Abstract
The annotation of novel plant genes is frequently based on sequence and structural similarity to known protein motifs. Understanding the biological function of these genes is dependent on identifying conditions under which they are activated, however. The resurrection plant, Xerophyta humilis is a good model system for identifying and characterizing genes which are important for desiccation tolerance. Desiccation induced-1 (dsi-1VOC), a previously uncharacterized plant gene, is up-regulated during desiccation in leaves, roots, and seeds in X. humilis. The X. humilis desiccation induced-1 gene, Xhdsi-1VOC, shares structural homology with the vicinal oxygen chelate (VOC) metalloenzyme superfamily. Proteins in this superfamily share little sequence similarity, but are characterized by a common βαβββ structural fold. A number of plant orthologues of XhDsi-1VOC have been identified, including Arabidopsis thaliana At1g07645, which is currently annotated as a glyoxalase I-like gene, and many ESTs derived from seed cDNA libraries. Xhdsi-1VOC and its orthologues do not, however, contain the glutathione and zinc binding sites conserved in glyoxalase I genes. Furthermore, expression of Xhdsi-1VOC in yeast failed to rescue a yeast glyoxalase I mutant. Messenger RNA transcripts for At1g07645 accumulate during seed maturation, but are not induced by water loss, salt or mannitol stress in vegetative tissue in Arabidopsis. It is concluded that dsi-1VOC is a seed-specific gene in desiccation-sensitive plants that is activated by water loss in vegetative tissues in the resurrection plant X. humilis and plays an important role in allowing plant tissues to survive loss of 95% of their relative water content.
Keywords: At1g07645, Desiccation, Dsi-1VOC, glyoxalase, resurrection plant, vicinal oxygen chelate metalloenzyme, VOC metalloenzyme, Xerophyta
Introduction
Desiccation tolerance in vascular plants is common in seeds, spores, and pollen but there are only 350 species (0.2% of total flora) that have the ability to survive desiccation of their vegetative tissue (Proctor and Tuba, 2002; Ingram and Bartels, 1996). These plants are known as resurrection plants as they are able to survive a loss of 95% of their relative water content and resume biological functions in existing tissue upon rehydration (Gaff, 1971, 1977; Michel et al., 1994; Ramanjalu and Bartels, 2002). The monocotyledonous resurrection plant Xerophyta humilis, which is indigenous to southern Africa, has been used as a model system to study the molecular basis of desiccation tolerance in vegetative tissue.
X. humilis is poikilochlorophyllous, and during periods of water loss, it avoids much of the free radical damage induced by photosynthesis by degrading chlorophyll, down-regulating expression of subunits of photosystem II and photosystem I, and dismantling the thylakoid membranes into small vesicles in response to desiccation (Sherwin and Farrant, 1996; Collett et al., 2003, 2004). In addition to reducing the damage caused by photosynthesis under stress, X. humilis and other resurrection plants have to maintain the integrity of cellular and subcellular organization and prevent oxidative damage induced by deregulated metabolic processes during both desiccation and the subsequent period of rehydration (Proctor and Tuba, 2002; Farrant, 2007).
Several studies have shown that standard anti-oxidant enzymes such as ascorbate peroxidase, glutathione reductase, and superoxide dismutase remain viable during desiccation in desiccation-tolerant resurrection plants, compared to the same enzymes in their sister desiccation-sensitive species (Illing et al., 2005; Farrant, 2007). Clearly these enzymes must be protected in some way. Several studies have shown that concentrations of compatible solutes such as sucrose, the raffinose family of oligosaccharides (Ghasempour et al., 1998; Whittaker et al., 2001, 2004; Illing et al., 2005; Moore et al., 2007; Peters et al., 2007) and proline (Vander Willigen et al., 2004) increase during desiccation and that these are likely to play an important role in the protection of membranes and proteins (for the current understanding of these roles, see Berjak et al., 2007; Farrant, 2007). Furthermore, proteins belonging to the late embryo abundant (LEA) families are highly expressed in both desiccated seed and vegetative tissue in response to desiccation (Illing et al., 2005). Several researchers have speculated that LEA proteins play an important role in the physiological protection of important proteins against denaturation during desiccation. In addition to the protection of housekeeping antioxidants, membranes, and functional proteins, cells require proteins for the repair of damage induced during desiccation and the detoxification of metabolic by-products. The thiol specific antioxidant 1-Cys peroxiredoxin, for instance, is apparently active only during the desiccation phase of orthodox seed development (Stacy et al., 1996) and has also been reported to be induced during desiccation of vegetative tissues of the resurrection plant Xerophyta viscosa (Mowla et al., 2002). We predict that other proteins that are active in the desiccation phase of orthodox seed development are similarly present in the dry vegetative tissue of desiccation-tolerant plants. The characterization of novel seed-specific genes from desiccation-sensitive plants that are expressed in vegetative tissue in X. humilis in response to desiccation, and which might be important for allowing cells to survive extreme water loss, is of particular interest.
As a first step in characterizing the transcriptome in desiccated X. humilis leaves, a small-scale microarray screen was completed using 424 annotated cDNA randomly selected from a 10 900 cDNAs derived from X. humilis leaf and root libraries (Collett et al., 2004). A number of differentially expressed cDNAs were identified, including 55 cDNAs that were up-regulated in leaves during desiccation. Several of these cDNAs were homologues of genes that were already known to be expressed in the desiccated tissue of resurrection plants. These included cDNAs annotated as enzymes that synthesize osmoprotectants such as aldose reductase and galactinol synthase, as well as protective proteins including several LEA proteins and dehydrins. (Collett et al., 2004). However, in addition to these familiar faces, there were many novel cDNAs which showed significant similarity to Arabidopsis genes that were either annotated as genes of unknown function, or were annotated on the basis of their structural similarity to known enzymes. Given the potential importance of detoxifying enzymes in desiccation survival, one of these cDNAs, HC205, was selected for further study, as it shared significant similarity with an Arabidopsis gene At1g07645, annotated as a glyoxalase-1 like protein in the database.
Changes in environmental factors such as water loss, temperature extremes, and high light, perturb metabolic pathways in photosynthesis and respiration in plants, resulting in an increase in active oxygen species and glycating agents (Noctor and Foyer, 1998; Mittler et al., 2004). The former induce cellular damage such as lipid peroxidation, protein degradation, modification and damage of DNA (Fridovich, 1986). The latter modify proteins, nucleic acids and basic phospholipids, producing advanced glycation end-products (Thornalley, 2003). Active oxygen species include superoxide (), hydroxyl radicals (OH.), hydrogen peroxide (H2O2), and singlet oxygen (Noctor and Foyer, 1998; Dutilleul et al., 2003). Glycation agents include oxoaldehydes, such as glyoxal formed by lipid peroxidase and glyoxalate oxidation (Freire et al., 2003), and methylglyoxal, a triosephosphate by-product of glycolysis and the breakdown product of threonine and acetone (Freire et al., 2003; Thornalley, 2003).
Methylglyoxal is detoxified to hydroxycarboxylic acids by the glyoxalase system which is found in animals, plants, and bacteria (Thornalley, 2003). Methylglyoxal and reduced glutathione react spontaneously to form a hemithioacetal that is recognized by glyoxalase I (also known as lactoylglutathione lyase), and is then isomerized to S-D-lactoylglutathione. In the presence of H2O, the S-D-lactoylglutathione is catalysed by glyoxalase II (hydroxy-acylglutathione hydrolase) into D-lactic acid, and the reduced glutathione is recycled (Martins et al., 2001; Thornalley, 2003).
Glyoxalase I has been shown to be up-regulated in plants in response to environmental stresses (Veena et al., 1999; Martins et al., 2001; Singla-Pareek et al., 2003). For example, glyoxalase I mRNA transcripts and protein levels increase in Brassica juncea (Veena et al., 1999) and tomato (Espartero et al., 1995) when the plants are exposed to salt stress, osmotic stress, and heavy metals. Over-expression of the B. juncea glyoxalase I in a tobacco transgenic line showed a significant increase in tolerance compared to wild-type plants when treated with methyglyoxal and sodium chloride (Veena et al., 1999). In addition, over-expression of either glyoxalase I or II in tobacco plants showed higher tolerance to salt and methyglyoxal than untransformed control plants (Singla-Pareek et al., 2003). Interestingly, when both glyoxalase I and II are over-expressed in the same plant, they act synergistically, increasing tolerance levels above that of the individual genes (Singla-Pareek et al., 2003). It is speculated that glyoxalase I is up-regulated in plants that are exposed to salt stress because glycolytic activity increases due to an increased in demand for ATP (Veena et al., 1999).
Glyoxalase I is a member of the vicinal oxygen chelate (VOC) superfamily. The VOC superfamily consists of proteins that have a paired βαβββ structure fold (Armstrong, 2000; Rife et al., 2002). Several enzymes and proteins have been assigned to the VOC superfamily on the basis of the similarity in their three-dimensional structures which have been solved using X-ray crystallography. Most of these proteins are classified as metalloenzymes, and the cavities within their tertiary structures, which contain similar metal co-ordination sites, have been identified (Armstrong, 2000). It is thought that these cavities facilitate the interaction between the substrate and the metals via the vicinal oxygen atoms of the substrate, hence the name VOC (Armstrong, 2000). These metalloenzymes catalyse diverse types of reactions, however. Members of the VOC superfamily include glyoxalase I, the bleomycin-resistant protein, fosfomycin A and X resistance proteins, mitomycin resistance protein D, methylmalonl-CoA epimerase, and extradiol dioxygenases (Eltis et al., 1993; Kita et al., 1999; Armstrong, 2000; McCarthy et al., 2001; Vetting et al., 2004). The bleomycin-resistant protein is different from the other members of the VOC family; it is not an enzyme nor does it contain metal binding sites, but instead contains a hydrophobic cavity for bleomycin adhesion. The bleomycin-resistant protein inhibits damage induced by bleomycin, a glyocopeptide antibiotic from Streptomyces verticillus (Kumagai et al., 1999). It has been suggested that the paired βαβββ structure present in this diverse set of proteins is derived from an ancestral single βαβββ module, which underwent gene duplication and fusion to generate a metal binding scaffold early in evolution (Bergdoll et al., 1998; McCarthy et al., 2001).
Given the potential importance of novel detoxifying enzymes in desiccation tolerance, the characterization of HC205, and its A. thaliana orthologue, At1g07645, which are annotated as glyoxalase I-like proteins in Genbank is reported here. Despite its name, our functional analysis shows that this protein is unable to complement a glyoxalase I mutant in yeast. Furthermore, conserved amino acids that form the binding sites for zinc and glutathione in glyoxalase I are absent. Structural comparisons firmly place the protein encoded by HC205 and its plant orthologues in the VOC superfamily. In the desiccation-sensitive plant A. thaliana, At1g07645 mRNA transcripts are expressed at high levels in mature seeds, but are not detectable in vegetative tissues, even when they are exposed to abiotic stresses. By contrast, HC205 is abundantly expressed in seeds, roots, and leaves of X. humilis during a cycle of desiccation. The name desiccation induced-1VOC (dsi-1VOC) has been coined to describe HC205 and its plant orthologues.
Materials and methods
Plant material and stress conditions
X. humilis plants were collected from Barakalalo National Park (Limpopo Province, South Africa) and were maintained in trays in a greenhouse with no supplemental lighting, shading or temperature control. Plants were desiccated by withholding water, allowing the plants to dry naturally to an air-dry state (≤5% relative water content (RWC)) under ambient conditions in the greenhouse in a cycle lasting from 30 January 2007 to 12 February 2007. Plants were kept dry for 2 weeks before rehydration by irrigation of the soil. During this sampling period, relative humidity ranged from 35% to 75% on a daily basis, temperature ranged from 15 °C to 30 °C, and daylight intensity ranged from 200–1000 μmol m−2 s−1. Leaves and roots were harvested at different stages in the desiccation and rehydration time-course. Twenty leaves were harvested from different plants within a tray. Each leaf was split in half, one half was weighed, immediately frozen in liquid nitrogen, and stored at –70 °C until further analyses were performed as described below. The other half was used to determine the leaf's relative water content as described by Dace et al. (1998). The half leaves with the same relative water content were pooled in order to obtain 0.1 g of tissue for RNA extraction. 0.2 g of root material collected from different plants within the tray at the different dehydrated–rehydration time-courses were cut into 2 cm segments, the cut roots were mixed. 0.1 g was frozen in liquid nitrogen and stored at –70 °C while the other 0.1 g was used to determine the relative water content.
Mature X. humilis seeds were collected under greenhouse conditions from flowers that developed regularly in response to a drop in ambient temperature followed by high light conditions (Laura Roden, personal communication). Flowers were hand pollinated. Seeds were shaken onto white paper for collection following flower senescence and natural pod desiccation. These mature dry seeds were stored at 4 °C until required. The seeds used in the experiments were collected over a period of 2 years.
A. thaliana was used as a model of a desiccation-sensitive plant. 0.5 g of wild-type A. thaliana seeds were surface-sterilized in 70% ethanol (7 min) followed by 10% bleach containing 0.02% Triton (15 min). Seeds were thoroughly rinsed five times in sterile distilled water and resuspended in 0.1% sterile agar. The seeds were plated on plant nutrient agar (Haughn and Somerville, 1986) in sealed Petri dishes and were germinated under 16 h light (100 μmol m−2 s−1)/8 h dark at 25 °C. Two weeks after germination, plants were used for stress treatments. For an osmotic and salt stress, they were transferred to Petri dishes containing 150 mM NaCl or 300 mM mannitol. As controls, the plants were transferred to Petri dishes containing water. For dehydration stress, plants were left in plant nutrient agar but lids were removed. The control for this experiment was maintenance of sealed growth conditions. Roots and leaves were harvested from 0.1 g of plantlet tissue after 4 h and 8 h of each treatment, frozen immediately in liquid nitrogen, and stored at −70 °C. A biological repeat was performed a few weeks later by germinating seed and subjecting the 2-week-old seedlings to the above stresses. Separate batches of A. thaliana seed were used for all biological repeats.
Isolation of total RNA
Total RNA from leaves and roots of X. humilis was isolated using 1 ml of TriReagent per 150 mg tissue (Molecular Research Centre Inc, Cincinnati, USA) following manufacturer's recommendations with the modification of adding 0.01 g of polyvinylpolypyrrolidone (PVPP) per 1 ml of TriReagent to inactivate polyphenolics. The phenol–chloroform phase was stored at –20 °C for protein extraction. Total RNA from vegetative tissue of A. thaliana was also isolated using TriReagent with the exception that PVPP was omitted in the extraction. Total RNA from batches of pooled seed of both A. thaliana and X. humilis was extracted using a method described by Wan and Wilkins (1994).
cDNA synthesis was obtained from 2.5 μg of total RNA by using 200 units of Superscript III reverse transcriptase enzyme (Life Technologies, USA) following the manufacturer's instructions. The cDNA yield was checked by using the NanoDropR ND-1000 spectrophotometer (NanoDrop Technologies, Inc, DE).
Northern blot analysis
Twenty micrograms of total RNA isolated from roots and leaves at different stages of desiccation and rehydration were run on a 1% agarose formaldehyde denaturing gel (Ausubel et al., 1987) then transferred to Hybond™-N+ membrane (Amersham Biosciences, UK) by capillary blotting, using 20× SSC. The membrane was dried at 80 °C for 10 min and fixed by a UV cross linker (Amersham Life Science, UK). To check the success of the RNA transfer, the membrane was stained in methylene blue [0.2% w/v in 0.3 M sodium acetate (pH 5.5)].
The membrane was prehybridized at 42 °C for 16 h in hybridization buffer containing 0.1% SDS, 50% formamide, 5× SSC, 50 mM NaPO4, pH 6.8, 0.1% sodium pyrophosphate, 5× Denhardt's solution, and 50 μg ml−1 heat-denatured salmon sperm DNA. α-32P labelled probe was added and incubated for a further 16 h at 42 °C. The probe consisted of HC205 digested with EcoRI and XhoI to release cDNA. HC205 was labelled with α-32P using the Megaprime™ DNA labelling kit (Amersham Biosciences, UK) and purified using SigmaSpin™ Post-Reaction clean up columns (Sigma, Germany) following the manufacturer's instructions.
Conditions for washes were 2× SSC, 0.1% SDS for 30 min at room temperature; 1× SSC, 0.1% SDS for 30 min at room temperature, and 0.2× SSC, 0.1% SDS for 30 min at 55 °C. The membrane was then wrapped in plastic and exposed to a hyperfilm™-βmax, high performance autoradiography film (Amersham, UK). Northern blots were repeated twice on data collected from two independent cycles of desiccation and rehydration.
Primer design
Forward and reverse degenerate primers were designed to the conserved regions of the X. humilis HC205 (Genbank Accession number AY570978) gene and the A. thaliana orthologue (At1g07645) to confirm the presence of mRNA transcripts encoded by HC205 and its orthologue At1g07645 (Table 1). 18S rRNA primers were designed to nucleotide sequences conserved between the X. humilis and A. thaliana 18S rRNA genes. As a positive control for abiotic stress treatment, primers were designed to the Arabidopsis LEA2 gene, At1g76180 which is known to be activated by salt, mannitol, and dehydration stress (Table 1). Primers were also designed to ubiquitin (At4g05320) as a positive control to check for the integrity of RNA (Table 1). All the primers were synthesized at the Molecular and Cell Biology Department, University of Cape Town.
Table 1.
RT-PCR primers used to identify mRNA transcripts for HC205/At1g07645, 18S rRNA, Arabidopsis LEA-2 (At1g76180), and Arabidopsis polyubiquitin 10 (At4g05320)
| Gene name | Forward primer | Reverse primer | Annealing temperature | No. cycles |
| HC205/At1g07645 | 5′-ACAGGTGGGG(AG)GAGCT(AG)GAG-3′ | 5′-ATTAACGTGGCTTCCGA(TG)GCG-3′ | 58 °C | 25 |
| 18S rRNA | 5′-CAGGCGCGCAAATTACCCAA-3′ | 5′-GCGACCATACTCCCCCGG-3′ | 54 °C | 20 |
| LEA 2 | 5′-GTACAAAGAGCGTGAAATCCGC-3′ | 5′-CTTCTCCTCTACGGACC-3′ | 56 °C | 30 |
| Ubiqutin | 5′-CAATTCTCTCTACCGTGA-3′ | 5′-CCATCTTCAAGTTGCTTTCCG-3′ | 55 °C | 32 |
Degenerate nucleotides are shown in parenthesis. The annealing temperatures and number of cycles used in the PCR reactions are summarized for each set of primers.
Reverse transcription polymerase chain reaction (RT-PCR)
The number of PCR cycles for amplification of each of the products was based on identifying the point at which the transcripts were amplified in a linear range (Table 1). This was established by removing a 10 μl aliquot from the PCR reactions at two cycle intervals from 16 cycles to 35 cycles. 250 ng of synthesized cDNA was used for the PCR reaction, which included 0.5 μM of each degenerate primer, 0.2 mM dNTP, 0.5 U Supertherm Taq (Hoffman-La Roche, USA), and 2.4 mM MgCl2. The following PCR conditions were standard for all the primers and only the annealing temperature was different for each set of primers. Initial denaturation temperature of 94 °C (1 min), followed by denaturation at 94 °C (30 s), and extension time of 72 °C (1 min). These experiments were repeated twice on independent biological samples.
Expression of HC205 recombinant protein in E. coli
The coding region of HC205 cDNA was amplified by PCR using a forward primer 5′-AGG GGA TCC ATG GCG AAT CT-3′ and reverse primer 5′-GAA TTC ACA GCT ACA TAC ACA T-3′. The underlined nucleic acids indicate the incorporation of BamHI and EcoRI restriction sites, respectively. The amplified HC205 PCR product was cloned into pGEM®-T Easy (Promega, USA) prior to being digested with BamHI and EcoRI restriction enzymes, purified, and cloned into the same sites in the pGEX-3X (Amersham Pharmacia Biotech, Sweden) expression vector. E. coli XL-1Blue transformed with the pGEX-3X:HC205 construct was grown at 37 °C in Luria broth medium containing ampicillin until the cell density reached 0.9 at OD600. Expression of recombinant HC205 was induced by adding 0.5 mM IPTG (Roche, USA) to the culture media for a period of 2 h at 30 °C. The culture was then spun down at 10 000 rpm at 4 °C in a bench-top centrifuge for 10 min and the resulting pellet was resuspended in PBS (147 mM NaCl, 2.7 mM KCl, 1.8 mM KH2PO4, 4.3 mM Na2HPO4.2H2O at pH 7.2). A 500 μl aliquot of culture was transferred to a centrifuge tube and the bacteria were lysed by the addition of 1 μg lyzozyme. The cells were incubated for 30 min at 4 °C before centrifuging for 10 min at 10 000 rpm (4 °C) to remove cellular debris. The protein concentration of the supernatant was determined by the Bradford assay (Bio-Rad Laboratories GmbH, Munchen).
Generation of anti-HC205/At1g07645 antibodies
A peptide (RRVDNSNRWGELESGE) that is conserved between the N-terminal domain of HC205 and the A. thaliana orthologue At1g07645, was synthesized and coupled to a KLH protein by the Princeton BioMolecules Corporation (USA). The peptide was used to raise anti-HC205 antibodies in rabbits (South African Vaccine Producers Ltd). The titre of the anti-HC205 antibodies was determined in an ELISA by the immunodetection of serial dilutions of anti-HC205 serum against 0.5 μg of peptide (not conjugated to the KLH protein). Serum collected from the rabbits before administering the antigen was used as a negative control (data not shown).
The specificity of the HC205 antibodies was checked by Western blot analysis of a crude E. coli cell extract of recombinant HC205 protein. Crude extracts prepared from either E. coli transformed with expression vector pGEX-3X only, or an induced culture of E. coli pGEX3-3X:HC205 were used as a negative controls.
Western blot analysis
X. humilis total protein from leaves and roots was extracted from the phenolic layer recovered from the RNA extraction in TriReagent, according to the manufacturer's recommendations (Molecular Research Centre Inc, Cincinnati, OH). 20 μg of total protein isolated from different stages of desiccation and rehydration in both roots and leaves were run on a 12% SDS–PAGE gel as described by Ausubel et al. (1987) then electroblotted onto PROTRANR nitrocellulose membrane (Schleicher and Schuell Biosciences GmbH, Germany). The nitrocellulose membrane was stained with Ponceau S (0.1% in 5% acetic acid) to confirm equal loading and transfer of protein samples.
For Western blot analysis, the membrane was incubated in blocking buffer, TBS (50 mM TRIS-Cl at pH 7.6, 150 mM NaCl) containing 5% fat-free milk powder at 4 °C for 16 h. The membrane was then transferred to TBS containing 2% fat-free milk powder and the HC205 anti-serum at a dilution of 1/1000 at 4 °C for 16 h. The membrane was then washed with 1× TBS containing 0.1% Tween-20 (Sigma, USA) and incubated at room temperature for 1 h with the anti-rabbit-HRP secondary antibody (Sigma, USA) at a dilution of 1/10 000. Detection was carried out by using a LumiGloR Reserve Western Blot Kit (KPL, USA); and exposing the membrane to high performance autoradiography film (Hyperfilm™-βmax Amersham, UK) for 1 min. Western blots were repeated twice on independent biological samples.
Methylglyoxal resistance studies in E. coli
For the methyglyoxal viability/resistance assay, an E. coli culture containing pGEX-3X:HC205 plasmid was grown for 16 h at 37 °C to an absorbance reading A600 of 0.9. Two microlitre of the culture were serially diluted and these dilutions were spotted on Luria agar plates containing different concentrations of methylglyoxal (0 mM–10 mM), 100 μg ml−1 ampicillin, and 0.5 mM of IPTG (Roche, USA) to activate the promoter (modified from Veena et al., 1999). The plates were incubated at 37 °C overnight, and the tolerance to the different concentrations of methylglyoxal was measured by growth of E. coli on the agar plates.
Complementation studies in yeast
A S. cerevisiae glyoxalase I mutant (glo1Δ) together with its isogenic wild-type strain were donated by Dr Yoshiharu Inoue (University of Kyoto, Japan). The genotype of the strains are as follows:
YPH250: MATa Trp1-Δ1 his-Δ200 leu2-Δ1 lys2-801 ade2-101 ura3-5
YPH250 (glo1Δ): MATa Trp1-Δ1 his-Δ200 leu2-Δ1 lys2-801 ade2-101 ura3-5, glo1Δ::HIS3.
The ability of HC205 to complement the yeast glyoxalase I mutant was tested. The HC205 coding region from pGEM-T-Easy was cloned into the BamHI and EcoRI cloning site of the pYES2 yeast expression vector (Invitrogen, Life Technologies, USA). The pYES2: HC205 construct was purified from E. coli XL-1 Blue using the High Pure Plasmid Isolation Kit (Roche, USA) and was then transformed into glo1Δ yeast cells by electroporation as described by Adams et al. (1997).
The transformed yeast cells were grown in Synthetic Minimal Medium containing 6.7 g nitrogen base without amino acids (Difco Laboratories, Inc, USA), 0.77 g complete synthetic medium without uracil (BIO 101 Systems, USA) per litre, 2% glucose and incubated at 30 °C for 3 d. A single colony was inoculated into 10 ml Synthetic Minimal Medium and grown until exponential phase. Three microlitres of the cell culture was spotted in duplicate on YPG (1% yeast extract, 2% peptone, and 2% agar) plates containing different concentrations of methylglyoxal (0 mM, 1 mM, 2 mM, 5 mM, and 10 mM) and galactose to induce the promoter. The glyoxalase I mutant and wild-type S. cerevisiae transformed with the parental vector were used as controls.
Bioinformatics
The Genbank non-redundant protein sequences (nr), est_other databases and the TIGR Gene Indices database (http://compbio.dfci.harvard.edu/tgi/) were searched by BLASTP, BLASTN, and TBLASTX to identify HC205 orthologues. The predicted amino acid sequences of HC205 orthologues were aligned by ClustalW in MEGA4 (Tamura et al., 2007), and was used to identify conserved protein motifs and to construct a Neighbor–Joining, phylogenetic tree with bootstrap values, using the Jones–Taylor–Thornton matrix. Full species names and Accession numbers for HC205 orthologues are listed in the supplementary data at JXB online. Protein structural motifs in the predicted HC205 and At1g07645 amino acid sequences were identified using PROSITE (Bairoch et al., 1997) submitted via the PredictProtein server (http://www.predictprotein.org) (Rost et al., 2004). This server was also used to submit sequences to PROF, a secondary structure prediction tool (Rost and Sander, 1993). A cut-off of reliability index value >4 was used to predict the presence of β sheets and α helices in HC205 and At1g07645. The PSIPRED server (http://bioinf.cs.ucl.ac.uk/psipred/) was used to submit the HC205 and At1g07645 amino acid sequences to mGenTHREADER, a fold recognition tool that aligns a protein sequence against a known three-dimensional structure from the Protein Data Base (PDB) (McGuffin and Jones, 2003; Bryson et al., 2005). The three-dimensional structures of HC205 and At1g07645 were predicted by the EsyPred3D server (http://www.fundp.ac.be/sciences/biologie/urbm/bioinfo/esypred/) by homology modelling (Lambert et al., 2002), and were visualized in PyMol (DeLano, 2002).
Results
X. humilis HC205 is a member of a novel gene family in plants
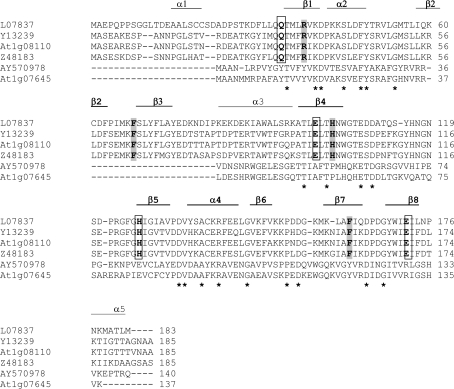
The X. humilis HC205 cDNA was originally isolated as a desiccation-upregulated gene via microarray analysis (Collett et al., 2004; Genbank accession number AY570978) and encodes a 140 amino acid protein with a predicted molecular weight of 15.6 kDa. A BLASTP search of the non-redundant protein sequences (nr) database (NCBI) indicated that HC205 was homologous to Oryza sativa NP_001049720 (Os03g0277500) (E-value 1e−45) and Arabidopsis NP_973779 (At1g07645) (E-value 2e−45). The latter is annotated as a glyoxalase family I protein and/or a lactolylglutathione lyase family protein in Genbank. Sequence alignment with known glyoxalase I genes, including one glyoxalase I (At1g08110) from A. thaliana, however, shows that HC205 shares limited similarity with these genes (12–16% amino acid identity) and lacks those regions that are important for glyoxalase I activity, as identified from the protein crystal structure of the human glyoxalase I (Cameron et al., 1997). The amino acids that make up the conserved zinc-binding and the glutathione binding sites in glyoxalase I enzymes are absent in HC205 and the Arabidopsis orthologue, At1g07645 (Fig. 1).
Fig. 1.
ClustalW alignment of amino acid sequences of X. humilis HC205 (AY570978) and the A. thaliana orthologue (At1g07645), with known glyoxalase I genes, including L07837 (Homo sapiens), Y13239 (Brassica juncea), At1g08110 (A. thaliana), and Z48183 (Lycopersicon esculentum). The asterisks represent amino acids that are identical in all of the six sequences. The βαβββ structural repeat determined in the X-ray crystal structure of the human glyoxalase I protein is indicated. This structural repeat is represented by β1α2β2β3β4 in the first domain (α1−β4), and by β5α4β6β7β8 in the second domain (β5−α6) (Cameron et al., 1997). Conserved amino acids that constitute the glutathione binding sites of glyoxalase I are highlighted in grey. Conserved amino acids which form the zinc binding sites are in bold and are boxed.
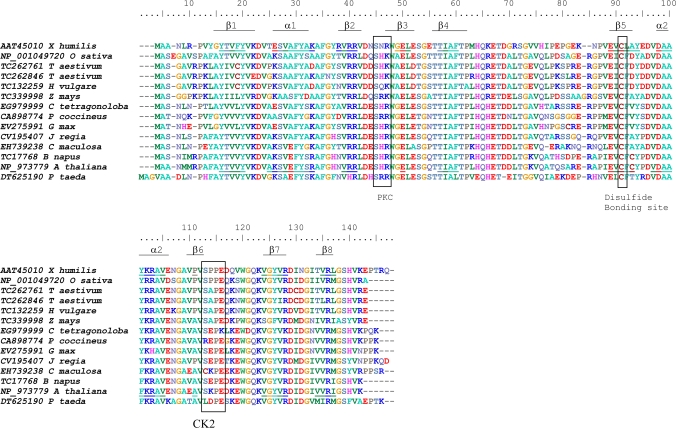
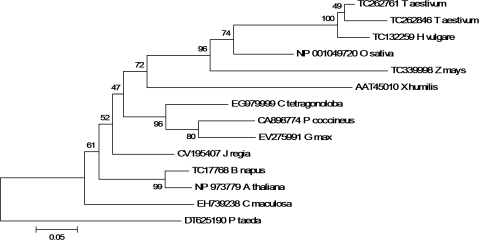
Several plant EST sequences with high similarity to the X. humilis HC205 gene were identified by a TBLASTX search against the TIGR and Genbank gene indices database (see Tables S1 and S2 at JXB online). Interestingly, many of these ESTs were originally identified in libraries constructed from seed or seed-related tissues. A multiple amino acid sequence alignment of HC205 and its plant orthologues shows that a protein kinase C phosphorylation site and a casein kinase II phosphorylation site are conserved across all the HC205 plant orthologues, included the Loblolly pine (Pinus taeda) (Fig. 2). A single cysteine, which is predicted to form a disulphide bond by DISULFIND (Vullo and Frasconi, 2004) is conserved in all plant orthologues (Fig. 2). In contrast to many other genes, HC205 orthologues are present as a single copy in plants, with the exception of Triticum aestivum where two paralogues of HC205 are represented in the EST database (Fig. 3). Dsi-1VOC appears to have evolved early in the plant lineage as a clear orthologue (CN203139, E-value=5e−30) was identified in the Tortula ruralis EST library (see Table S2 at JXB online). The ancient gene probably evolved from ancestral eubacteria, as the closest homologues to this gene family are found in bacteria, for example, the Roseobacter sp. CCS2 glyoxalase family protein (ZP_01750924) which shares 58% similarity with X. humilis HC205 (E-value=2e−23).
Fig. 2.
ClustalW alignment of predicted amino acid sequences of plant dsi-1VOC orthologues identified in the Genbank and TIGR EST databases, including X. humilis, rice (Oryza sativa), wheat (Triticum aestivum), barley (Hordeum vulgare), maize (Zea mays), guar (Cyamopsis tetragonoloba), runner bean (Phaseolus coccineus), soybean (Glycine max), walnut (Juglans regia), spotted knapweed (Centaurea maculosa), oilseed rape (Brassica napus), Arabidopsis thaliana, and Loblolly pine (Pinus taeda). Boxed amino acids demarcate conserved phosphorylation sites for protein kinase C (PKC) and protein casein kinase II (CK2) identified by PROSITE, and a potential disulphide bonding site predicted by DISULFIND. Amino acids in the X. humilis and A. thaliana dsi-1VOC orthologues which are predicted to fold into the βαβββ structural repeats characteristic of the VOC superfamily, are underlined.
Fig. 3.
Phylogenetic relationship amongst plant dsi-1VOC orthologues including X. humilis, rice (Oryza sativa), wheat (Triticum aestivum), barley (Hordeum vulgare), maize (Zea mays), guar (Cyamopsis tetragonoloba), runner bean (Phaseolus coccineus), soybean (Glycine max), walnut (Juglans regia), spotted knapweed (Centaurea maculosa), oilseed rape (Brassica napus), Arabidopsis thaliana, and Loblolly pine (Pinus taeda). A Neighbor–Joining tree based on the alignment of full-length amino acid sequences shown in Fig. 2, was constructed using the Jones–Taylor–Thornton matrix. Bootstrap values are given at each node. The Loblolly pine (Pinus taeda) sequence was used to root the tree. Genbank and TIGR EST database accession numbers are indicated.
HC205 is a member of theVOC metalloenzyme superfamily
Structural orthologues of HC205 identified using mGenTHREADER were all members of the vicinal oxygenase chelate superfamily and included divergent enzymes from Arabidopsis thaliana, human glyoxalase I, and a number of bacterial antibiotic resistance proteins and enzymes (Table 2). Although these proteins shared little amino acid sequence similarity with HC205 (<22%), they had significant structural homology (P value <10e−09). Proteins encoded by HC205 and its Arabidopsis orthologue, At1g07645 were predicted by PROF (Rost and Sander, 1993) to fold into two distinct βαβββ domains, a common structural feature of members of the VOC superfamily (Fig. 2). HC205 was re-named desiccation-induced-1VOC, dsi-1VOC to reflect the induction of HC205 by desiccation in X. humilis and its membership of the VOC superfamily.
Table 2.
Structural homologues of HC205 identified by mGenThreader recognition software via the PredictProtein server, ranked according to their best net scores
| Net score | P-value | Pdb ID | Species | Gene name | Metal ions | Sequence identity |
| 0.860 | 1e−09 | 1xy7 | Arabidopsis thaliana | At5g48480, unknown protein | None | 21.5% |
| 0.841 | 2e−09 | 1ecs | Klebsiella pneumoniae | Bleomycin resistance protein | Ca2+ | 20.8% (A chain) |
| 0.838 | 3e−09 | 1qip | Homo sapiens | Glyoxalase I | Zn2+ | 19.3% |
| 0.829 | 3e−09 | 2i7r | Streptococcus pneuomoniae | Unknown function | None | 17.4% |
| 0.828 | 4e−09 | 1kll | Streptoymces lavendulae | Mitomycin resistance protein | None | 20.3% |
| 0.826 | 4e−09 | 1sqd | Arabidopsis thaliana | At1g06570; oxidoreductase; dihydroxybiphenyl dioxygenase | Fe2+ | 20.0% |
| 0.825 | 4e−09 | 1nki | Pseudomonas aeraginosa | Fosfomycin resistance protein | K+, Mn2+ | 13.4% |
| 0.809 | 7e−09 | 1r9c | Mesorhizobium loti | Fosfomycin resistance protein | Mn2+ | 14.4% |
| 0.801 | 9e−09 | 1mpy | Pseudomonas putida | Catechol 2,3 dioxygenase | Fe2+ | 12.9% |
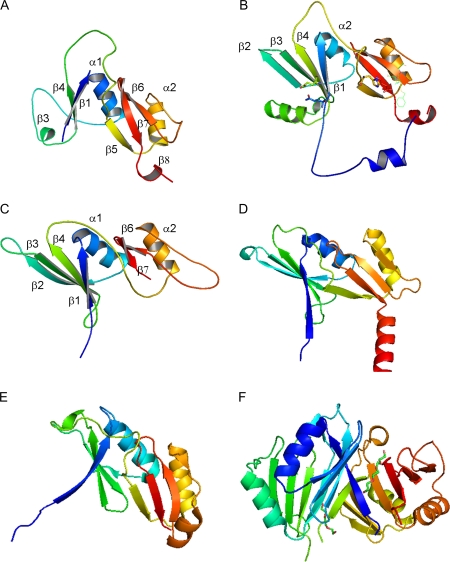
The tertiary structure of Xhdsi-1VOC was predicted by the EsyPred 3D server which selected the human glyoxalase I structure (1FRO.pdb) for homology modelling of Xhdsi-1VOC (Fig. 4A). Unlike other structurally characterized members of the VOC superfamily, the first βαβββ repeat which was predicted by PROF, is not clearly folded. However, the second βαβββ repeat shows a similar structure to human glyoxalase I, and other members of the VOC superfamily for which structures have been solved (Fig. 4B, D–F). The EsyPred 3D server selected the bacterial glyoxalase I protein (2C21.pdb) from Leishmania major for modelling of the Arabidopsis orthologue, Atdsi-1VOC encoded by At1g07645. Unlike Xhdsi-1VOC, the first βαβββ repeat is predicted to be clearly structured in Atdsi-1VOC (Fig. 4C).
Fig. 4.
Schematic representation of the three-dimensional structures of monomers of (A) Xhdsi-1VOC (B), human glyoxalase I (1FRO.pdb) (C) Atdsi-1VOC (D) Leishmania major glyoxalase I protein (2C21.pdb) (E) unknown Arabidopsis protein encoded by At5g48480 (1XY7.pdb), and (F) Klebsiella pneumoniae bleomycin resistance protein (1ECS.pdb). The tertiary structure of Xhdsi-1VOC was predicted using the A chain of human glyoxalase I (B) as a template, while the tertiary structure of Atdsi-1VOC was predicted using the A chain of bacterial glyoxalase I (D) as a template, via the EsyPred3D server. The unknown Arabidopsis protein (E) and bleomycin resistant protein (F) were selected as the closest structural matches to Xhdsi-1VOC by mGenThreader. The monomers have been colour ramped according to residue number, starting with blue (N-terminus) and finishing with red (C-terminus). The first βαβββ repeat of the human glyoxalase I protein is numbered in (B), while the predicted α-helices and β-sheets for Xhdsi-1VOC and Atdsi-1VOC are annotated in (A) and (C), respectively.
Xhdsi-1VOC (HC205) confers low levels of methylglyoxal tolerance in E. coli
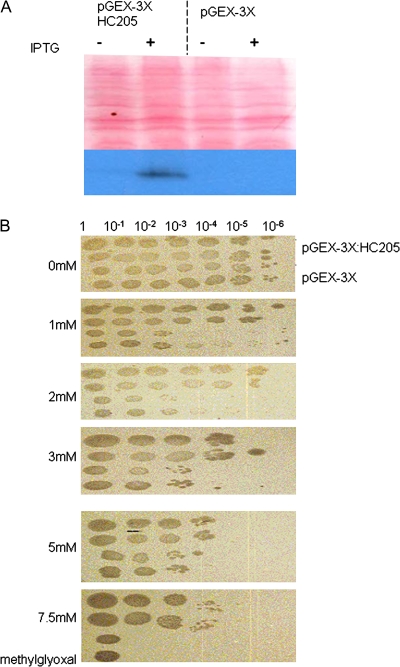
Because Xhdsi-1VOC (HC205) and Atdsi-1VOC (At1g07645) have high predicted structural homology to glyoxalase I, it was investigated whether over-expression of Xhdsi-1VOC would increase tolerance to methylglyoxal in E. coli. The expression of recombinant GST:HC205 in crude extracts of E. coli was first confirmed by Western blot analysis. An antibody generated to Xhdsi-1VOC was affinity purified and was shown to recognize recombinant Xhdsi-1VOC protein specifically in a crude extract of E. coli (Fig. 5A). E. coli expressing recombinant Xhdsi-1V VOC from pGEX-3X:HC205 were a 1000-fold more viable than the control strain to 1 mM and 2 mM methylglyoxal (Fig. 5B). At higher concentrations (up to 7.5 mM methyglyoxal), E. coli (pGEX-3X: HC205) was consistently 10–100-fold more viable than the control strain, and at 10 mM methyglyoxal, E. coli cells containing both the experimental and control constructs were unable to grow (data not shown). Expression of recombinant Xhdsi-1VOC (HC205) therefore confers low-level tolerance of up to 7.5 mM methyglyoxal, in E. coli.
Fig. 5.
(A) Western blot of crude extracts from E. coli pGEX-3X:HC205 or pGEX-3X, in the presence or absence of IPTG. The upper panel shows the nitrocellulose membrane with the transferred protein, stained with Ponceau S; the lower panel is a Western blot using anti-Dsi-1VOC antibodies, of the same membrane, showing the presence of recombinant XhDsi-1VOC when expression of pGEX-3X: HC205 is induced by IPTG in E. coli. (B) E. coli methylglyoxal resistance assay in which serial dilutions of the E. coli transformed with either pGEX-3X or pGEX-3X:HC205 were spotted in duplicate on IPTG plates containing the following concentrations of methylglyoxal: 0 mM (A), 1 mM (B), 2 mM (C), 3 mM (D), 5 mM (E), 7.5 mM (F). (This figure is available in colour at JXB online.)
Ectopic expression of Xhdsi-1VOC (HC205) is lethal in yeast
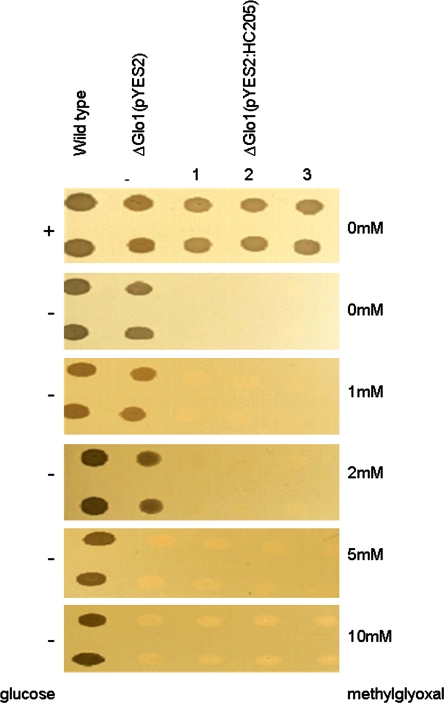
Xhdsi-1VOC (HC205) was also tested for its ability to complement a S. cerevisiae glyoxalase I mutant glo1Δ. Wild-type yeast tolerated concentrations of up to 10 mM methylglyoxal, whereas the glo1Δ mutant failed to grow in media containing more than 2 mM methylglyoxal (Fig. 6). However, ectopic expression of recombinant Xhdsi-1VOC (HC205) in the glo1Δ, mutant was lethal, and this strain failed to grow even in the absence of methylglyoxal (Fig. 6).
Fig. 6.
Methylglyoxal viability assays in wild-type S. cerevisiae yeast, in the S. cerevisiae glyoxalase I mutant transformed with the pYES2 vector (Δglo1(pYES2)), and in the S. cerevisiae glyoxalase I mutant transformed with the pYES2:HC205 recombinant vector (Δglo1(pYES2:HC205)), grown in triplicate (1–3). The addition (+) or absence (–) of glucose which suppresses expression from the pYES2 vector is indicated in the left hand side panel. Galactose was added to the medium in the absence of glucose to activate expression of the pYES2 vector. Concentration of methylglyoxal added to the growth media is indicated in the right hand side panel. (This figure is available in colour at JXB online.)
A comparison of Xhdsi-1VOC and At1g07645 expression in X. humilis and A. thaliana
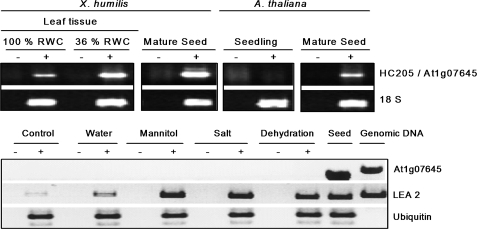
The origins of the EST libraries which contained orthologues of Xhdsi-1VOC suggested that these genes are expressed during seed development and germination in desiccation-sensitive plants (see Tables S1 and S2 at JXB online). This was tested in a RT-PCR study in which the expression of Dsi-1VOC was compared in X. humilis and in A. thaliana (Fig. 7). Dsi-1VOC mRNA transcripts were present at low levels in hydrated leaf tissue, and at higher levels in desiccating leaves and mature, dry seed in X. humilis. By contrast, mRNA transcripts of the Xhdsi-1VOC orthologue, At1g07645, were absent in unstressed A. thaliana 3-week-old seedlings but were present in dry mature seed (Fig. 7A). Exposure of 2-week-old Arabidopsis seedlings to abiotic stresses, including mannitol, salt, and dehydration lead to activation of expression of LEA-2 mRNA transcripts which are known to be induced by abiotic stress (Illing et al., 2005). However, although At1g07645 mRNA transcripts were present in mature seed, they were absent in the seedlings exposed to abiotic stresses. It is concluded that At1g07645 mRNA transcripts are expressed in A. thaliana seed, but are not activated in seedlings in response to abiotic stress.
Fig. 7.
RT-PCR analysis of HC205/At1g07645 mRNA transcript abundance in seed and leaf tissues of X. humilis and A. thaliana. (A) Products of amplification of HC205 mRNA transcripts in X. humilis leaf tissue (100% and 36% RWC) and in mature dry seeds, and amplification of At1g07645 mRNA transcripts in 3-week-old seedlings and mature dry seeds of A. thaliana are indicated. Amplification of 18S rRNA transcripts are shown as a control for relative abundance of sample RNA. (B) Amplification of At1g07645, LEA2, and ubiquitin mRNA transcripts in mature seeds and 2-week-old A. thaliana seedlings exposed to mannitol, salt, and dehydration stress for 4 h. Controls include A. thaliana seedlings prior to stress treatments. Amplification of LEA 2 mRNA transcripts was included as a positive control to show activation of the abiotic stress response. Ubiquitin mRNA transcripts are expected to be expressed at similar levels in all samples, and were included to control for starting sample RNA concentrations. For each RT-PCR reaction, (+) indicates the presence, and (–) the absence, of reverse transcriptase in the cDNA synthesis reactions.
Expression profile of Xhdsi-1VOC in X. humilis vegetative tissue during desiccation and rehydration
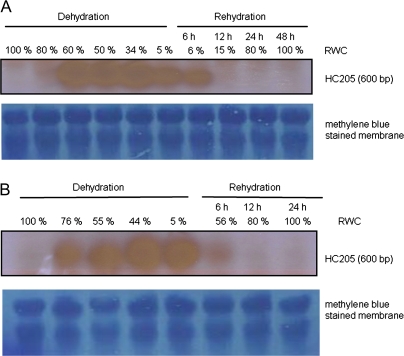
Expression patterns of Xhdsi-1VOC mRNA transcripts and protein in leaves and roots were compared during a cycle of desiccation and rehydration in X. humilis to characterize the changes in Xhdsi-1VOC expression in response to water loss and subsequent rehydration further. Northern blot analysis of total RNA extracted from X. humilis vegetative tissue shows that Xhdsi-1VOC mRNA transcripts were not detectable in hydrated leaves or roots (Fig. 8). The presence of 600 bp mRNA transcripts encoded by Xhdsi-1VOC was first detected at low levels in leaf tissue of 80% RWC and increased substantially between 80% and 60%, whereafter levels appeared to remain constant up to 34% RWC. Xhdsi-1VOC mRNA transcript abundance declined slightly in fully desiccated leaves (5% RWC). In roots, Xhdsi-1VOC mRNA transcripts were also absent in hydrated tissue but were abundant in 76% RWC root tissue and appeared to reach a steady-state thereafter until full desiccation (<5% RWC). Roots rehydrate more rapidly than leaves, and after 6 h their RWC had reached 56%, compared to 6% in leaves. The rate of disappearance of Xhdsi-1VOC mRNA transcripts in leaves and roots after watering followed the same trend. After 12 h of rehydration, Xhdsi-1VOC mRNA transcripts were absent in both rehydrated leaves and roots.
Fig. 8.
Northern blot analysis of HC205 at different stages of desiccation and rehydration in leaves (A) and roots (B) of X. humilis. The time-scale of rehydration is shown relative to the change in RWC in leaves and roots. Equal loading and transfer of total RNA in each sample is shown by methylene blue staining of the membranes. (This figure is available in colour at JXB online.)
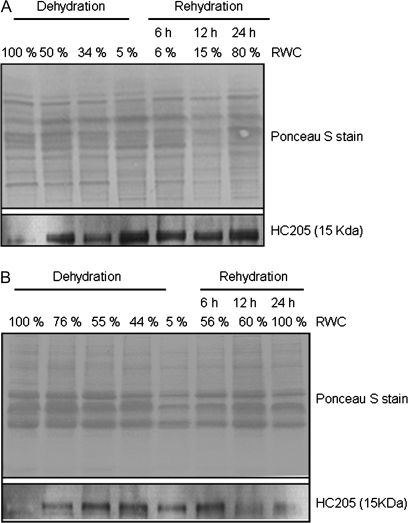
The anti-Dsi-1VOC antibodies recognized a 15 kDa protein in total protein extracts from X. humilis desiccating leaves and roots which agrees with the predicted size of Dsi-1VOC (Fig. 9). Western analysis showed that Dsi-1VOC was strongly expressed in roots and leaves following desiccation and immunoreactive protein bands were clearly visible in leaf and root samples of 50% and 76% RWC, respectively. Unlike mRNA transcripts which were rapidly turned over following rehydration, Dsi-1VOC protein was stably expressed throughout the desiccation and rehydration cycle (Fig. 9).
Fig. 9.
Western blot analysis of total protein extracts from X. humilis leaves (A) and roots (B) at different stages of desiccation and rehydration. The upper panels show the Ponceau S-stained nitrocellose membrane confirming equal loading and transfer of each experimental sample. The lower panel shows the Western blot result following the probing of these membranes with anti-Dsi-1VOC antibodies.
Discussion
The vicinal oxygenase chelate (VOC) superfamily is a diverse group of proteins, many of which are metalloenzymes. Members of this family are unrelated in terms of sequence, but are united by their common tertiary structure which includes two or more βαβββ folds, represented by the glyoxalase/bleomycin resistance protein/dioxygenase InterPro domain (IPR004360). Most of the members of the VOC superfamily function as dimers, although some members have sufficient repeats to function as monomers (Table 3). Several genes in Arabidopsis, including At1g07645, have been predicted to be members of this superfamily on the basis of their structural homology to the IPR004360 domain, and have been annoted as ‘lactoylglutathione lyase family protein/glyoxalase I family protein; similar to glyoxalase family protein’ in the Arabidopsis Information Resource (TAIR) (www.arabidopsis.org) database.
Table 3.
Characteristics of members of the VOC superfamily
| Name | Abbreviation | Organism | Metal ion | Co-enzyme | Function | Functional unit | Reference |
| Bleomycin resistance protein | BRP | Fungi and eubacteria | No | Sequesters bleomycin and related compounds(no degradation or transformation) | Homodimer*1 | Bergdoll et al., 1998 | |
| 2,3-Dihydroxy-biphenyl 1,2-dioxygenase (estradiol dioxygenase | DHBD | Eubacteria | Fe2+ | Microbial degradation of aromatic compounds (e.g. degrades biphenyl and polychlorinated biphenyls) | Monomer *1 | Bergdoll et al., 1998 | |
| Glyoxalase I (small) | GLO | Eubacteria, plants and animals | Zn2+ | glutathione | Isomerization reaction: glutathione-dependent inactivation of toxic methylglyoxal | Homodimer*1 | Bergdoll et al., 1998 |
| Glyoxalase I (large) | GLO | Fungi | Zn2+ | glutathione | Isomerization reaction: glutathione-dependent inactivation of toxic methylglyoxal | Monomer*2 | Thornalley, 2003 |
| Fosfomycin resistance protein | FosA | Eubacteria | Mn2+ | glutathione | Inactivation of the antibiotic fosfomycin by nucleophilic opening of epoxide ring | Homodimer*3 | Rigsby et al., 2007 |
| Methylmalonyl-CoA eprimerase | MMCE | Eubacteria, Archea and animals | Co2+ | Epimerization reaction: catalyses conversion of (2R)-methylmalonyl-CoA to (2S)-methylmalonyl-CoA. Methylmalonyl-CoA is a metabolic intermediate in several degradation pathways (e.g. lipids and branched amino acids) and biosynthetic pathways (e.g. important polyketide antibiotics) | Homodimer*4 | McCarthy et al., 2001 |
A novel plant gene, Xhdsi-1V°C has been characterized which was first identified as a gene (HC205; AY570978) that was up-regulated in X. humilis leaves during desiccation (Collett et al., 2004). HC205 is an orthologue of the Arabidopsis gene At1g07645. In this paper, evidence is presented that HC205 and At1g07645 are unlikely to have glyoxalase activity, but encode a protein that is a novel member of the VOC superfamily and that is expressed at high levels in desiccated tissue, hence the proposed name Dsi-1VOC.
Although X. humilis Dsi-1VOC and its Arabidopsis orthologue, At1g07645 have low sequence identity with other glyoxalase I genes, they do share high structural homology with members of the vicinal oxygen chelate (VOC) superfamily, which includes glyoxalase I genes (Table 2). XhDsi-1V°C is predicted to fold into two βαβββ domains (Fig. 2), and is predicted by mGenTHREADER to have the highest structural homology to an Arabidopsis gene At5g48480, which, although of unknown function, has had its tertiary structure determined (1XY7.pdb). At5g48480 is also a member of the glyoxalase/bleomycin resistance protein/dihydroxybiphenyl dioxygenase superfamily (SSF54593). mGenTHREADER identified several other members of the VOC superfamily which are structurally similar to XhDsi-1VOC, including the Klebsiella pneumoniae bleomycin resistance protein, and human glyoxalase I (Table 3). The former has no enzymatic activity and sequesters the antibiotic bleomcyin. The latter is part of the glyoxalase system found in animals, plants, and bacteria that detoxifies 2-oxoaldehydes to hydroxycarboxylic acids, and has an affinity for a particular oxoaldehyde, methyglyoxal (Thornalley, 2003). The EsyPred Server selected human glyoxalase I (1FRO.pdb) for homology modelling of XhDsi-1VOC and a bacterial glyoxalase I (2C21.pdb) for homology modelling of its Arabidopsis orthologue, At1g07645 (Fig. 4). Although XhDsi-1VOC and At1g07645 were predicted to share the highest structural similarity with glyoxalase I, they lack the conserved reduced glutathione binding sites and metal binding sites that are essential for the activity of this enzyme. Glyoxalase I detoxifies methyglyoxal which is the by-product of three pathways, namely, the breakdown of triosephosphates in glycolysis, the catabolism of threonine, and the catabolism of acetone. Desiccation has been shown to induce changes in many metabolic pathways, including those involved in glycolysis, sugar and amino acid metabolism in seeds and vegetative tissues of resurrection plants (Bewley and Black, 1994; Kermode, 1995; Vertucci and Farrant, 1995; Martinelli et al., 2007; Whittaker et al., 2007), and therefore it was reasoned that XhDsi-1VOC might be a specialized enzyme for detoxifying methylglyoxal during desiccation.
When over-expressed in E. coli, XhDsi-1VOC conferred low-level resistance to methylglyoxal (7.5 mM). This is approximately three times lower than the levels of tolerance conferred by the Brassica juncea glyoxalase I in E. coli (i.e. concentrations of 25 mM) (Veena et al., 1999). Therefore, although HC205 does increase tolerance to methylglyoxal in E. coli, it is not as efficient as other plant glyoxalase I enzymes. Unlike other glyoxalase I genes, ectopic expression of XhDsi-1VOC in yeast proved to be lethal, and did not confer tolerance to methylglyoxal in a yeast glyoxalase I mutant. Taking all this evidence together, the lack of conserved reduced glutathione binding and metal binding sites, the low efficiency of conferring tolerance to methylglyoxal in E. coli, and the lethality in yeast, suggests that XhDsi-1VOC is not a glyoxalase, and encodes a novel enzyme in the vicinal oxygenase superfamily.
Expression of dsi-1VOC orthologues is associated with seeds in desiccation-sensitive plants. In angiosperms (e.g. maize, wheat) and gymnosperms (e.g. spruce, pine) mRNA transcripts of orthologues of XhDsi-1VOC are represented in EST libraries derived from developing and germinating seed (see Tables S1 and S2 at JXB online). In Arabidopsis, At1g07645 mRNA transcripts are not transcribed in response to abiotic stress treatment, but are expressed at high levels in mature, desiccated seed (Fig. 7). In contrast, in X. humilis, expression of Xhdsi-1VOC mRNA transcripts is activated in both leaves and roots in response to desiccation, in addition to being expressed in mature seeds (Figs 7, 8).
Xhdsi-1VOC mRNA transcripts are up-regulated early in the desiccation cycle in vegetative tissue and are already abundant in X. humilis roots at 76% RWC (Fig. 8). Transcriptional activation of Xhdsi-1VOC in response to water loss may be slower in leaves, with lower levels in leaves at 80% RWC, but with abundant expression at 60% RWC. While mRNA transcripts remain abundant until both leaves and roots are fully desiccated (i.e. <5% RWC), they are rapidly turned over once the plants are watered. A striking difference is that the levels of Xhdsi-1VOC mRNA transcripts decline rapidly in roots within the first 6 h of rehydation. However, X. humilis roots rehydrate at a much faster rate than leaves, which is evident from their respective RWC contents at 6 h after rehydration. Norwood et al. (2003) have observed this rapid rehydration of roots in the resurrection plant Craterostigma plantigineum, with concomitant earlier initiation of metabolism in roots than in leaves.
The activation of expression of Xhdsi-1VOC protein during desiccation of root and leaf tissues correlates with the mRNA profiles (Fig. 9). However, unlike the mRNA transcripts, the XhDsi-1VOC protein persists during rehydration in both tissues for at least 24 h (Fig. 9). Similar to the pattern seen for the mRNA transcripts, the levels of XhDsi-1VOC protein decline more rapidly in roots and, after 24 h, expression levels are substantially lower in roots compared to leaves. Once again, this could reflect the faster increase in RWC in roots compared to leaves. The expression data, namely the EST library representation analysis, the Arabidopsis RT-PCR comparative analysis, and the X. humilis Northern blots, suggests that Dsi-1VOC plays an important role in seed maturation and germination in desiccation-sensitive plants, and in the survival of desiccation and subsequent rehydration in the vegetative tissue of desiccation-tolerant plants such as X. humilis.
In addition to being regulated at the transcriptional level, XhDsi-1VOC may be regulated post-translationally by phosphorylation. A protein kinase C and a protein casein kinase II phosphorylation site are conserved in both angiosperm and gymnosperm Dsi-1VOC orthologues (Fig. 2). The activity of Dsi-1VOC may be further regulated by dimerization. Homodimers are the functional units of all the smaller members of the VOC superfamily which have two predicted βαβββ domains (Table 3). Homology modelling of Dsi-1VOC predicts that although the first N-terminal βαβββ is unstructured when compared to human glyoxalse I, the second domain folds in a similar path to the second human βαβββ glyoxalase I domain (Fig. 4). In glyoxlase I, this second domain forms the cavity required for interaction with those methylglyoxal-like analogues which were used in determining the 3D structures of human glyoxalase I (Fig. 4). We predict, on the basis of this evidence, that XhDsi-1VOC is likely to function as a homodimer, or possibly even a heterodimer during desiccation.
Additional evidence supporting this hypothesis comes from the report that an orthologue of XhDsi-1VOC (TC132259; see Table S1 at JXB online) in mature barley seed was reduced by the addition of exogenous thioredoxin h (Maeda et al., 2004). Dsi-1VOC is predicted to have a single conserved cysteine in the second βαβββ domain which could form a disulphide bond (Fig. 2). Dsi-1VOC would therefore have to dimerize with itself or another member of the VOC superfamily before it could be a substrate for reduction by thioredoxin h in the experiment reported by Maeda et al. (2004). Reduction of the Dsi-1VOC barley orthologue by exogenous thioredoxin h was not detectable in germinating seed and the authors concluded that the Dsi-1VOC barley orthologue was rapidly reduced by endogenous thioredoxins during germination (Maeda et al., 2004). It is speculated that XhDsi-1VOC is similarly inactivated by endogenous thioredoxins in leaves and roots during rehydration in X. humilis.
Dsi-1VOC orthologues lack the conserved metal binding and glutathione binding sites that are conserved in all VOC family members which have enzymatic activity (Table 3). However, XhDsi-1VOC could have a similar mode of action to the bleomycin resistance protein, and sequester some metabolite that builds up during desiccation, and which is required for rehydration. The reduction of disulphide bonds between dimers of Dsi-1VOC by thioredoxin during rehydration in the vegetative tissue of desiccation-tolerant plants, or seed germination in desiccation-sensitive plants, could release this metabolite. Ectopic expression of XhDsi-1VOC in yeast may be lethal because it sequesters the metabolite and because the metabolite is essential for cellular activity under normal growth conditions.
It is proposed that X. humilis has acquired the ability to activate this ‘seed-specific’ gene in response to drying in vegetative tissue (roots and leaves). A similar finding has been reported by Illing et al. (2005) who used expression data from microarrays to show that certain seed-specific LEA and antioxidant mRNA transcripts, which were expressed in the desiccated leaves of X. humilis, were expressed only in the mature seeds of Arabidopsis, and not in the vegetative tissue under abiotic stress conditions. Mowla et al. (2002) have similarly reported the characterization of a seed-specific antioxidant gene, 1-cys peroxiredoxin, which is activated in the desiccated leaves of the closely related resurrection plant, Xerophyta viscosa. These experimental findings support the hypothesis proposed by Oliver et al. (2000) that the ability of vegetative tissues of angiosperm resurrection plants to survive desiccation is a consequence of the activation of genes that are important for conferring desiccation tolerance in seeds, in leaves, and in roots.
Desiccation tolerance in angiosperms has polyphyletic origins having evolved independently in several angiosperm lineages (Oliver et al., 2000). X. humilis is a poikilochlorophyllous resurrection plant, which breaks down chlorophyll, dismantles its chloroplasts and down-regulates expression of photosynthetic genes in response to desiccation (Collett et al., 2003). It remains to be determined whether the activation of Dsi-1VOC in leaves and roots in response to water loss is restricted to poikilochlorophyllous resurrection plants such as X. humilis, or whether activation of this seed-specific gene in vegetative tissue in response to water loss has evolved in other angiosperm resurrection plants.
Supplementary data
Lists of plant orthologues of XhDsi-1VOC identified by screening the NCBI and TIGR EST databases by TBLASTX search are provided as supplementary tables associated with this article and are available at JXB online. Genbank Accession numbers of XhDsi-1VOC orthologues used in phylogenetic analysis are listed in the Supplementary data.
Supplementary Table S1. Percentage similarity and E-values for TBLASTX search of full-length orthologues of HC205 present in the plant gene indices (TC) in TIGR gene indices database (http://compbio.dfci.harvard.edu/tgi/). The tissue origins of libraries in which these ESTs are present are summarized. At1g07645 is represented by TC302237 in the TIGR A. thaliana EST gene index.
Supplementary Table S2. Percentage similarity and E-values for TBLASTX search of HC205 full-length orthologues in the Genbank EST_other database. The tissue origins of libraries in which these ESTs are present are summarised.
Supplementary Material
Acknowledgments
The National Research Fund (NRF), Equity Development Programme, and the University of Cape Town International Scholarships, funded this work. We thank the Barakalalo National Park for allowing collection of the X. humilis plants and Archie Corfield and Les Cousins for assisting in the collection thererof. Dr Yoshiharu Inoue from the University of Kyoto is acknowledged for donating the S. cerevisiae glyoxalase I mutant (glo1Δ) together with its isogenic wild-type strain and Lara Donaldson and Dr Katherine Denby for providing us with Arabidopsis thaliana seed. Finally, we would like to acknowledge Karen Cooper and Bridget Hamann for collection of X. humilis seed.
References
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Transformation of yeast. Methods in yeast genetics. A Cold Spring Harbor Laboratory Course Manual; 1997. pp. 41–52. [Google Scholar]
- Ausubel MF, Brent R, Kingston ER, Moore D, Seidman GJ, Smith AJ, Struhl K. Preparation and analysis of RNA. Current protocols in molecular biology. 1994 Vol. 1. Library of Congress cataloguing in publication data. [Google Scholar]
- Armstrong NR. Mechanistic diversity in a metalloenzyme superfamily. Biochemistry. 2000;39:13625–13632. doi: 10.1021/bi001814v. [DOI] [PubMed] [Google Scholar]
- Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1997. Nucleic Acids Research. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergdoll M, Eltis LD, Cameron AD, Dumas P, Bolin JT. All in the family: structural and evolutionary relationships among three modular proteins with diverse functions and variable assembly. Protein Science. 1998;7:1661–1670. doi: 10.1002/pro.5560070801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berjak P, Farrant JM, Pammenter NW. Seed desiccation-tolerance mechanisms. In: Jenks MA, Wood AJ, editors. Plant desiccation tolerance. Oxford: Blackwell Publishing; 2007. pp. 151–192. [Google Scholar]
- Bewley JD, Black M. Seeds: physiology of development and germination. 2nd edn. New York: Plenum Press; 1994. [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Research. 2005;33 doi: 10.1093/nar/gki410. (Web Server issue) W36–W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron AD, Olin B, Ridderström M, Mannervik B, Jones TA. Crystal structure of human glyoxalase I: evidence for gene duplication and 3D domain swapping. The EMBO Journal. 1997;16:3386–3395. doi: 10.1093/emboj/16.12.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett H, Butowt R, Smith J, Farrant J, Illing N. Photosynthetic genes are differentially transcribed during the dehydration–rehydration cycle in the resurrection plant, Xerophyta humilis. Journal of Experimental Botany. 2003;54:2593–2595. doi: 10.1093/jxb/erg285. [DOI] [PubMed] [Google Scholar]
- Collett H, Shen A, Gardner M, Farrant JM, Denby K, Illing N. Towards transcripts profiling of desiccation tolerance in Xerophyta humilis: construction of a normalized 11k X. humilis cDNA set and microarray expression analysis of 424 cDNAs in response to dehydration. Physiologia Plantarum. 2004;122:39–53. [Google Scholar]
- Dace H, Sherwin HW, Illing N, Farrant JM. Use of metabolic inhibitors to elucidate mechanisms of recovery from desiccation stress in the resurrection plant Xerophyta humilis. Plant Growth Regulation. 1998;24:171–177. [Google Scholar]
- DeLano WL. The PyMOL molecular graphics system. Palo Alto, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- Dutilleul C, Garmier M, Noctor G, Mathieu C, Chetrit P, Foyer CH, De Paepe R. Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signalling and diurnal regulation. The Plant Cell. 2003;15:1212–1226. doi: 10.1105/tpc.009464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltis LD, Hofmann B, Hecht HJ, Lunsdorf H, Timmis KN. Purification and crystallization of 2,3 dihydroxybiphenyl 1,2-dioxygenase. Journal of Biological Chemistry. 1993;268:2727–2732. [PubMed] [Google Scholar]
- Espartero J, Sanchez-Aguayo I, Pardo JM. Molecular characterization of glyoxalase I from a higher plant; up-regulation by stress. Plant Molecular Biology. 1995;29:1223–1233. doi: 10.1007/BF00020464. [DOI] [PubMed] [Google Scholar]
- Farrant JM. Mechanisms of desiccation tolerance in Angiosperm resurrection plants. In: Jenks MA, Wood AJ, editors. Plant desiccation tolerance. Oxford: Blackwell Publishing; 2007. pp. 51–90. [Google Scholar]
- Freire AP, Ferreira A, Gomes R, Cordeiro C. Anti-glycation defences in yeast. Biochemical Society Transactions. 2003;31:1409–1412. doi: 10.1042/bst0311409. [DOI] [PubMed] [Google Scholar]
- Fridovich I. Biological effects of the superoxide radical. Archives of Biochemistry and Biophysics. 1986;15:1–11. doi: 10.1016/0003-9861(86)90526-6. [DOI] [PubMed] [Google Scholar]
- Gaff DF. Desiccation tolerant flowering plants in Southern Africa. Science. 1971;174:1033–1034. doi: 10.1126/science.174.4013.1033. [DOI] [PubMed] [Google Scholar]
- Gaff DF. Desiccation tolerant vascular plants of Southern Africa. Oecologia. 1977;31:95–109. doi: 10.1007/BF00348713. [DOI] [PubMed] [Google Scholar]
- Ghasempour HR, Gaff DF, Williams RPW, Gianello RD. Contents of sugars in leaves of drying desiccation-tolerant flowering plants, particularly grasses. Plant Growth Regulation. 1998;24:185–191. [Google Scholar]
- Haughn GW, Somerville C. Sulfonylurea-resistant mutant of Arabidopsis thaliana. Molecular and General Genetics. 1986;204:430–434. [Google Scholar]
- Illing N, Denby KJ, Collett H, Shen A, Farrant JM. The signature of seeds in resurrection plants: a molecular and physiological comparison of desiccation tolerance in seed and vegetative tissue. Integrative and Comparative Biology. 2005;45:771–787. doi: 10.1093/icb/45.5.771. [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annual Reviews of Plant Physiology and Plant Molecular Biology. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Kermode A. Regulatory mechanisms in the transition from seed development to germination: interactions between the embryo and the seed environment. In: Kigel J, Galili G, editors. Seed development and germination. New York: Marcel Dekker; 1995. pp. 273–332. [Google Scholar]
- Kita A, Kita S, Fujisawa I, Inaka K, Ishida T, Horiike K, Nozaki M, Miki K. An archetypical extradiol-cleaving catecholic dioxygenase: the crystal structure of catechol 2, 3- dioxygenase (metapyrocatechase) from Pseudomonas putida mt-2. Structure Fold Design. 1999;7:25–34. doi: 10.1016/s0969-2126(99)80006-9. [DOI] [PubMed] [Google Scholar]
- Kumagai T, Maruyama M, Matoba Y, Kawano Y, Sugiyama M. Crystallization and preliminary X-ray diffraction studies of bleomycin-binding protein encoded on the transposon Tn5. Acta Crystallography Section D Biological Crystallography. 1999;55:1095–1097. doi: 10.1107/s0907444999002875. [DOI] [PubMed] [Google Scholar]
- Lambert C, Leonard N, De Bolle X, Depiereux E. ESyPred3D: Prediction of proteins 3D structures. Bioinformatics. 2002;18:1250–1256. doi: 10.1093/bioinformatics/18.9.1250. [DOI] [PubMed] [Google Scholar]
- Maeda K, Finnie C, Svensson B. Cy5 maleimide labelling for sensitive deletions of free thiols in native protein extracts: identification of seed proteins targeted by barley thioredoxin isoform. Biochemical Journal. 2004;378:497–507. doi: 10.1042/BJ20031634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli T, Whittaker A, Masclaux-Daubresse C, Farrant JM, Brilli F, Loreto F, Vazzana C. Evidence for the presence of photorespiration in desiccation-sensitive leaves of the C4 ‘resurrection’ plant Sporobolus stapfianus during dehydration stress. Journal of Experimental Botany. 2007;58:3929–3939. doi: 10.1093/jxb/erm247. [DOI] [PubMed] [Google Scholar]
- Martins AM, Mendes P, Cardeiro C, Freire AP. In situ kinetic analysis of glyoxalase I and glyoxalase II in Saccharomyces cerevisiae. European Journal of Biochemistry. 2001;268:3930–3936. doi: 10.1046/j.1432-1327.2001.02304.x. [DOI] [PubMed] [Google Scholar]
- McCarthy AA, Baker HM, Shewry SC, Patchett ML, Baker EN. Crystal structure of methylmalonyl-coenzyme A epimerase from P. shermanii: a novel enzymatic function on an ancient metal binding scaffold. Structure. 2001;9:637–646. doi: 10.1016/s0969-2126(01)00622-0. [DOI] [PubMed] [Google Scholar]
- McGuffin JL, Jones TD. Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics. 2003;19:874–881. doi: 10.1093/bioinformatics/btg097. [DOI] [PubMed] [Google Scholar]
- Michel D, Furini A, Salamini F, Bartels D. Structure and regulation of an ABA- and desiccation-responsive gene from the resurrection plant Craterostigma plantagineum. Plant Molecular Biology. 1994;24:549–560. doi: 10.1007/BF00023553. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends in Plant Sciences. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Moore JP, Hearshaw M, Ravenscroft N, Lindsey GG, Farrant JM, Brandt WF. Desiccation-induced ultrastructural and biochemical changes in the leaves of the resurrection plants Myrothamnus flabellifolia. Australian Journal of Botany. 2007;55:482–491. [Google Scholar]
- Mowla SB, Thompson JA, Farrant JM, Mundree SG. A novel stress-inducible antioxidant enzyme identified from the resurrection plant Xerophyta humilis Baker. Planta. 2002;215:716–726. doi: 10.1007/s00425-002-0819-0. [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer HC. Ascorbate and glutathione: keeping active oxygen under control. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- Norwood M, Toldi O, Richter A, Scott P. Investigation into the ability of roots of the poikilohydric plant Craterostigma plantagineum to survive dehydration stress. Journal of Experimental Botany. 2003;54:2313–2321. doi: 10.1093/jxb/erg255. [DOI] [PubMed] [Google Scholar]
- Oliver MJ, Tuba Z, Mishler BD. The evolution of vegetative desiccation tolerance in land plants. Plant Ecology. 2000;151:85–100. [Google Scholar]
- Peters SW, Mundree SG, Thomson JA, Farrant JM, Keller F. Protection mechanism in the resurrection plant Xerophyta viscosa (Baker): Both sucrose and raffinose oligosaccharides (RFOs) accumulate in leaves in response to water deficit. Journal of Experimental Botany. 2007;58:1947–1956. doi: 10.1093/jxb/erm056. [DOI] [PubMed] [Google Scholar]
- Proctor MC, Tuba Z. Poikilohydry and homoihydry: antithesis or spectrum of possibilities? New Phytologist Review. 2002;156:327–349. doi: 10.1046/j.1469-8137.2002.00526.x. [DOI] [PubMed] [Google Scholar]
- Ramanjulu S, Bartels D. Drought and desiccation-induced modulation of gene expression in plants. Plant, Cell and Environment. 2002;25:141–151. doi: 10.1046/j.0016-8025.2001.00764.x. [DOI] [PubMed] [Google Scholar]
- Rife CL, Pharris RE, Newcomer ME, Armstrong RN. Crystal structure of a genomically encoded fosfomycin resistance protein (FosA) at 1.19 A resolution by MAD phasing off the L-III edge of T1(+) Journal of the American Chemical Society. 124:11001–11003. doi: 10.1021/ja026879v. [DOI] [PubMed] [Google Scholar]
- Rigsby RE, Brown DW, Dawson E, Lybrand TP, Armstrong RN. A model for glutathione binding and activation in the fosfomycin resistance protein, FosA. Archives of Biochemistry and Biophysics. 2007;464:277–283. doi: 10.1016/j.abb.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. Journal of Molecular Biology. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu J. The PredictProtein Server. Nucleic Acids Research. 2004;32 doi: 10.1093/nar/gkh377. (Web Server issue), W321–W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwin HW, Farrant JM. Differences in rehydration of three desiccation tolerant angiosperm species. Annals of Botany. 1996;78:703–710. [Google Scholar]
- Singla-Pareek SL, Reddy MK, Sopory SK. Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proceedings of the National Academy of Sciences, USA. 2003;100:12672–14677. doi: 10.1073/pnas.2034667100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacy RAP, Munthe E, Steinum T, Sharma B, Reidunn AB. A peroxiredoxin antioxidant is encoded by a dormancy-related gene, Per1, expressed during late development in the aleurone and embryo of barley grains. Plant Molecular Biology. 1996;31:1205–1216. doi: 10.1007/BF00040837. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thornalley PJ. Glyoxalase I-structure, function and a critical role in the enzymatic defence against glycation. Biochemical Society Transactions. 2003;31:1343–1348. doi: 10.1042/bst0311343. [DOI] [PubMed] [Google Scholar]
- Vander Willigen C, Pammenter NW, Mundree SG, Farrant JM. Mechanical stabilization of desiccation vegetative tissues of the resurrection grass Eragrostis nindensis: does a TIP 3:1 and/or compartmentalization of subcellular components and metabolites play a role? Journal of Experimental Biology. 2004;55:651–661. doi: 10.1093/jxb/erh089. [DOI] [PubMed] [Google Scholar]
- Veena RSV, Sopory SK. Glyoxalase 1 from Brassica juncea: molecular cloning and its overexpression confer tolerance in transgenic tobacco under stress. The Plant Journal. 1999;17:385–395. doi: 10.1046/j.1365-313x.1999.00390.x. [DOI] [PubMed] [Google Scholar]
- Vetting WM, Wackett PL, Que L, Lipscomb DJ, Ohlendorf HD. Crystallographic comparison of manganese and iron-dependent Homoprotocatechuate 2, 3-dioxygenase. Journal of Bacteriology. 2004;186:1945–1958. doi: 10.1128/JB.186.7.1945-1958.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertucci CW, Farrant JM. Acquisition and loss of desiccation tolerance. In: Kigel J, Galili G, editors. Seed Development and Germination. New York: Marcel Dekker; 1995. pp. 237–271. [Google Scholar]
- Vullo A, Frasconi P. Disulfide connectivity prediction using recursive neural networks and evolutionary information. Bioinformatics. 2004;20:653–659. doi: 10.1093/bioinformatics/btg463. [DOI] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.) Analytical Biochemistry. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- Whittaker A, Bochicchio A, Vazzana C, Lindsey G, Farrant J. Changes in leaf hexokinase activity and metabolite levels in response to drying in the desiccation-tolerant species Sporobolus stapfianus and Xerophyta viscosa. Journal of Experimental Botany. 2001;52:961–969. doi: 10.1093/jexbot/52.358.961. [DOI] [PubMed] [Google Scholar]
- Whittaker A, Martinelli T, Bochicchio A, Vezzana C, Farrant J. Comparison of sucrose metabolism during rehydration of desiccation-tolerant and desiccation-sensitive leaf material of Sporobolus stapfianus. Physiologia Plantarum. 2004;122:11–20. [Google Scholar]
- Whittaker A, Martinelli T, Farrant JM, Bochicchio A, Vazzana C. Sucrose phosphate synthase activity and the co-ordination of carbon partitioning during sucrose and amino acid accumulation in desiccation-tolerant leaf material of the C4 resurrection plant Sporobolus stapfianus during dehydration. Journal of Experimental Botany. 2007;58:3775–3787. doi: 10.1093/jxb/erm228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.