopsis CP12::GUS fusion constructs and plant transformation
For each of the CP12 genes, the genomic sequences upstream of the initiating ATG were amplified using PCR and cloned into the pBI201 plasmid, containing the coding region of the β-glucuronidase (GUS; ∼1.94 kb) gene and NOS terminator sequence (Jefferson et al., 1987). The primers used for CP12-1 were forward 5′-AAGCTTAATATCAGGAAAAATATAGGTAGTGG and reverse 5′-GGACCAGGATTTTGATGGAGGAGAAAAGT to amplify a 502 bp fragment; for CP12-2 a 306 bp fragment was amplified using forward 5′-AGCTTTTGACTTTTCCTTAAACAGTGTGG and reverse 5′-GGATCCCTTTGGCTGGAGAAGGTACACG; and an 891 bp size fragment for CP12-3 using forward 5′-AAGCTTTATGAGTGTACGTTTGGTTTTAAGTTTAGT and reverse 5′-GGATCCAATTTCGTTTCTCTTCTTCGTCTTT. The upstream regions of each of the CP12 genes were cloned into the pGEM-T-easy vector and sequenced. The primers were designed with the HindIII site at the 5′ end and a BamHI site at the 3′ end, and these sites were used to transfer the CP12 promoter sequences into the pBI201 plasmid which was then introduced into Agrobacterium tumefaciens GV3101 by electroporation. These constructs were then transferred into Arabidopsis using the floral dipping method as described (Clough and Bent, 1998). Kanamycin-resistant seedlings were selected and transferred to soil for self-pollination and propagation. T2 generation plants were used for GUS assays.
Histochemical GUS assays
For GUS staining, plant tissue was immersed in GUS assay buffer (1% Triton X-100, 0.5 mM X-Glc, 100 mM phosphate buffer) and vacuum infiltrated. Samples were kept at 37 °C overnight, and tissues were cleared using 70% ethanol. Photographic images were captured using a digital camera (Canon, Power Shot A95).
Bioinformatic analysis of CP12 sequences
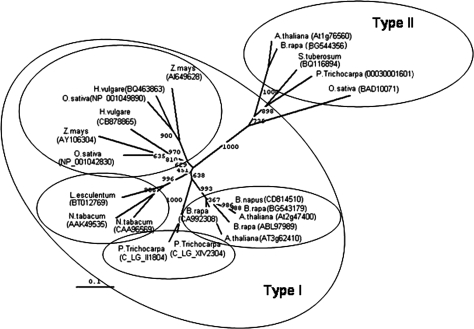
CP12 sequences were identified using the BLASTP and T-BLASTN search programs available at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) together with the ‘Proteins from AGI, Total Genome’ data set found in the TAIR database (http://www.arabidopsis.org/Blast/). Multiple sequence alignments were produced using the ClustalX program (Thompson et al., 1997). The radial cladogram that shows the phylogenetic relationship between the CP12 proteins was created by using the Neighbor–Joining option present on ClustalX. Bootstrap values from 1000 pseudo-replicates were used for the creation of the cladogram. The tree file was saved in PHYLIP format (.phb extension) and opened with the TreeView program (Page, 1996) to obtain the phylogenetic tree.
Microarray data available through Genevestigator (https://www.genevestigator.ethz.ch; Zimmermann et al., 2004) for CP12-1, CP12-2, and CP12-3 were analysed. In each case, treatment versus control signal values at corresponding time points are reported and, where three data points were available, standard errors were determined. Details of growth conditions and treatment regimes are available through Genevestigator.
Results
CP12-related proteins have been found only in photosynthetic organisms and are highly conserved across all groups, including cyanobacteria, algae, and lower and higher plants. Phylogenetic analysis of higher plant CP12 proteins has revealed that there are three CP12-like sequences which form two distinct groups, one with the closely related proteins CP12-1 (At2g47400) and CP12-2 (At3g62410) (Type I), and another with CP12-3 (At1g76560) (Type II) (Fig. 1). The Type I CP12 proteins form a monocot and a eudicot subgroup. In the eudicots, the CP12-1- and CP12-2-like proteins are grouped together within each of the families. In contrast, in the monocot group, the CP12-1- and CP12-2-like proteins form two subgroups. This suggests that duplication of the CP12-1- and CP12-2-like genes occurred after separation of the monocot and dicot lineages.
Fig. 1.
Phylogenetic relationship of the CP12 genes. Cladogram showing the relationship of the CP12 genes in terrestrial plants produced using the Neighbor–Joining option on the ClustalX program. Bootstrap values from 1000 pseudo-replicates were used for the creation of the cladogram. The tree file was saved in PHYLIP format (.phb extension) and opened with the TreeView program (Page, 1996) to obtain the phylogenetic tree.
Tissue-specific and developmental expression of the CP12 genes
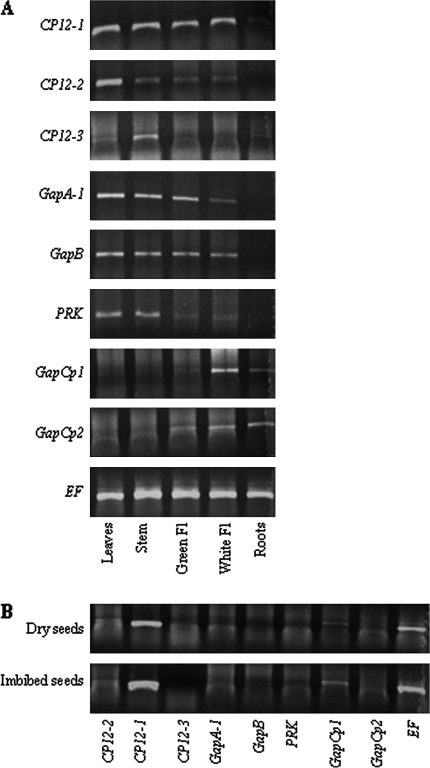
Total RNA was isolated from leaves, stems, flowers, and roots of 6-week-old, soil-grown, Arabidopsis plants and subjected to RT-PCR. This analysis revealed that the level of CP12-1 transcripts was similar in all organs tested (leaves, stem, and flowers) with the exception of the roots, where the expression was only just detectable (this was confirmed by increasing the number of cycles from 24 to 28, data not shown) (Fig. 2A). In contrast, the level of CP12-2 expression was greatest in leaves, with significantly lower levels of transcripts in the stem and flowers, and no detectable expression in root tissue. No CP12-3 transcript was found in any tissues examined using the RT-PCR (24 cycles) conditions that revealed CP12-1 and CP12-2 expression. However, by increasing the number of cycles to 45, CP12-3 transcripts were detected in the stem and to a lesser extent in leaves and roots (Fig. 2A). The expression patterns for PRK and plastid GAPDH genes were also determined. The highest levels of expression of the Calvin cycle genes GAPA-1, GAPB, and PRK were in chloroplast-containing tissues; leaves, stems, and green flowers, with some expression of GAPA-1 and GAPB also evident in white flowers. The transcripts for the plastid GAPCp-type genes (GAPCp1 and GAPCp2) are below the level of detection in leaves and stems when 24 cycles of amplification were used, but expression of both of these transcripts was observed in white flowers and roots (Fig. 2A). In both dried and imbibed seeds, significant levels of CP12-1 gene expression were detected; the only other transcript detected in the group being investigated was GAPCp1 (Fig. 2B).
Fig. 2.
RT-PCR expression analysis in different Arabidopsis organs. (A) Leaves, stems, flowers, and roots of 6-week-old plants grown in soil. (B) Dry and imbibed (24 h) seeds. Non-saturating cycling parameters were determined for each primer set. CP12-1, CP12-2, GapA-1, GapB, PRK, GapCp1, and GapCp2 fragments were amplified by 24 cycles; CP12-3 was amplified by 45 cycles. The elongation factor (EF) was used as a control and was amplified by 24 cycles. Immature (Green Fl) and mature (White Fl) flowers; results are representative of two independent experiments.
Differential expression of the CP12 genes in response to light and dark
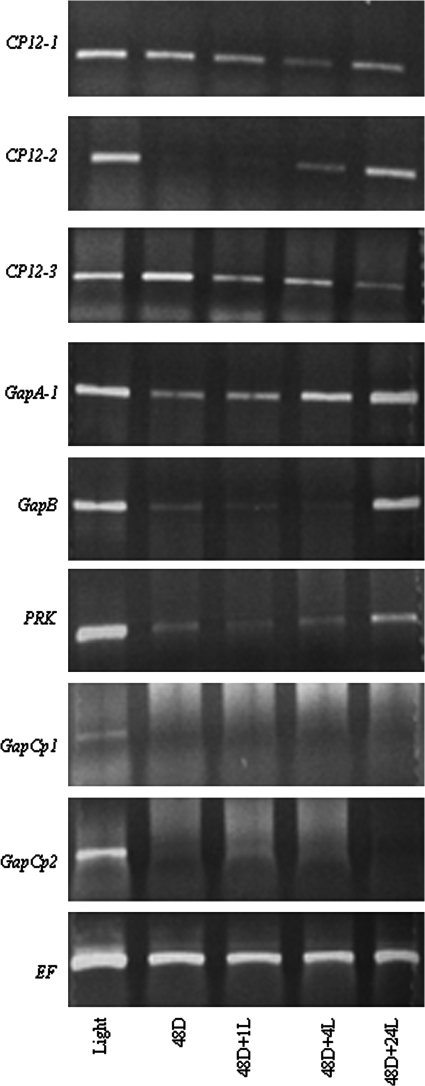
Arabidopsis plants were grown for a period of 3 weeks under a normal light–dark regime (14 h light/10 h dark) before being transferred to the dark for 48 h, following which the plants were re-illuminated for 1, 4, or 24 h. Tissue was sampled at each of these time points and total RNA extracted and subjected to RT-PCR analysis. Very little change in the CP12-1 transcripts was evident after 48 h in the dark, but transcript levels declined during the 1 h and 4 h re-illumination; however, after 24 h in the light, CP12-1 transcripts had increased but remained lower than the control (Fig. 3). In contrast, the CP12-2 transcript was below the level of detection after 48 h in the dark. However, after only 1 h of illumination, the level of transcript increased, and after 24 h levels had returned to that of control plants. Expression of GAPA-1, GAPB, and PRK transcripts responded to changes in the light/dark status in a similar manner to that of the CP12-2 gene. The response of the CP12-3 transcript was similar to that of CP12-1, with little change in expression over the time course studied (Fig. 3). A higher level of expression of the CP12-3 transcripts was observed in this experiment compared with that shown in Fig. 2A. This was due to the fact that all of the above-ground plant material was used in this experiment and higher levels of expression of the CP12-3 transcripts have been detected in stems and basal parts of the leaves (see Figs 2A, 6, and Supplementary Fig. S3 at JXB online).
Fig. 3.
Differential expression of CP12-1, CP12-2, CP12-3, GAPA-1, GAPB, PRK, GapCp1, and GapCp2 under different light and dark regimes. Arabidopsis plants were grown for 4 weeks in soil under 14 h light/10 h dark conditions, transferred to complete darkness for 48 h, and then illuminated for 1, 4, and 24 h. RNA was extracted from whole seedlings (excluding roots) and RT-PCR analysis was carried out as in Fig.1; results are representative of three independent experiments Fig. 2.
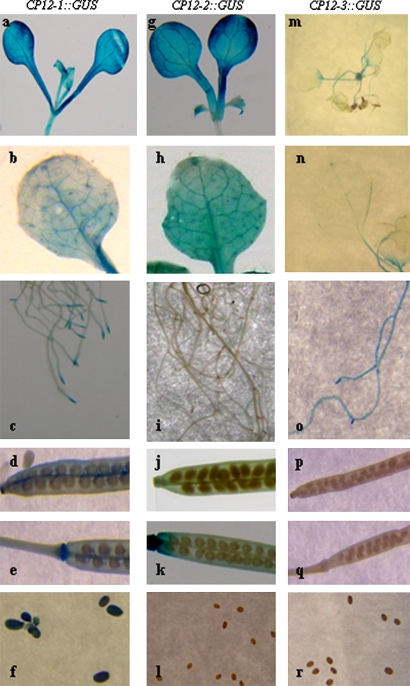
Fig. 6.
Localization of CP12-1::GUS, CP12-2::GUS, and CP12-3::GUS activity in transgenic Arabidopsis plants. Plants were germinated and grown in soil under 14 h light/10 h dark conditions in a controlled-environment room and samples taken throughout the life cycle of the plants. Cotyledons (a, g), young seedling (m), rosette leaves (b, h, n), roots (c, i, o), siliques (d, e, j, k, p, q), and seeds (f, l, r). Data shown are representative of results from a minimum of three independent lines for each of the CP12::GUS constructs.
Differential effects of environmental stress on the expression of the CP12-1, -2, and -3 genes
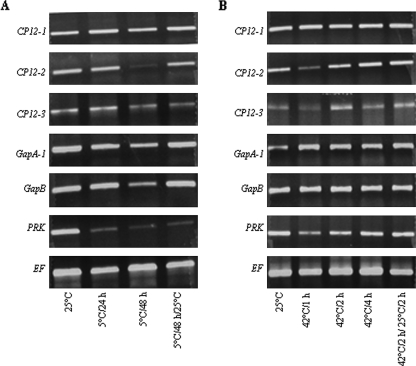
To explore the possibility that the CP12 proteins play a role in the regulation of plastid metabolism in response to environmental stress, the effect of low and high temperature shifts and anaerobic conditions were examined. No major effect on the transcript levels of the CP12-1 or CP12-3 genes was evident in response to a shift to 5 °C for 24 h or 48 h (Fig. 4). In contrast, after 48 h at 5 °C, a large decrease in CP12-2 transcript level was observed and this correlated with a reduction in the transcript for both GAPA-1 and GAPB. The most dramatic effect of low temperature was seen in the case of the PRK transcript which was only just detectable after 24 h at 5 °C and did not increase on return to 25 °C for 24 h. Increasing temperature also had little effect on expression of the CP12-1, GAPA-1, or GAPB genes. A small decrease in CP12-2, CP12-3, and PRK transcript levels was evident after 1 h at 42 °C, but this recovered to control levels while the plants were kept at 42 °C (Fig. 4).
Fig. 4.
Expression analysis of CP12-1, CP12-2, CP12-3, GAPA-1, GAPB, and PRK genes of Arabidopsis in response to temperature. Arabidopsis plants were grown in soil for 4 weeks under 14 h light/10 h dark, 25 °C, transferred to (A) 5 °C for 24 h or 48 h, and then returned to 25 °C for 24 h; and (B) 42 °C for 1, 2, and 4 h, and then returned to 25 °C for 2 h. RT-PCR analysis was carried out as described in Fig. 2; results are representative of two independent experiments.
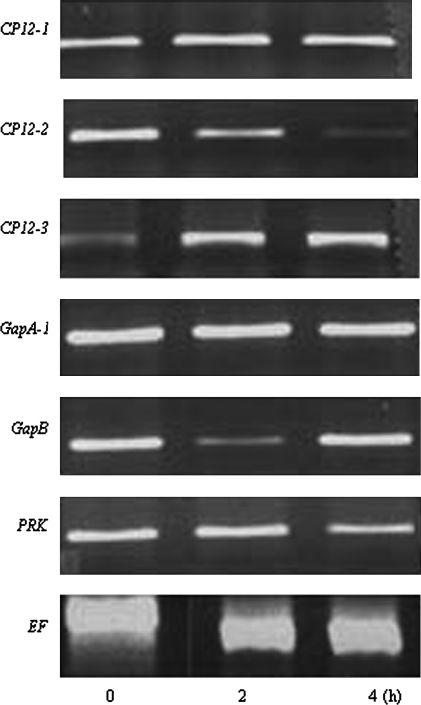
To simulate anaerobic conditions, Arabidopsis seedlings grown on Petri dishes for 12 d were submerged in sterile double-distilled water for either 2 h or 4 h. RT-PCR analysis showed that there was no change in the transcript levels of either the CP12-1 or GAPA-1 genes (Fig. 5). The expression of the CP12-2 transcript progressively decreased during the 2–4 h anaerobic treatment, and the PRK transcript showed a similar trend. The GAPB transcript decreased significantly after 2 h anaerobic treatment but had returned to the control level at the 4 h time point. Uniquely, the transcript level of CP12-3 increased and remained high throughout the experiment (Fig. 5). This increase in expression was also observed in the CP12-3::GUS plants where blue staining was evident after 4 h anaerobic treatment (Supplementary Fig. S1 at JXB online).
Fig. 5.
Expression analysis of CP12-1, CP12-2, CP12-3, GAPA-1, GAPB, and PRK genes of Arabidopsis in response to anaerobic conditions. Arabidopsis seedlings were grown on agar (1/2 MS and 1% sucrose) for 2 weeks and then immersed in water for 2 h or 4 h prior to sampling for RNA. RT-PCR analysis was carried out as described in Fig. 2; results are representative of three independent experiments.
GUS expression analysis of the CP12 genes in Arabidopsis
To gain a more detailed picture of the tissue-specific expression of the CP12 genes, a transgenic approach using promoter–GUS fusions was undertaken. The upstream regions of the CP12-1, -2, and -3 genes were isolated from Arabidopsis genomic DNA using gene-specific primers, were placed upstream of the GUS reporter gene, and transferred to Arabidopsis plants using the floral dip method of transformation. Putative transformants were selected as kanamycin-resistant seedlings and then tested for GUS activity. A number of GUS-positive lines were identified for each of the CP12 genes, and a minimum of three independent CP12::GUS transgenic lines were used in each of the expression studies reported below.
Tissue-specific and light-dependent CP12-1::GUS, CP12::GUS and CP12-3::GUS activity:
CP12-1::GUS-expressing lines showed GUS staining in the hypocotyl, cotyledons, and mature rosette leaves (Fig. 6a, b). Notably and in keeping with RT-PCR analysis, expression of CP12-1::GUS was also observed in root tips and lateral roots (Fig. 6c). GUS staining was also evident at the tip and the base of the silique, in the funiculus of the CP12-1::GUS lines (Fig. 6d, e), and in the mature seeds. The most intense staining was in the seed coat and micropyle (Fig. 6f). A high level of GUS staining was observed in the cotyledons and the mature leaves of CP12-2::GUS lines (Fig. 6g, h) and at the base and tip of the silques (Fig. 6j, k). However, in the CP12-2::GUS lines, no GUS staining was present in the hypocotyl, the roots, or seeds (Fig. 6g, i, l). In the CP12-3::GUS plants, GUS activity was detected throughout the roots, in stems, and at low levels in leaves (Fig. 6m–o), but no expression was detected in the seeds or siliques (Fig. 6p–r).
The expression of the CP12-1, CP12-2, and CP12-3 genes was also investigated in etiolated seedlings. Seeds of the CP12::GUS transgenic lines were germinated in the dark and grown for a period of 14 d before being transferred into the light for between 1 h and 24 h. GUS activity was detected in the cotyledons of the dark-grown etiolated seedlings in all of the CP12-1::GUS lines but not in either the CP12-2::GUS or CP12-3::GUS plants (Supplementary Fig. S2 at JXB online). Following 1 h of illumination, GUS activity in the CP12-2::GUS lines was similar to that seen in the CP12-1::GUS plants. In contrast, no detectable level of staining was evident in the CP12-3::GUS plants even after 24 h in the light (Supplementary Fig. S2 at JXB online).
CP12-1::GUS, CP12-2::GUS, and CP12-3::GUS expression in flowers:
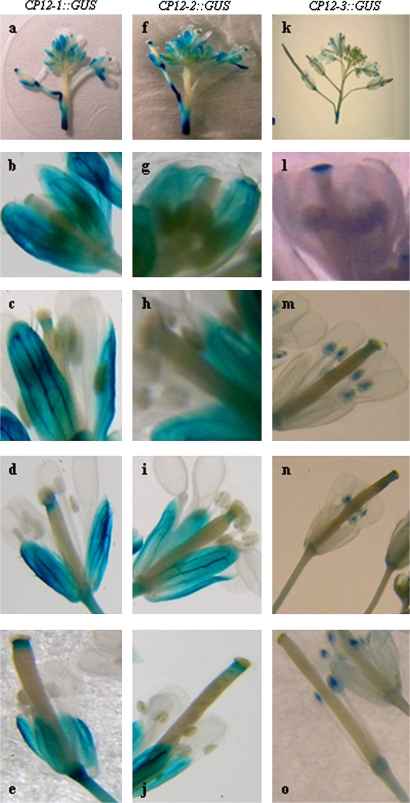
RT-PCR analysis had indicated that both CP12-1 and CP12-2 transcripts were present in floral tissues. In keeping with this, GUS activity was detected in the sepals and the style in all of the CP12-1::GUS and CP12-2::GUS lines (Fig. 7a, f). Interestingly, the intensity of the GUS staining in the style increased as the flowers matured (Fig. 7b–e, g–j). Notably no GUS staining was detected in the petals, stigma, anthers, or filaments at any stage of flower development in any of the CP12-1::GUS or CP12-2::GUS lines. In floral tissues of the CP12-3::GUS lines, GUS activity was restricted to the stigma and the anthers, with no staining detected in the style, petals, or sepals (Fig. 7k), and in this case the level of GUS staining in the stigma reduced as the flowers matured (Fig. 7l–o).
Fig. 7.
Localization of CP12-1::GUS, CP12-2::GUS, and CP12-3::GUS activity in developing flowers of Arabidopsis transgenic plants. Plants were germinated and grown in soil under 14 h light/10 h dark conditions in a controlled-environment room, and tissues were analysed following the appearance of the flowers. Inflorescences (a, f, k), flowers at stage 13 (b, g, l), stage 14 (c, d, h, i, m, n), and stage 15 (e, j, o). Data shown are representative patterns of expression obtained from a minimum of three independent lines for each of the CP12::GUS constructs.
Expression of the CP12 gene family as revealed by gene profiling data
Investigation of the expression of the Arabidopsis CP12 gene family using the publicly available micorarray data obtained from Genevestigator (http://www.genevestigator.etch.ch; (Zimmermann et al., 2004) supports both the RT-PCR and GUS analysis data presented herein (Supplementary Fig. S3 at JXB online). These data revealed that the level of expression of CP12-1 was higher than that of CP12-2 and CP12-3 and it is expressed in all tissues of seedlings and mature plants. In contrast, CP12-2 is predominantly expressed in green photosynthetic tissues such as cotyledons, sepals, and leaves. In most tissues the expression level of CP12-3 was negligble in comparison with that of either CP12-1 or CP12-2 (Supplementary Fig. S3 at JXB online).
Discussion
Evolutionary history of CP12 genes
Previously it was shown that the CP12 proteins in Arabidopsis are encoded by a small gene family (Marri et al., 2005a). The current bioinformatic analysis has revealed that in the plant species where a complete genome sequence is available, including the monocots and the eudicots, three CP12 genes are present. The CP12 proteins that are most similar to the closely related Arabidopsis CP12-1 and CP12-2 proteins form one group (Type I) and those most like the Arabidopsis CP12-3 protein form a second group (Type II). The CP12-1- and CP12-2-like proteins within a eudicot family are more similar to each other than either the CP12-1 or CP12-2 protein between families. This suggests that the CP12-1 and CP12-2 genes may have arisen as a result of a recent gene duplication. In contrast, in the monocots, the CP12-1- and CP12-2-like proteins from different species form two groups. This suggests that the gene duplication that resulted in the production of these two closely related proteins must have taken place independently after the divergence of the monocot and eudicots. This is an interesting observation indicating that in the angioperms there is a functional requirement for two Type I CP12 proteins.
Co-ordinate expression of the CP12 gene family with PRK and the plastid GAPDH isozymes
Analysis of publicly available microarray data (Supplementary Fig. S3 at JXB online), together with the RT-PCR expression studies reported herein, has shown that the three Arabidopsis CP12 genes are differentially expressed. As expected from a previous study, the tissue-specific and light-dependent expression pattern of CP12-2 was found to be most closely correlated with that of GapB and PRK, while expression of the CP12-1 gene remained constant and was most closely correlated with that of GAPA-1. These results are in keeping with role of CP12 in the formation of the multiprotein complex with PRK and GAPDH in the chloroplast stroma (Wedel et al., 1997; Scheibe et al., 2002; Graciet et al., 2004; Marri et al., 2005a; Trost et al., 2006). However, under anaerobic treatment, the correlation between expression of CP12-2 and that of PRK, GAP-A1, or GAPB was not evident. Furthermore, CP12-1 transcripts were detected in roots and seeds, and this was supported by the pattern of staining observed in the CP12-1::GUS plants. Interestingly, expression in the roots of the CP12-1::GUS lines was restricted to the tip and the newly emerging lateral roots, indicating a role for the CP12-1 protein in meristematic tissue and cells undergoing differentiation. The expression profile of the CP12-1 genes indicates that, this protein may play a role in metabolism during early seedling growth and in roots. Detectable levels of the CP12-3 transcript were also found in roots, and in this case GUS staining was observed throughout the roots of the CP12-3::GUS plants. No expression of GAPA-1, GAPB, or PRK was found in roots or seeds, suggesting that the function of CP12-1 and -3 proteins in these tissues is unlikely to involve the PRK and GAPDH proteins. However, transcripts for GAPCp1 and GAPCp2 were detected in roots, and that of GAPCp1 in seeds. The role of the GAPCp protein has not been fully elucidated, but it has been suggested that in non-photosynthetic plastids this form of GAPDH may be NAD dependent and may play a role in energy production during dark metabolism (Backhausen et al., 1998; Petersen et al., 2003). It is tempting to speculate that CP12-1 and CP12-3 proteins may interact with this form of GAPDH in these tissues. However, at present there are no data on interactions with GAPCp proteins to support this hypothesis.
The expression of the CP12-3 gene transcript increased in leaves of plants subjected to anaerobic treatment. Analysis of the CP12-3::GUS lines showed that increased expression of the CP12-3 gene occurred throughout the leaf. It has been shown previously that reduction of internal oxygen levels triggers a number of responses including changes in gene expression and metabolism (Geigenberger, 2003). The high expression level of the CP12-3 gene under these conditions may indicate that the CP12-3 protein is involved in regulating metabolism in response to changes in oxygen status. A role for the chloroplast NADP-GAPDH, encoded by the GAPA and GAPB genes, in the provision of NADP+ required for glycolysis and ATP production during anaerobic metabolism has been proposed (Yang et al., 1993). Although a transient decrease in the level of the GAPB gene transcript was observed in the plants, after 4 h treatment the levels of both the GAPA-1 and GAPB genes were similar to that of the control.
The CP12 gene family is expressed in reproductive tissues
Analysis of the transgenic Arabidopsis plants expressing the CP12-1, -2, and -3 promoter::GUS constructs revealed tissue-specific patterns of expression of these genes in floral tissues. Although RT-PCR had detected transcripts for the CP12 genes in flowers, the tissue-specific and developmental patterns of expression revealed in this study were unexpected. In the CP12-1::GUS and CP12-2::GUS lines, GUS staining was seen in the style and the sepals, with no GUS staining detected in any other part of the flower. Of particular note was the observation that in both the CP12-1::GUS and CP12-2::GUS lines, the intensity of the GUS staining increased during floral maturation. GUS expression was also observed in the flowers of the CP12-3::GUS plants. In contrast to the pattern observed in the CP12-1::GUS and CP12-2::GUS plants, GUS staining in the CP12-3::GUS lines was restricted to the stigma and the anthers. The GUS staining detected in the cells of the stigmatic surface in young flowers (stage 13) decreased as the flowers matured. Although the physiological significance of these observations remains to be resolved, these results are interesting and suggest a novel function for the CP12 proteins in metabolic processes that take place in floral tissues. Very little is known about the role of metabolism during the fertilization process, but what is clear is that the energetic demands of the tissues involved will increase. Furthermore, the oxygen status of the pistil and the pollen grains is known to be low, and this is likely to require a switch in metabolism. There is now some evidence that changes in metabolites such as the four-carbon amino acid GABA (γ-aminobutyric acid) and production of reactive oxygen species may be an important part of the guidance system directing the pollen tube to the ovary (Palanivelu et al., 2003; Dong et al., 2005; McInnis et al., 2006). The present data suggest that it is possible that the CP12 proteins have a role in the redox modulation of metabolic process that occur prior to and leading up to pollination and fertilization.
The ability of the CP12-1 and -2 proteins to form a complex with chloroplastic PRK and GAPDH has been shown to be regulated via changes in the redox state of the two cysteine pairs, most probably mediated by the chloroplastic thioredoxin f, although a role for thioredoxin m cannot be ruled out at this stage (Marri et al., 2008; Howard et al., 2008). In addition to the well described role of the thioredoxin f and m proteins in chloroplasts, it has been shown recently that both of these forms of chloroplast thioredoxin are expressed in a wide range of non-photosynthetic tissues including roots, seed, flowers, and pollen grains (de Dios Barajas-Lopez et al., 2007; Traverso et al., 2008). This raises the intriguing possibility that the CP12 proteins may function together with specific thioredoxin isoforms to form a redox network regulating metabolic processes in plastids of both photosynthetic and heterotrophic plant tissues in response to changes in the availability of reducing power. Clearly the present analysis provides information only on patterns of gene expression, and therefore many questions still remain about the function of the CP12 protein family.
The high degree of similarity between the CP12-l- and CP12-2-like proteins together with the overlapping expression patterns of the genes encoding these proteins suggests that they may be functionally redundant. However, differences in the spatiotemporal expression of these genes provided evidence that the CP12-1 protein may have a role in roots and seeds and possibly during early germination. Furthermore, the analyses of tissue-specific expression in GUS-expressing transgenic lines suggest that the CP12 proteins may have a novel role in the fertilization process. These data suggest that the function of the CP12 proteins in higher plants is unlikely to be restricted to the regulation of the PRK and GAPDH activity in the Calvin cycle. Further support for this comes from a recent study demonstrating that the Chlamydomonas CP12 can bind to plastid aldolase (Erales et al., 2008). Interestingly, in all tissues and conditions studied here, expression of one of the CP12 genes correlates with expression of at least one member of the GAPDH gene family and therefore it is possible that the CP12 proteins are capable of interacting with one or more of the different GAPDH proteins in different tissues.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Histochemical staining of two CP12-3::GUS lines after anaerobic treatment.
Fig. S2. GUS expression analysis in the CP12-1::GUS, CP12-2::GUS, and CP12-3 ::GUS transgenic lines in dark-grown seedlings.
Fig. S3. Expression profiling data for the Arabidopsis CP12 genes.
Supplementary Material
Acknowledgments
The authors would like to thank Dr JC Lloyd, Essex University for critical reading of the manuscript, and Dr T Lawson for the photography. This work was supported by an ORS award and University of Essex studentship (PS) and BBSRC grant P19403 (DK).
References
- Baalmann E, Scheibe R, Cerff R, Martin W. Functional studies of chloroplast glyceraldehyde-3-phosphate dehydrogenase subunits A and B expressed in Escherichia coli: formation of highly active A(4) and B-4 homotetramers and evidence that aggregation of the B-4 complex is mediated by the B subunit carboxy terminus. Plant Molecular Biology. 1996;32:505–513. doi: 10.1007/BF00019102. [DOI] [PubMed] [Google Scholar]
- Backhausen JE, Emmerlich A, Holtgrefe S, Horton P, Nast G, Rogers JJM, Muller-Rober B, Scheibe R. Transgenic potato plants with altered expression levels of chloroplast NADP-malate dehydrogenase: interactions between photosynthetic electron transport and malate metabolism in leaves and in isolated intact chloroplasts. Planta. 1998;207:105–114. [Google Scholar]
- Boggetto N, Gontero B, Maberly SC. Regulation of phosphoribulokinase and glyceraldehyde 3-phosphate dehydrogenase in a freshwater diatom, Asterionella formosa. Journal of Phycology. 2007;43:1227–1235. doi: 10.1111/j.1529-8817.2008.00600.x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- de Dios Barajas-Lopez J, Serrato AJ, Olmedilla A, Chueca A, Sahrawy M. Localization in roots and flowers of pea chloroplastic thioredoxin f and thioredoxin m proteins reveals new roles in nonphotosynthetic organs. Plant Physiology. 2007;145:946–960. doi: 10.1104/pp.107.105593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Kim ST, Lord EM. Plantacyanin plays a role in reproduction in Arabidopsis. Plant Physiology. 2005;138:778–789. doi: 10.1104/pp.105.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nature Reviews Molecular Cell Biology. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Erales J, Avilan L, Lebreton S, Gontero B. Exploring CP12 binding proteins revealed aldolase as a new partner for the phosphoribulokinase/glyceraldehyde 3-phosphate dehydrogenase/CP12 complex—purification and kinetic characterization of this enzyme from Chlamydomonas reinhardtii. FEBS Journal. 2008;275:1248–1259. doi: 10.1111/j.1742-4658.2008.06284.x. [DOI] [PubMed] [Google Scholar]
- Gardebien F, Thangudu RR, Gontero B, Offmann B. Construction of a 3D model of CP12, a protein linker. Journal of Molecular Graphics and Modelling. 2006;25:186–195. doi: 10.1016/j.jmgm.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Geigenberger P. Response of plant metabolism to too little oxygen. Current Opinion in Plant Biology. 2003;6:247–256. doi: 10.1016/s1369-5266(03)00038-4. [DOI] [PubMed] [Google Scholar]
- Graciet E, Gans P, Wedel N, Lebreton S, Camadro JM, Gontero B. The small protein CP12: a protein linker for supramolecular complex assembly. Biochemistry. 2003;42:8163–8170. doi: 10.1021/bi034474x. [DOI] [PubMed] [Google Scholar]
- Graciet E, Lebreton S, Gontero B. Emergence of new regulatory mechanisms in the Benson–Calvin pathway via protein–protein interactions: a glyceraldehyde-3-phosphate dehydrogenase/CP12/phosphoribulokinase complex. Journal of Experimental Botany. 2004;55:1245–1254. doi: 10.1093/jxb/erh107. [DOI] [PubMed] [Google Scholar]
- Howard TP, Metodiev M, Lloyd JC, Raines CA. Thioredoxin-mediated reversible dissociation of a stromal multiprotein complex in response to changes in light availability. Proceedings of the National Academy of Sciences, USA. 2008;105:4056–4061. doi: 10.1073/pnas.0710518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions—β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton S, Andreescu S, Graciet E, Gontero B. Mapping of the interaction site of CP12 with glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii—functional consequences for glyceraldehyde-3-phosphate dehydrogenase. FEBS Journal. 2006;273:3358–3369. doi: 10.1111/j.1742-4658.2006.05342.x. [DOI] [PubMed] [Google Scholar]
- Marri L, Sparla F, Pupillo P, Trost P. Co-ordinated gene expression of photosynthetic glyceraldehyde-3-phosphate dehydrogenase, phosphoribulokinase, and CP12 in Arabidopsis thaliana. Journal of Experimental Botany. 2005a;56:73–80. doi: 10.1093/jxb/eri020. [DOI] [PubMed] [Google Scholar]
- Marri L, Trost P, Pupillo P, Sparla F. Reconstitution and properties of the recombinant glyceraldehyde-3-phosphate dehydrogenase/CP12/phosphoribulokinase supramolecular complex of Arabidopsis. Plant Physiology. 2005b;139:1433–1443. doi: 10.1104/pp.105.068445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marri L, Trost P, Trivelli X, Gonnelli L, Pupillo P, Sparla F. Spontaneous assembly of photosynthetic supramolecular complexes as mediated by the intrinsically unstructured protein CP12. Journal of Biological Chemistry. 2008;283:1831–1838. doi: 10.1074/jbc.M705650200. [DOI] [PubMed] [Google Scholar]
- McInnis SM, Desikan R, Hancock JT, Hiscock SJ. Production of reactive oxygen species and reactive nitrogen species by angiosperm stigmas and pollen: potential signalling crosstalk? New Phytologist. 2006;172:221–228. doi: 10.1111/j.1469-8137.2006.01875.x. [DOI] [PubMed] [Google Scholar]
- Oesterhelt C, Klocke S, Holtgrefe S, Linke V, Weber APM, Scheibe R. Redox regulation of chloroplast enzymes in Galdieria sulphuraria in view of eukaryotic evolution. Plant and Cell Physiology. 2007;48:1359–1373. doi: 10.1093/pcp/pcm108. [DOI] [PubMed] [Google Scholar]
- Page RDM. TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Brass L, Edlund AF, Preuss D. Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell. 2003;114:47–59. doi: 10.1016/s0092-8674(03)00479-3. [DOI] [PubMed] [Google Scholar]
- Penfield S, Josse EM, Kannangara R, Gilday AD, Halliday KJ, Graham IA. Cold and light control seed germination through the bHLH transcription factor SPATULA. Current Biology. 2005;15:1998–2006. doi: 10.1016/j.cub.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Petersen J, Brinkmann H, Cerff R. Origin, evolution, and metabolic role of a novel glycolytic GAPDH enzyme recruited by land plant plastids. Journal of Molecular Evolution. 2003;57:16–26. doi: 10.1007/s00239-002-2441-y. [DOI] [PubMed] [Google Scholar]
- Pohlmeyer K, Paap BK, Soll J, Wedel N. CP12: a small nuclear-encoded chloroplast protein provides novel insights into higher-plant GAPDH evolution. Plant Molecular Biology. 1996;32:969–978. doi: 10.1007/BF00020493. [DOI] [PubMed] [Google Scholar]
- Raines CA, Lloyd JC, Dyer TA. Molecular-biology of the C3-photosynthetic carbon-reduction cycle. Photosynthesis Research. 1991;27:1–14. doi: 10.1007/BF00029971. [DOI] [PubMed] [Google Scholar]
- Raines CA, Paul MJ. Products of leaf primary carbon metabolism modulate the developmental programme determining plant morphology. Journal of Experimental Botany. 2006;57:1857–1862. doi: 10.1093/jxb/erl011. [DOI] [PubMed] [Google Scholar]
- Scheibe R, Wedel N, Vetter S, Emmerlich V, Sauermann SM. Co-existence of two regulatory NADP-glyceraldehyde 3-P dehydrogenase complexes in higher plant chloroplasts. European Journal of Biochemistry. 2002;269:5617–5624. doi: 10.1046/j.1432-1033.2002.03269.x. [DOI] [PubMed] [Google Scholar]
- Singh P. Molecular analysis of the Arabidopsis CP12 gene family. PhD dissertation. University of Essex. 2007 [Google Scholar]
- Sparla F, Marri L, Trost P, Trivelli X, Gonnelli L. Assembly of supramolecular complexes of Calvin cycle enzymes as mediated by the intrinsically unstructured protein CP12. Photosynthesis Research. 2007;91:235–235. [Google Scholar]
- Sparla F, Pupillo P, Trost P. The C-terminal extension of glyceraldehyde-3-phosphate dehydrogenase subunit B acts as an autoinhibitory domain regulated by thioredoxins and nicotinamide adenine dinucleotide. Journal of Biological Chemistry. 2002;277:44946–44952. doi: 10.1074/jbc.M206873200. [DOI] [PubMed] [Google Scholar]
- Sun N, Ma LG, Pan DY, Zhao HY, Deng XW. Evaluation of light regulatory potential of Calvin cycle steps based on large-scale gene expression profiling data. Plant Molecular Biology. 2003;53:467–478. doi: 10.1023/B:PLAN.0000019071.12878.9e. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. Intrinsically unstructured proteins. Trends in Biochemical Sciences. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- Traverso JA, Vignols F, Cazalis R, Serrato AJ, Pulido P, Sahrawy M, Meyer Y, Cejudo FJ, Chueca A. Immunocytochemical localization of Pisum sativum TRXs f and m in non-photosynthetic tissues. Journal of Experimental Botany. 2008;59:1267–1277. doi: 10.1093/jxb/ern037. [DOI] [PubMed] [Google Scholar]
- Trost P, Fermani S, Marri L, Zaffagnini M, Falini G, Scagliarini S, Pupillo P, Sparla F. Thioredoxin-dependent regulation of photosynthetic glyceraldehyde-3-phosphate dehydrogenase: autonomous vs. CP12-dependent mechanisms. Photosynthesis Research. 2006;89:263–275. doi: 10.1007/s11120-006-9099-z. [DOI] [PubMed] [Google Scholar]
- Wedel N, Soll J. Evolutionary conserved light regulation of Calvin cycle activity by NADPH-mediated reversible phosphoribulokinase/CP12/glyceraldehyde-3-phosphate dehydrogenase complex dissociation. Proceedings of the National Academy of Sciences, USA. 1998;95:9699–9704. doi: 10.1073/pnas.95.16.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedel N, Soll J, Paap BK. CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants. Proceedings of the National Academy of Sciences, USA. 1997;94:10479–10484. doi: 10.1073/pnas.94.19.10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YJ, Kwon HB, Peng HP, Shih MC. Stress responses and metabolic-regulation of glyceraldehyde-3-phosphate dehydrogenase genes in Arabidopsis. Plant Physiology. 1993;101:209–216. doi: 10.1104/pp.101.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Genevestigator GW. Arabidopsis microarray database and analysis toolbox (vol 136, pg 2621, 2004) Plant Physiology. 2004;136:4335–4335. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.