Abstract
The tripeptide glutathione is a major antioxidant and redox buffer with multiple roles in plant metabolism. Glutathione biosynthesis is restricted to the cytosol and the plastids and the product is distributed to the various organelles by unknown mechanisms. In the present study immunogold cytochemistry based on anti-glutathione antisera and transmission electron microscopy was used to determine the relative concentration of glutathione in different organelles of Arabidopsis thaliana leaf and root cells. Glutathione-specific labelling was detected in all cellular compartments except the apoplast and the vacuole. The highest glutathione content was surprisingly not found in plastids, which have been described before as a major site of glutathione accumulation, but in mitochondria which lack the capacity for glutathione biosynthesis. Mitochondria of both leaf and root cells contained 7-fold and 4-fold, respectively, higher glutathione levels than plastids while the density of glutathione labelling in the cytosol, nuclei, and peroxisomes was intermediate. The accuracy of the glutathione labelling is supported by two observations. First, pre-adsorption of the anti-glutathione antisera with glutathione reduced the density of the gold particles in all organelles to background levels. Second, the overall glutathione-labelling density was reduced by about 90% in leaves of the glutathione-deficient Arabidopsis mutant pad2-1 and increased in transgenic plants with enhanced glutathione accumulation. Hence, there was a strong correlation between immunocytochemical and biochemical data of glutathione accumulation. Interestingly, the glutathione labelling of mitochondria in pad2-1 remained very similar to wild-type plants thus suggesting that the high mitochondrial glutathione content is maintained in a situation of permanent glutathione-deficiency at the expense of other glutathione pools. High and constant levels of glutathione in mitochondria appear to be particularly important in cell survival strategies and it is predicted that mitochondria must have highly competitive mitochondrial glutathione uptake systems. The present results underline the suggestion that subcellular glutathione concentrations are not controlled by a global mechanism but are controlled on an individual basis and it is therefore not possible to conclude from global biochemical glutathione analysis on the status of the various organellar pools.
Keywords: Arabidopsis, glutathione, immunocytochemistry, transmission electron microscopy
Introduction
The tripeptide glutathione (γ-glutamyl-cysteinyl-glycine) is the major non-protein thiol in plant cells and fulfils multiple functions. As an antioxidant it plays important protective roles by detoxifying reactive oxygen species (ROS), which could otherwise damage cellular components, either directly by scavenging them or indirectly through the ascorbate–glutathione cycle (Noctor and Foyer, 1998; Tausz et al., 2004). Glutathione also protects proteins from irreversible modifications induced by ROS or reactive nitrogen species by forming reversible disulphide bonds between the cysteine thiol groups of the protein and glutathione (Giustarini et al., 2004; Dixon et al., 2005; Lindermayr et al., 2005). In addition, various herbicides and xenobiotics are detoxified by GSH-S-transferases that catalyse the conjugation of the herbicide to glutathione (Schröder, 2001; Edwards et al., 2005; DeRidder and Goldsbrough, 2006; Kopriva, 2006). Glutathione serves as a precursor for the synthesis of phytochelatins which contribute to heavy metal tolerance (Rennenberg, 2001; Kopriva, 2006). Glutathione is a key regulator of redox signalling which, at different levels, controls gene expression and contributes to cell survial (Foyer et al., 2001; Schäfer and Buertner, 2001; Maughan and Foyer, 2006). Glutathione is also involved in sulphur metabolism where it is thought to play roles in the uptake, assimilation, transport, and storage of reduced sulphur (Foyer and Rennenberg, 2000; Kopriva and Rennenberg, 2004; Kopriva, 2006).
Glutathione synthesis in plants seems to take place exclusively in plastids and the cytosol in two ATP-dependent steps. The first step of glutathione synthesis is the formation of γ-glutamyl cysteine catalysed by γ-glutamyl cysteine synthetase (GSH1). This step takes place either exclusively in plastids, as demonstrated in leaves of Arabidopsis thaliana (Wachter et al., 2005), or in both plastids and the cytosol in some other plant species (Noctor et al., 1998, 2002; Kopriva, 2006). In a second step, catalysed by glutathione synthetase (GSH2), glycine is added to γ-glutamyl cysteine to form glutathione. This step takes place in plastids and the cytosol (Noctor et al., 2002; Sugiyama et al., 2004). GSH1 (At4g23100) and GSH2 (At5g27380) are encoded by single copy genes in Arabidopsis. Mutational knock-outs in GSH1 in Arabidopsis were shown to block glutathione production completely and resulted in a lethal phenotype (Cairns et al., 2006). In conclusion, there seems to be only one pathway for glutathione synthesis in plants and the necessary components of this pathway are restricted to plastids and the cytosol. Thus, all other cellular compartments depend on the import of glutathione from the cytosol.
Glutathione degradation catalysed by γ-glutamyl transpeptidase, which transfers glutamate from glutathione to other dipeptides, occurs at the plasmalemma, at the tonoplast or within vacuoles and the apoplast (Foyer et al., 2001; Storozhenko et al., 2002; Shaw et al., 2005; Ohkama-Ohtsu et al., 2007a, b). Another pathway of glutathione degradation is facilitated by a carboxypeptidase that has been detected within the vacuoles of barley leaves (Wolf et al., 1996) and removes glycine from glutathione. The remaining dipeptides are then metabolized by a dipeptidase to the component amino acids (Foyer et al., 2001).
In the last few years some progress was made in studying the subcellular localization of glutathione in plant cells, thus helping to understand glutathione metabolism better as well as its protective roles within single cells and organelles, especially during pathogen attack (Kuzinak and Sklodowska, 2005; Zechmann et al., 2005, 2007). With biochemical methods, glutathione was detected within the plastids of spinach leaves (Hartmann et al., 2003), within mitochondria and peroxisomes of pea leaves (Jiménez et al., 1997, 1998) and within mitochondria, peroxisomes, and plastids of tomato leaves (Kuzinak and Sklodowska, 2001, 2004, 2005). In addition, glutathione was detected in the apoplast of oat and barley plants (Vanacker et al., 1998a, b, c) and Arabidopsis plants (Ohkama-Ohtsu et al., 2007a). Nevertheless, biochemical methods are based on the isolation of organelles from large amounts of plant material. These methods suffer from two intrinsic problems. First, the isolation procedure can lead to contamination problems of non-organelle-specific glutathione. Second, because of the lengthy procedure it is unclear how well the obtained results reflect the in vivo situation (Noctor et al., 2002; Chew et al., 2003). With light and confocal microscopy and the use of the fluorescent probes monochloro- and monobromobimanes, glutathione was detected in the cytosol and within nuclei of different plant species (Fricker et al., 2000; Meyer and Fricker, 2000; Meyer et al., 2001; Müller et al., 2005). Due to limitations of this technique (Fricker and Meyer, 2001), it was not possible to gain more detailed information about the subcellular distribution of glutathione besides high concentrations of glutathione in the cytoplasm and the absence of glutathione in vacuoles and cell walls. An alternative to the above-mentioned methods is the detection of glutathione and its precursors with electron microscopical techniques which has been applied to the analysis of subcellular glutathione distribution in Cucurbita pepo (Müller et al., 2004; Zechmann et al., 2006b).
The aim of the present study was to adapt these techniques to leaf and root material of Arabidopsis in order to obtain more information about the subcellular distribution of glutathione in single cells of this well-established model plant. To confirm the accuracy of the applied antibodies, and to understand how alterations in glutathione synthesis affect compartment-specific glutathione contents, these methods were also applied on different Arabidopsis mutants with altered glutathione metabolism. For these experiments the Arabidopsis GSH1-mutant pad2-1, which contains about 80% less glutathione than the wild type, and two transgenic lines overexpressing GSH1 with increased glutathione levels were used (Parisy et al., 2007). These mutants were chosen because these alterations in glutathione metabolism did not affect their growth phenotypes nor the ultrastructure of root and leaf cells. The present results confirm the accuracy of the glutathione labelling procedure and, surprisingly, identified mitochondria and not plastids as a hotspot of glutathione accumulation.
Materials and methods
Plant material and growth conditions
After stratification for 4 d at 4 °C, seeds of A. thaliana accession Col-0, the mutant line pad2-1 and two GSH1 overexpressing lines (35S::GSH1; Parisy et al., 2007) were grown in growth chambers with a 14/10 h day/night photoperiod. Day and night temperatures were 22 °C and 18 °C, respectively, the relative humidity was 60% and the plants were kept at 100% relative soil water content. Light intensity varied between 110 and 140 μmol m−2 s−1. Four weeks after stratification, root tips and samples from the youngest fully developed rosette leaf were harvested 2 h after the onset of the light period and prepared for electron microscopy. Leaves at this stage were approximately 2 cm in length and 0.7 cm in width.
Sample preparation for electron microscopy
Root tips and small leaf samples (about 1.5 mm2) from at least three different plants were cut on a modelling wax plate either in a drop of (i) 3% glutardialdehyde in 0.06 M Sørensen phosphate buffer (pH 7.2) for ultrastructural investigations or (ii) in a drop of 2.5% paraformaldehyde/0.5% glutardialdehyde in 0.06 M Sørensen phosphate buffer (pH 7.2) for cytohistochemical analysis. Samples were then transferred into glass vials and fixed for 90 min at room temperature (RT) in the above-mentioned media.
For ultrastructural analysis samples were then rinsed in buffer (4 times for 15 min each) and post-fixed in 1% osmium tetroxide in 0.06 M Sørensen phosphate buffer for 90 min at RT. The samples were then dehydrated in a graded series of increasing concentrations of acetone (50%, 70%, 90%, and 100%). Pure acetone was then exchanged for propylene oxide and the specimens were gradually infiltrated with increasing concentrations of Agar 100 epoxy resin (30%, 60%, and 100%) mixed with propylene oxide for a minimum of 3 h per step. Samples were finally embedded in pure, fresh Agar 100 epoxy resin (Agar Scientific Ltd, Stansted, UK) and polymerized at 60 °C for 48 h.
For cytohistochemical investigations samples were rinsed in buffer (4 times for 15 min each) after fixation and then dehydrated in increasing concentrations of acetone (50%, 70%, and 90%) for 2 times 10 min each. Subsequently, specimens were gradually infiltrated with increasing concentrations of LR-White resin (30%, 60%, and 100%; London Resin Company Ltd., Berkshire, UK) mixed with acetone (90%) for a minimum of 3 h per step. Samples were finally embedded in pure, fresh LR-White resin and polymerized at 50 °C for 48 h in small plastic containers under anaerobic conditions. Ultrathin sections (80 nm) were cut with a Reichert Ultracut S ultramicrotome. For ultrastructural investigations sections were post-stained for 5 min with lead citrate and for 15 min with uranyl acetate at RT before they were observed with a Philips CM10 TEM. For cytohistochemical investigations sections remained either unstained or were stained for 15 s with uranyl acetate at RT.
Cytohistochemical investigations
Immunogold labelling of glutathione was done with ultrathin sections on nickel grids. The ideal dilution of the primary and secondary antibody was determined in preliminary studies by evaluating the labelling density after a series of labelling experiments. The final dilution of the primary and secondary antibody used in this study showed a minimum of background labelling outside the sample with a maximum of specific labelling inside the sample. For cytohistochemical analysis samples were blocked with 2% bovine serum albumin (BSA) in phosphate-buffered saline (PBS, pH 7.2) for 20 min at RT. The excess of blocking solution on the grids was sucked off with filter paper and the samples were then treated with the primary antibody (anti-glutathione rabbit polyclonal IgG; Chemicon International, California) diluted 1:50 in PBS containing 1% goat serum for 2 h at RT. After a short rinse in PBS (3 times for 5 min) the samples were incubated with a 10 nm gold-conjugated secondary antibody (goat anti rabbit IgG, British BioCell International, Cardiff, www.british-biocell.co.uk) diluted 1:50 in PBS for 90 min at RT. After a short wash in PBS (3 times for 5 min) and distilled water (2 times for 5 min) labelled grids were either immediately observed in a Philips CM10 transmission electron microscope (TEM) or post-stained with uranyl-acetate (15 s). Post-staining with uranyl acetate was applied to facilitate the distinction of different cell structures enabling a clearer identification of the investigated organelles.
The selectivity and proper affinity of the primary antibody against glutathione has been tested with competition assays in tissue sections using bona fide GSH and GSSG as the target and a range of displacers. No measurable glutardialdehyde-fixed tissue cross-reactivity against L-alanine, γ-aminobutyrate, 1-amino-4-guanidobutane (AGB), D/L-arganine, D/L-aspartate, L-citrulline, L-cysteine, D/L-glutamate, K/L-glutamine, glycine, L-lysine, L-ornithine, L-serine, taurine, L-threonine, L-tryptophan, or L-tyrosine was detected. In addition, the antibody did not bind to glutathionylated proteins on Western blot experiments nor to glutathionylated proteins in sections fixed with formaldehyde (see technical note for glutathione antibody on the website of the manufacturer: www.immunologics.com). Therefore a reaction of the antibody with glutathionylated proteins bound by glutardialdehyde to the protein matrix of the tissue on ultrathin section seems very unlikely. The antibody does not discriminate between free reduced and oxidized glutathione (according to Signature Immunologics Inc.).
Several negative controls were made to confirm the specificity of the immunogold procedure. Negative controls were treated either with (i) gold conjugated secondary antibody (goat anti-rabbit IgG) without prior incubation of the section with the primary antibody, (ii) non-specific secondary antibody (goat anti-mouse IgG), and (iii) primary antibody pre-adsorbed with an excess of glutathione for 2 h at RT prior to labelling of the sections. For the latter a solution containing 10 mM of glutathione was incubated with 0.5% glutardialdehyde for 1 h. The excess of glutardialdehyde was then saturated by incubation for 30 min in a solution of 1% (w/v) BSA. The resulting solution was used to saturate the anti-GSH-antibodies for 2 h prior to its use in the immunogold labelling procedure described above.
Quantitative analysis of immunogold labelling
Micrographs of randomly photographed immunogold labelled sections of mesophyll cells and root tips were digitized and gold particles were counted automatically using the software package Cell D with the particle analysis tool (Olympus, Life and Material Science Europa GmbH, Hamburg, Germany) in different visually identified cell structures (mitochondria, plastids, nuclei, peroxisomes, and the cytosol). Due to the low amount of gold particles found in the apoplast, vacuoles, endoplasmic reticulum (ER), and dictyosomes of samples treated with the primary antibody against glutathione, no statistical evaluation of the gold particle density was made for these compartments. For statistical evaluation at least three different samples from roots and leaves were examined for each tissue type (root tip, mesophyll). A minimum of 20 (peroxisomes) to 60 (other cell structures) sectioned cell structures of at least 15 different cells throughout the block were analysed for gold particle density. In addition, gold particle density was evaluated in leaf cells exclusively located at the edge of the block and compared with results obtained from cells throughout the block. The obtained data were statistically evaluated using Statistica (Stat-Soft, USA, 1994) and presented as the number of gold particles μm−2. Unspecific background labelling was determined on 30 different sections (outside the specimen) from five different samples and subtracted from the obtained values inside the sample. Unspecific background labelling was around 0.1 gold particles μm−2. For all statistical analyses the non-parametric Kruskal–Wallis test followed by a post hoc comparison according to Conover was used (Bortz et al., 2000). P <0.05 was regarded as significant.
Isolation of mitochondria and immunoblotting
Mitochondria from 4-week-old A. thaliana plants accession Col-0, were isolated as previously described (Werhahn et al., 2001; Kruft et al., 2001). Ten μg of protein were loaded per lane and separated by SDS–PAGE and transferred on a nitrocellulose membrane (Schleicher and Schuell, Dassel, Germany) or the gel was stained with Coomassie Blue. Before probing with anti-glutathione antibodies, the blot was incubated for 90 min in 0.06 M Sørensen phosphate buffer (pH 7.2) containing 2.5% paraformaldehyde/0.5% glutardialdehyde as used for sample preparation for transmission electron microscopy. Blots were probed overnight at 4 °C with polyclonal anti-glutathione antiserum (1:250) or monoclonal anti-pyruvate dehydrogenase (PDH) antiserum (1:5000) raised against maize mitochondrial PDH, followed by incubation for 2 h at RT with the secondary antibody [respectively, goat anti-rabbit (1:2000) or goat anti-mouse (1:500); Dako, Glostrup, Denmark] linked to alkaline phosphatase. Then the two blots were assayed for alkaline phosphatase activity with sodium 5-bromo-4-chloro-3-indolyl-phosphate and 4-nitroblue tetrazolium chloride for the same length of time.
Results
The aim of the present work was to determine by immunocytochemical methods the subcellular distribution of glutathione in leaves and roots of Arabidopsis. To this end the Arabidopsis accession Col-0 and mutant Arabidopsis plants with globally decreased or increased glutathione accumulation were used. The glutathione-deficient mutant pad2-1 accumulates only 20% of the wild-type glutathione levels while transgenic plants overexpressing γ-glutamyl cysteine synthetase (GSH1; OE2 and OE3) show increased glutathione levels (Parisy et al., 2007). The low glutathione of the pad2-1 mutant had no detectable effect on the ultrastructure of leaf and root cells. (see Supplementary Fig. S1 at JXB online). Cells of the wild type and of mutant plants showed a dense cytosol with well-preserved organelles.
Subcellular distribution of glutathione
Glutathione-labelling was detected in all cell compartments of leaves and roots from Arabidopsis plants except in vacuoles and the apoplast where gold particle density was below the level of detection (Figs 1A, B, 2A–C). Glutathione was also frequently detected within the lumen of the endoplasmic reticulum (ER) and at its membranes (Fig. 1A inset). Omission of the primary antibody or the use of a non-specific secondary antibody in the labelling protocol reduced the immunogold staining to background levels (data not shown). Most importantly, pre-adsorption of the anti-GSH antibody with an excess of free GSH also reduced the density of gold particles to background levels in sections of cells from Col-0, pad2-1, and the GSH1 overexpressing lines (Figs 1C, D, 2E) thus indicating that the observed immunogold labelling appears to be solely linked to glutathione.
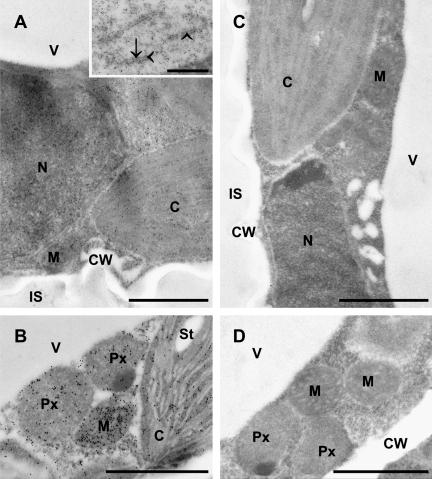
Fig. 1.
Transmission electron micrographs of mesophyll cells from Arabidopsis leaves after immunogold labelling of glutathione. (A, B) Cells of the wild-type Col-0 showing the highest amounts of gold particles in mitochondria (M), followed by the nucleus (N), the cytosol, peroxisomes (Px), and the chloroplasts (C) with or without starch (St). No or only few gold particles were detected in vacuoles (V), the cell walls (CW), and within the intercellular space (IS). Inset in (A) shows a close up of an endoplasmic reticulum with gold particles bound to glutathione at the membranes (arrows) and within its lumen (arrowheads). (C, D) Negative controls (the anti-glutathione antibody was pre-adsorbed with an excess of free glutathione prior to its application as stated in material and methods) of cells from Col-0 (C) and the GSH1 overexpressing line OE2 (D) showing no or only few gold particles in the individual compartments. Sections were post-stained with uranyl acetate for 15 s. Bars: 1 μm for (A), (B), (C), and (D) and 0.5 μm for inset.
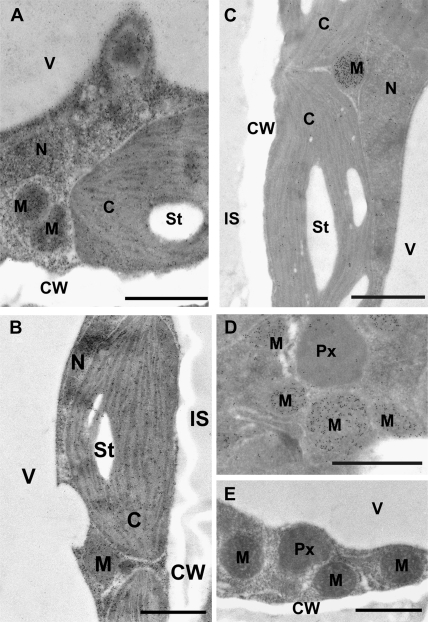
Fig. 2.
Transmission electron micrographs showing a comparison of glutathione labelling density between mesophyll cells from leaves of the wild type and different mutants of Arabidopsis plants. Different amounts of gold particles can be observed between the Arabidopsis wild-type Col-0 (A), the GSH1 overexpressing line OE2 (B), and the pad2-1 mutant (C, D). Note that cells of the GSH1 overexpressing line (B) show, in general, higher amounts of gold particles bound to glutathione when compared to Col-0. By contrast, cells of the pad2-1 mutant (C, D) show fewer gold particles bound to glutathione, except in mitochondria where gold particle density was similar to Col-0 (A). Pre-adsorption of the anti-glutathione antibody with an excess of free glutathione prior to its application (for details see Materials and methods) resulted in the complete absence of gold particles bound to glutathione in all cell compartments of the pad2-1 mutant (E). C, chloroplasts; St, starch; CW, cell walls; IS, intercellular spaces; M, mitochondria; N, nuclei; Px, peroxisomes; V, vacuoles. Sections were post-stained with uranyl acetate for 15 s. Bars: 1 μm.
In cells of leaves and roots from Col-0 the highest levels of glutathione were detected in mitochondria which contained 7-fold and 4-fold, respectively, higher glutathione contents than plastids, which showed the lowest levels of glutathione (Fig. 1A, B; Table 1). In leaves, nuclei contained the second highest amount of gold particles bound to glutathione followed by the cytosol and peroxisomes. In roots, the cytosol contained the second highest amount of gold particles bound to glutathione followed by nuclei. Gold particle density in leaves was higher in nuclei (3.2-fold), the cytosol (1.7-fold), and mitochondria (1.4-fold), but slightly lower in plastids when compared with the same organelles in roots of wild-type plants (Table 1). No significant difference in glutathione labelling density was found between cells located at the very edge of the tissue block and cells further inside the tissue block (Table 1). Thus, a possible delayed fixation of cells further away from the edge of the block did not influence the subcellular distribution of glutathione.
Table 1.
Subcellular glutathione contents
| Gold particles bound to glutathione μm−2 | ||
| Cell structures | Younger leaves | Roots |
| Cytosol | 200±11 d (188±11 d) | 117±5 f |
| Plastids | 73±2 h (74±2 h) | 84±3 g |
| Mitochondria | 491±25 a (506±19 a) | 361±15 b |
| Nuclei | 291±51 c (328±7 bc) | 90±5 g |
| Peroxisomes | 148±7 e (151±7 e) | n.d. |
| Vacuoles | 0 | n.d. |
| IS | 0 | n.d. |
Values are means with standard errors and document the amounts of gold particles bound to glutathione μm−2 in different cell compartments of cells from younger leaves and roots. Values in parenthesis represent the mean values (and standard errors) of gold particles bound to glutathione in cells only at the edge of the sample block, whereas the other values were obtained from cells throughout the sample block. Significant differences between the samples are indicated by different lowercase letters, samples which are significantly different from each other have no letter in common. P <0.05 was regarded as significant as analysed by the Kruskal–Wallis test followed by post hoc comparison according to Conover. n >20 for peroxisomes and n >60 for other cell structures. n.d., not determined. IS, intercellular spaces.
Immunolabelling experiments with pad2-1 resulted in a global decrease of gold particle density of about 90% when compared with the wild type. The correlation of the reduced GSH content of pad2-1 which accumulates around 20% of wild-type amounts of GSH and the observed reduction of anti-GSH antibody-dependent immunolabelling in pad2-1 strongly supports the GSH-specificity of the immunolabelling experiments. In both leaves and roots, the pad2-1 mutant showed, compared to the wild-type Col-0, a 72–92% decrease in gold particle density in all cell compartments investigated except mitochondria (Figs 2C, D, 3).
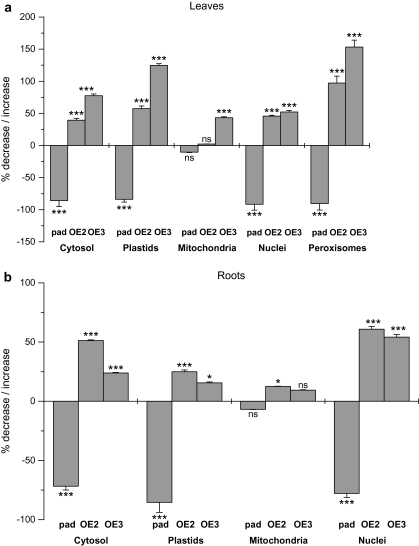
Fig. 3.
Summary of glutathione labelling results. Graphs show a comparison of glutathione labelling densities between Col-0, pad2-1 and GSH1 overexpressing lines. Values are means ±standard errors and document the percentage of increase or decrease of the amount of gold particles μm−2 bound to glutathione in different cell compartments of leaves (A) and roots (B) of the Arabidopsis mutant pad2-1 (pad) and the GSH1 overexpressing lines (OE2, OE3) compared with the Arabidopsis wild-type Col-0 (values given in Table 1). Significant differences in gold particle density within organelles between Col-0 and the mutants were calculated with the Mann Whitney U-test. ns if P >0.05, * P <0.05, ** P <0.01, and *** P <0.001. nd, not determined; n >20 for peroxisomes and n >60 for other cell structures.
Surprisingly, gold particle density in mitochondria in leaves and roots of the pad2-1 mutant remained at wild-type levels. In leaves, glutathione contents were decreased by about 92% in nuclei, 91% in peroxisomes, 86% in the cytosol, and 84% in plastids. In roots, gold particle density was also strongly decreased in plastids (86%), nuclei (78%), and the cytosol (72%). The complemented lines (OE2 and OE3) showed an increase in glutathione content in most cell compartments when compared to the control and to pad2-1 (Figs 2B, 3). In leaves, gold particle density was increased between 2-fold and 2.5-fold in peroxisomes, 1.6-fold and 2.3-fold in plastids, 1.5-fold in nuclei, 1.4-fold and 1.8-fold in the cytosol, and up to 1.4-fold in mitochondria depending on the complemented line (Fig. 3). Similar results were found in roots where the complemented lines showed an increase in glutathione content up to 1.6-fold in nuclei and in the cytosol, up to 1.3-fold in plastids, and up to 1.1-fold in mitochondria depending on the complemented line (Fig. 3).
Immunoblotting
The wild-type level of glutathione labelling in mitochondria of the pad2 mutant was a surprising finding. Even though the anti-glutathione antiserum does not cross-react with various substances with similar chemical structure there was the possibility that the anti-glutathione antibody could react with a sub-population of mitochondria-specific glutathione-tagged proteins. To test this possibility, a mitochondrial fraction from Arabidopsis Col-0 leaves was isolated and tested by immunoblotting with an antiserum specific for the mitochondrial protein PDH (mitochondrial pyruvate dehydrogenase) and the anti-glutathione antiserum (see Supplementary Fig. S2 at JXB online). The immunoblot of Fig. S2b shows that, compared to the crude extract, the mitochondrial fraction is highly enriched for the mitochondrial protein PDH. No signals were found on immuoblots developed with the anti-glutathione antiserum (see Supplementary Fig. S2c at JXB online). Apparently, there is no major mitochondrial protein fraction reacting with the anti-glutathione antiserum that could be responsible for the high immunogold labelling observed in mitochondria.
Discussion
The subcellular distribution of glutathione was analysed in detail by high-resolution immunogold cytochemistry in cells of leaves and roots of Arabidopsis plants. The accuracy of the observed labelling is supported by the results of pre-adsorption experiments of the anti-glutathione antibody with glutathione and the immunoblotting experiments. In addition, analysis of plants with altered glutathione levels revealed a strong correlation between biochemically determined total glutathione levels and the immunolabelling data. Glutathione labelling density was strongly decreased in roots and leaves of the pad2-1 mutant which accumulates around five times less glutathione compared to the wild type Col-0 (Parisy et al., 2007). In accordance with biochemical results (Parisy et al., 2007), transgenic plants with increased glutathione biosynthesis showed increased glutathione labelling. Therefore these results confirm that the cytohistochemical labelling approach for glutathione is specific for the respective compound and can be used for their relative quantification in all cell compartments of Arabidopsis.
Nevertheless, one could argue that glutathione is redistributed by diffusion during the sample preparation procedure, thus leading to a false conclusion about the subcellular distribution of glutathione. This seems very unlikely as the labelling density of glutathione at the edge of the sample, which gets into contact with the fixative immediately, was similar to what was found in cells closer to the centre of the block (block sizes were about 1.5 mm2), which were fixed with a delay. In addition, no glutathione was detected in the apoplast and in vacuoles. If a redistribution of glutathione occurs during the present procedure, a labelling in these cell compartments would be a logical consequence. Also, the use of paraformaldehyde and glutardialdehyde as fixatives is well established for immunocytochemical analysis of glutathione and has also been successfully used for the detection of glutathione in animal cells (Hjelle et al., 1994; Huster et al., 1998). Paraformaldehyde penetrates the tissue with rates that are five times faster than glutardialdehyde, which penetrates the tissue in rates of about 1 mm h−1 and irreversible fixes the cross-links made by paraformaldehyde (Bozzola and Russell, 1999). Therefore, the present method guarantees a relatively rapid fixation of the whole tissue block.
Even though the mutants showed an accumulation or depletion of glutathione, no ultratructural differences were found between cells of Col-0 plants and the mutants (see Supplementary Fig. S1 at JXB online). Ultrastructural changes and changes in the phenotype, due to oxidative damage, as described in transgenic tobacco plants overexpressing a chloroplast-targeted GSH1 (Creissen et al., 1999) were not found in the GSH1 overexpression lines nor in the pad2-1 mutant. In addition, the growth phenotype did not differ between the mutants and Col-0. Similar results have been obtained for poplar plants overexpressing GSH1 in the chloroplast which also did not develop any differences in the phenotype, despite changes in glutathione metabolism (Noctor et al., 1998). It can be concluded that the glutathione deficiency of pad2-1 and the increased glutathione content of the GSH1-overexpressing transgenic lines did not cause any detectable ultrastructural damage.
The subcellular distribution of gold particles bound to glutathione found in cells of Col-0 was similar to what was found in Cucurbita pepo plants (Müller et al., 2004) and extends the current knowledge about the distribution of glutathione in cells of Arabidopsis plants made with confocal laser scanning microscopy to a higher level of resolution (Meyer and Fricker, 2000; Meyer et al., 2001). Whereas the latter method was limited to the detection of cytoplasmic glutathione of Arabidopsis root and suspension-cultured cells, the present study revealed that glutathione is present in all cellular compartments except the apoplast and vacuoles of root and leaf cells. The highest levels of glutathione were detected in mitochondria, whereas plastids showed the lowest densities of glutathione-specific gold particles. That glutathione contents in mitochondria are higher than in other cellular compartments has also been shown in Cucurbita pepo (Müller et al., 2004), in animal tissue (Huster et al., 1998), and cultured mammalian cells (Söderdahl et al., 2003).
Surprisingly, glutathione contents in mitochondria of the pad2-1 mutant remained statistically unchanged, when compared to Col-0, whereas all other cell compartments showed a strong decrease of glutathione levels of up to 91.5% in the nuclei of leaves. Immunoblotting experiments revealed that the high gold particle density in mitochondria was not due to the binding of the glutathione antibody to mitochondrial proteins (e.g. mitochondria-specific glutathione-tagged proteins; see Supplementary Fig. S2 at JXB online). Therefore these results indicate that mitochondria of pad2-1 plants can accumulate wild-type levels of glutathione, despite severe glutathione deficiency in all other cell compartments. It has previously been shown with pharmacological experiments using the GSH1 inhibitor buthionine sulphoximine (BSO), that mitochondria of Cucurbita pepo plants could maintain glutathione levels while glutathione was depleted in all other compartments (Zechmann et al., 2006a). Similar results have been obtained from mammalian cells treated with BSO. Depletion of glutathione in the cytoplasm started with BSO treatments as low as 0.001 mM while a 100-fold higher BSO-concentration was needed to deplete glutathione in mitochondria (Green et al., 2006). Apparently, plant and mammalian mitochondria have the capacity to maintain high glutathione levels under conditions of induced glutathione depletion. The present results with pad2-1 indicate that mitochondria can not only maintain glutathione levels in situations of accidental glutathione depletion but also accumulate wild-type levels of mitochondrial glutathione in a genetically glutathione-deficient plant. This finding indicates that mitochondria, which can not synthesize glutathione in Arabidopsis (Wachter et al., 2005), must have very competitive glutathione-uptake systems.
That mitochondria play, generally, an important role in redox buffering within plant cells has been recently described for Arabidopsis plants. In response to hydrogen peroxide, mitochondria were found to maintain the original redox status better than the cytosol, thus indicating that mitochondria have a higher capacity to buffer redox changes than the cytoplasm (Jiang et al., 2006). The observed high and stable levels of glutathione in mitochondria may play a key role in the detoxification of mitochondrially generated ROS especially under conditions of severe glutathione depletion (Armstrong and Jones, 2003; Green et al., 2006; Zechmann et al., 2006a). During such conditions an increase of ROS could lead to damage of mitochondrial membrane components resulting in the release of cytochrome C, which then could trigger caspase-dependent cell death (Fernández-Checa, 2003; Rodriguez-Enriquez et al., 2004). Thus, high and constant levels of glutathione in mitochondria might play a crucial role in cell survival strategies. In addition, glutathione in the mitochondria is involved in the protection of mitochondrial DNA and proteins from oxidative modification (Foyer and Noctor, 2005; Green et al., 2006; Rhoads and Subbaiah, 2007). Glutathionylation (the formation of mixed disulphides between the thiol group of the protein and glutathione) seems to play essential roles not only in the protection of mitochondrial proteins against oxidation but also for redox sensing and signalling, for regulating the activities of enzymes, transcription factors, and transporters (Hurd et al., 2005a, b).
With the exception of vacuoles and intercellular spaces, the lowest contents of glutathione in roots and leaves were found in plastids which, together with the cytosol, are considered to be the exclusive centres of glutathione synthesis (Noctor et al., 2002; Sugiyama et al., 2004; Wachter et al., 2005; Zechmann et al., 2006a). These results are contradictory to the widely accepted view that plastids contain the highest levels of glutathione (Noctor et al., 2002; Hartmann et al., 2003; Maughan and Foyer, 2006). The latter results are based on biochemical analysis of glutathione contents obtained after plastid isolation. Low glutathione content of chloroplasts has been already shown in Cucurbita pepo plants (Müller et al., 2004; Zechmann et al., 2006a) and has been calculated for young wheat leaves (Noctor et al., 2002), but was rejected by the authors as an artefact.
Glutathione was also detected in high levels in nuclei especially in leaves, suggesting a role of glutathione in this cell compartment. Glutathione in nuclei has also been found with light microscopical techniques after monochlorobimane staining in different plant species (Müller et al., 2005), but could not be clearly identified in cells of Arabidopsis (Meyer and Fricker, 2000; Meyer et al., 2001). The precise role of glutathione in nuclei of plant cells is unclear. It has been proposed for mammalian cells that nuclear glutathione protects DNA from oxidative modifications, as the latter were negatively correlated with reduced nuclear glutathione content (Green et al., 2006). Nevertheless, as the generation of large amounts of ROS in nuclei seems to be unlikely, high levels of glutathione in this cell compartment might play additional roles linked to maintaining reducing conditions, the control of redox signalling and gene transcription. In plants, glutathione was shown to be involved in the activation of transcriptional regulators, which control the expression of defence-related genes (Mou et al., 2003; Gomez et al., 2004).
In the present study, glutathione was frequently detected within the endoplasmic reticulum. The presence of glutathione within the ER has been demonstrated for animal tissue (Jessop and Bulleid, 2004) and indirectly for plant ER by using a ratiometric redox-sensitive GFP expressed in Arabidopsis thaliana (Meyer et al., 2007). In animal cells, the reduced form of glutathione is essential in the formation of native disulphide bonds within the ER lumen by maintaining ER oxidoreductases in a reduced state. Thus, allowing a correct folding of glycoproteins and secretory proteins, even under severe stress situations (Jessop and Bulleid, 2004). Whether glutathione fulfils a similar role in the ER of plant cells needs to be clarified.
It was not possible to detect free glutathione in the vacuoles in the present study. As it has also not been possible to detect free glutathione in vacuoles with biochemical and light microscopical techniques (Foyer and Rennenberg, 2000; Rennenberg, 2001; Müller et al., 2004, 2005), it seems that free glutathione does not accumulate within vacuoles. A similar situation was found in the apoplast where glutathione labelling was below the level of detection. In contrast to the present results, glutathione has been detected with biochemical methods in the apoplast of Arabidopsis plants (Ohkama-Ohtsu et al., 2007a) and also in oat and barley plants (Vanacker et al., 1998a, b). Nevertheless, the concentrations found in the apoplast were extremely low in comparison to the total glutathione pool of the investigated organ.
In summary, the present results give an insight into the subcellular distribution of glutathione in Arabidopsis plants. As glutathione contents in mitochondria were found to be highest and did not decrease in glutathione-deficient Arabidopsis, it seems that glutathione plays a key role in mitochondria. Plastids, which were so far considered to be the organelle with the highest glutathione concentration, contained the lowest levels of glutathione of all organelles with the exception of the vacuole and the apoplast. The present study also demonstrated that subcellular glutathione contents and the ratio of glutathione between organelles can vary greatly, especially during situations of extreme and permanent glutathione deficiency, without visible changes in the phenotype of the plant. As glutathione contents are also subject to change during plant development and strongly depend on growth and physiological conditions of plants (Tausz et al., 2004; Foyer and Noctor, 2005), differences in the subcellular distribution and of the ratio of organelle-specific glutathione can also be expected between plants under different environmental conditions/stresses and at different developmental stages. To study such organelle-specific changes of the glutathione pool during different developmental and environmental situations will be essential to clarify its role in the protection against oxidative stress, gene expression, redox signalling, and sensing on the cellular level and will contribute to a better understanding of the importance of compartment-specific glutathione metabolism for the development and growth of the whole plant.
Supplementary data
Supplementary Figures S1 and S2 can be found at JXB online.
Fig. S1. Ultrastructural comparison between Col-0 and pad2-1.
Fig. S2. Test of anti-glutathione antisera for cross-reactivity with mitochondrial proteins isolated from Arabidopsis plants.
Supplementary Material
Acknowledgments
We thank Laurence Drouard, CRNS, Institut de Biologie Moléculaire des Plantes, Strasbourg, France, for providing the monoclonal anti-PDH antiserum and the protocol for the isolation of mitochondria. This work was supported by the Austrian Science Fund (FWF, P18976 and P20619).
Glossary
Abbreviations
- ATP
adenosine triphosphate
- ER
endoplasmic reticulum
- GSH
reduced glutathione
- GSH1
γ-glutamyl cysteine synthetase
- GSH2
glutathione synthetase
- PDH
pyruvate dehydrogenase
- ROS
reactive oxygen species
- TEM
transmission electron microscope
References
- Armstrong JS, Jones DP. Glutathione depletion enforces the mitochondrial permeability transition and causes cell death in BcL-2 overexpressing HL60 cells. FASEB Journal. 2003;16:1263–1265. doi: 10.1096/fj.02-0097fje. [DOI] [PubMed] [Google Scholar]
- Bortz J, Lienert GA, Bohenke K. Verteilungsfreie Methoden in der Biostatistik. Berlin, Heidelberg, New York, Tokyo: Springer Verlag; 2000. [Google Scholar]
- Bozzola JJ, Russell D. Electron microscopy. Boston, Toronto, London, Singapore: Jones and Barlett Publishers; 1999. [Google Scholar]
- Chew O, Whelan J, Millar AH. Molecular definition of the ascorbate–glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defences in plants. Journal of Biological Chemistry. 2003;278:46869–46877. doi: 10.1074/jbc.M307525200. [DOI] [PubMed] [Google Scholar]
- Cairns NG, Pasternak M, Wachter A, Cobbett CS, Meyer AJ. Maturation of Arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiology. 2006;141:446–455. doi: 10.1104/pp.106.077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creissen G, Firmin J, Fryer M. Elevated glutathione biosynthetic capacity in the chloroplasts of transgenic tobacco plants paradoxically causes increased oxidative stress. The Plant Cell. 1999;11:1277–1291. doi: 10.1105/tpc.11.7.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRidder BP, Goldsbrough PB. Organ-specific expression of glutathione S-transferases and the efficacy of herbicide safeners in Arabidopsis. Plant Physiology. 2006;140:167–175. doi: 10.1104/pp.105.067199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Skipsey M, Grundy NM, Edwards R. Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiology. 2005;138:2233–2244. doi: 10.1104/pp.104.058917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R, Brazier-Hicks M, Dixon DP, Cummins I. Chemical manipulation of antioxidant defences in plants. Advances in Botanical Research. 2005;42:1–32. [Google Scholar]
- Fernández-Checa JC. Redox regulation and signaling lipids in mitochondrial apoptosis. Biochemical and Biophysical Research Communications. 2003;304:471–479. doi: 10.1016/s0006-291x(03)00619-3. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Oxidant and antioxidant signalling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant, Cell and Environment. 2005;28:1056–1071. [Google Scholar]
- Foyer CH, Rennenberg H. Regulation of glutathione synthesis and its role in abiotic and biotic stress defence. In: Brunold C, Rennenberg H, De Kok LJ, Stulen I, Davidian JC, editors. Sulfur nutrition and sulfur assimilation in higher plants. Bern: Paul Haupt Verlag; 2000. pp. 127–153. [Google Scholar]
- Foyer CH, Theodoulou FL, Delrot S. The functions of inter- and intracellular glutathione transport systems in plants. Trends in Plant Science. 2001;6:486–492. doi: 10.1016/s1360-1385(01)02086-6. [DOI] [PubMed] [Google Scholar]
- Fricker MD, May M, Meyer AJ, Sheard N, White NS. Measurement of glutathione levels in intact roots of Arabidopsis. Journal of Microscopy. 2000;198:162–173. doi: 10.1046/j.1365-2818.2000.00696.x. [DOI] [PubMed] [Google Scholar]
- Fricker MD, Meyer AJ. Confocal imaging of metabolism in vivo: pitfalls and possibilities. Journal of Experimental Botany. 2001;52:631–640. [PubMed] [Google Scholar]
- Giustarini D, Rossi R, Milzani A, Colombo R, Dalle-Donne I. S-glutathionylation: from redox regulation of protein functions to human diseases. Journal of Cellular and Molecular Medicine. 2004;8:201–212. doi: 10.1111/j.1582-4934.2004.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez LD, Noctor G, Knight MR, Foyer CH. Regulation of calcium signalling and expression by glutathione. Journal of Experimental Botany. 2004;55:1851–1859. doi: 10.1093/jxb/erh202. [DOI] [PubMed] [Google Scholar]
- Green RM, Graham M, O'Donovan MR, Chipman JK, Hodges NJ. Subcellular compartmentalization of glutathione: correlations with parameters of oxidative stress related to genotoxicity. Mutagenesis. 2006;21:383–390. doi: 10.1093/mutage/gel043. [DOI] [PubMed] [Google Scholar]
- Hartmann TN, Fricker MD, Rennenberg H, Meyer AJ. Cell-specific measurement of cytosolic glutathione in poplar leaves. Plant, Cell and Environment. 2003;26:965–975. doi: 10.1046/j.1365-3040.2003.01031.x. [DOI] [PubMed] [Google Scholar]
- Hjelle OP, Chaudhry FA, Ottersen OP. Antisera to glutathione: characterization and immunocytochemical application to the rat cerebellum. European Journal of Neuroscience. 1994;6:791–804. doi: 10.1111/j.1460-9568.1994.tb00990.x. [DOI] [PubMed] [Google Scholar]
- Huster D, Hjelle OP, Haug FM, Nagelhus EA, Reichelt W, Ottersen OP. Subcellular compartmentation of glutathione and glutathione precursors. A high resolution immunogold analysis of the outer retina of guinea pig. Anatomy and Embryology. 1998;198:277–287. doi: 10.1007/s004290050184. [DOI] [PubMed] [Google Scholar]
- Hurd TR, Filipovska A, Costa NJ, Dahm CC, Murphy MP. Disulphide formation on mitochondrial protein thiols. Biochemical Society Transactions. 2005a;33:1390–1393. doi: 10.1042/BST0331390. [DOI] [PubMed] [Google Scholar]
- Hurd TR, Costa NJ, Dahm CC, Beer SM, Brown SE, Filipovska A, Murphy MP. Glutathionylation of mitochondrial proteins. Antioxidants and Redox Signaling. 2005b;7:999–1010. doi: 10.1089/ars.2005.7.999. [DOI] [PubMed] [Google Scholar]
- Jessop CE, Bulleid NJ. Glutathione directly reduces an oxidoreductase in the endoplasmic reticulum of mammalian cells. Journal of Biological Chemistry. 2004;279:55341–55347. doi: 10.1074/jbc.M411409200. [DOI] [PubMed] [Google Scholar]
- Jiang K, Schwarzer C, Lally E, Zhang S, Ruzin S, Machen T, Remington SJ, Feldman L. Expression and characterization of a redox-sensing green fluorescent protein (reduction-oxidation-sensitive green fluorescent protein) in Arabidopsis. Plant Physiology. 2006;141:397–403. doi: 10.1104/pp.106.078246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez A, Hernández JA, del Río LA, Sevilla F. Evidence for the presence of the ascorbate–glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant Physiology. 1997;114:275–284. doi: 10.1104/pp.114.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez A, Hernández JA, Pastori G, del Río LA, Sevilla F. Role of the ascorbate–glutathione cycle of mitochondria and peroxisomes in the senescence of pea leaves. Plant Physiology. 1998;118:1327–1335. doi: 10.1104/pp.118.4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S. Regulation of sulfate assimilation in Arabidopsis and beyond. Annals of Botany. 2006;97:479–495. doi: 10.1093/aob/mcl006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Rennenberg H. Control of sulphate assimilation and glutathione synthesis: interaction with N and C metabolism. Journal of Experimental Botany. 2004;55:1831–1842. doi: 10.1093/jxb/erh203. [DOI] [PubMed] [Google Scholar]
- Kruft V, Eubel H, Jänsch L, Werhahn W, Braun H-P. Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiology. 2001;127:1694–1710. [PMC free article] [PubMed] [Google Scholar]
- Kuźniak E, Sklodowska A. Ascorbate, glutathione, and related enzymes in chloroplasts of tomato leaves infected by Botrytis cinerea. Plant Science. 2001;160:723–731. doi: 10.1016/s0168-9452(00)00457-x. [DOI] [PubMed] [Google Scholar]
- Kuźniak E, Sklodowska A. Comparison of two methods for preparing mitochondria from tomato leaves to study the ascorbate–glutathione cycle activity. Biologia Plantarum. 2004;48:537–542. [Google Scholar]
- Kuźniak E, Sklodowska A. Compartment-specific role of the ascorbate–glutathione cycle in the response of tomato leaf cells to Botrytis cinerea infection. Journal of Experimental Botany. 2005;56:921–933. doi: 10.1093/jxb/eri086. [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J. Proteomic identification of S-nitrosylated in Arabidopsis. Plant Physiology. 2005;137:921–930. doi: 10.1104/pp.104.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan S, Foyer CH. Engineering and genetic approaches to modulating the glutathione network in plants. Physiologia Plantarum. 2006;126:382–397. [Google Scholar]
- Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, Hell R. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. The Plant Journal. 2007;52:973–986. doi: 10.1111/j.1365-313X.2007.03280.x. [DOI] [PubMed] [Google Scholar]
- Meyer AJ, Fricker MD. Direct measurement of glutathione in epidermal cells of intact Arabidopsis roots by 2-photon laser scanning microscopy. Journal of Microscopy. 2000;198:174–181. doi: 10.1046/j.1365-2818.2000.00697.x. [DOI] [PubMed] [Google Scholar]
- Meyer AJ, May MJ, Fricker M. Quantitative in vivo measurement of glutathione in Arabidopsis cells. The Plant Journal. 2001;27:67–78. doi: 10.1046/j.1365-313x.2001.01071.x. [DOI] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113:935–944. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- Müller M, Zechmann B, Zellnig G. Ultrastructural localization of glutathione in Cucurbita pepo plants. Protoplasma. 2004;223:213–219. doi: 10.1007/s00709-003-0035-1. [DOI] [PubMed] [Google Scholar]
- Müller M, Zellnig G, Urbanek A, Zechmann B. Recent developments in methods intracellulary localizing glutathione within plant tissues and cells (a minivreview) Phyton (Horn) Austria. 2005;45:45–55. [Google Scholar]
- Noctor G, Arisi ACM, Jouanin L, Foyer CH. Manipulation of glutathione and amino acid biosynthesis in the chloroplast. Plant Physiology. 1998;118:471–482. doi: 10.1104/pp.118.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Foyer CH. Ascorbate and glutathione: keeping active oxygen under control. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:229–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- Noctor G, Gomez L, Vanacker H, Foyer CH. Interactions between biosynthesis, compartmentation, and transport in the control of glutathione homeostasis and signalling. Journal of Experimental Botany. 2002;53:1283–1304. doi: 10.1093/jexbot/53.372.1283. [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Radwan S, Peterson A, Zhao P, Badr AF, Xiang C, Oliver DJ. Characterization of the extracellular γ-glutamyl transpeptidases, GGT1 and GGT2, in Arabidopsis. The Plant Journal. 2007a;49:865–877. doi: 10.1111/j.1365-313X.2006.03004.x. [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Zhao P, Xiang C, Oliver DJ. Glutathione conjugates in the vacuole are degraded by γ-glutamyl transpeptidase GGT3 in Arabidopsis. The Plant Journal. 2007b;49:878–888. doi: 10.1111/j.1365-313X.2006.03005.x. [DOI] [PubMed] [Google Scholar]
- Parisy V, Poinssot B, Owsianowski L, Buchala A, Glazebrook J, Mauch F. Identification of PAD2 as a γ-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. The Plant Journal. 2007;49:159–172. doi: 10.1111/j.1365-313X.2006.02938.x. [DOI] [PubMed] [Google Scholar]
- Rennenberg H. Glutathione: an ancient metabolite with modern tasks. In: Grill D, Tausz M, De Kok LJ, editors. Significance of glutathione to plant adaptation to the environment. Dordrecht, Boston, London: Kluwer Academic Publishers; 2001. pp. 1–11. [Google Scholar]
- Rhoads DM, Subbaiah CC. Mitochondrial retrograde regulation in plants. Mitochondrion. 2007;7:177–194. doi: 10.1016/j.mito.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Enriquez S, He L, Lemasters JJ. Role of mitochondrial permeability transition pores in mitochondrial autophagy. International Journal of Biochemistry and Cell Biology. 2004;36:2463–2472. doi: 10.1016/j.biocel.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Schäfer FQ, Buettner GH. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radical Biology and Medicine. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- Schröder P. The role of glutathione and glutathione S-transferases in plant reaction and adaptation to xenobiotics. In: Grill D, Tausz M, De Kok LJ, editors. Significance of glutathione to plant adaptation to the environment. Dordrecht, Boston, London: Kluwer Academic Publishers; 2001. pp. 155–183. [Google Scholar]
- Shaw ML, Pither-Joyce MD, McCallum JA. Purification and cloning of a γ-glutamyl transpeptidase from onion (Allium cepa) Phytochemistry. 2005;66:515–522. doi: 10.1016/j.phytochem.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Söderdahl T, Enoksson M, Lundberg M, Holmgren A, Ottersen OP, Orrenius S, Bolcsfoldi G, Cotgreave IA. Visualization of the compartmentalization of glutathione and protein-glutathione mixed disulfides in cultured cells. FASEB Journal. 2003;17:124–126. doi: 10.1096/fj.02-0259fje. [DOI] [PubMed] [Google Scholar]
- Sugiyama A, Nishimura J, Mochizuki Y, Inagaki K, Sekiya J. Homoglutathione synthesis in transgenic tobacco plants expressing soybean homoglutathione synthetase. Plant Biotechnology. 2004;21:79–83. [Google Scholar]
- Storozhenko S, Belles-Boix E, Babiychuk E, Hérouart D, Davey MW, Slooten L, van Montagu M, Inzé D, Kushnir S. γ-glutamyl transpeptidase in transgenic tobacco plants. Cellular localization, processing, and biochemical properties. Plant Physiology. 2002;128:1109–1119. doi: 10.1104/pp.010887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tausz M, Šircelj H, Grill D. The glutathione system as a stress marker in plant ecophysiology: is a stress-response concept valid? Journal of Experimental Botany. 2004;55:1955–1962. doi: 10.1093/jxb/erh194. [DOI] [PubMed] [Google Scholar]
- Vanacker H, Carver TLW, Foyer CH. Pathogen-induced changes in the antioxidant status of the apoplast in barley leaves. Plant Physiology. 1998a;117:1103–1114. doi: 10.1104/pp.117.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacker H, Foyer CH, Carver TLW. Changes in apoplastic antioxidants induced by powdery mildew attack in oat genotypes with race non-specific resistance. Planta. 1998b;208:444–452. [Google Scholar]
- Vanacker H, Harbinson J, Ruisch J, Carver TLW, Foyer CH. Antioxidant defences of the apoplast. Protoplasma. 1998c;205:129–140. [Google Scholar]
- Wachter A, Wolf S, Steininger H, Bogs J, Rausch T. Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. The Plant Journal. 2005;41:15–30. doi: 10.1111/j.1365-313X.2004.02269.x. [DOI] [PubMed] [Google Scholar]
- Werhahn W, Niemeyer A, Jänsch L, Kruft V, Schmitz UK, Braun H-P. Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis. Identification of multiple forms of TOM20. Plant Physiology. 2001;125:943–954. doi: 10.1104/pp.125.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf AE, Dietz KJ, Schröder P. Degradation of glutathione S-conjugates by a carboxypeptidase in the plant vacuole. FEBS Letters. 1996;384:31–34. doi: 10.1016/0014-5793(96)00272-4. [DOI] [PubMed] [Google Scholar]
- Zechmann B, Müller M, Zellnig G. Intracellular adaptations of glutathione content in Cucurbita pepo (L.) induced by reduced glutathione and buthionine sulfoximine treatment. Protoplasma. 2006a;227:197–209. doi: 10.1007/s00709-005-0129-z. [DOI] [PubMed] [Google Scholar]
- Zechmann B, Zellnig G, Müller M. Changes in the subcellular distribution of glutathione during virus infection in Cucurbita pepo (L.) Plant Biology. 2005;7:49–57. doi: 10.1055/s-2004-830477. [DOI] [PubMed] [Google Scholar]
- Zechmann B, Zellnig G, Müller M. Immunocytochemical localization of glutathione precursors in plant cells. Journal of Electron Microscopy. 2006b;55:173–181. doi: 10.1093/jmicro/dfl022. [DOI] [PubMed] [Google Scholar]
- Zechmann B, Zellnig G, Urbanek-Krajnc A, Müller M. Artificial elevation of glutathione affects symptom development in ZYMV-infected Cucurbita pepo L. plants. Archives of Virology. 2007;152:747–762. doi: 10.1007/s00705-006-0880-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.