Abstract
Hormogonia are nongrowing filaments, motile by means of a gliding mechanism, that are produced by certain cyanobacteria. Their differentiation is induced by positive and negative factors for growth, such as deprivation of combined nitrogen (nitrogen stress induction [NSI]). In Nostoc punctiforme, they are also induced by the exudate (hormogonium-inducing factor [HIF]) of a symbiotic plant partner. Time course (0.5 to 24 h) transcription profiles were determined by DNA microarray assays for hormogonia of N. punctiforme following induction by HIF and NSI. Clustering analysis revealed both common and distinct transcriptional patterns for the two methods of induction. By 24 h, a common set of 1,328 genes was identified. This 24-h common set of genes arose by the transition of 474 genes from an 819-member common set of genes at 1 h after induction; 405 and 51 genes unique to the HIF and NSI groups at 1 h, respectively; and 398 genes differentially transcribed at later time points. The NSI hormogonia showed a transcriptional checkpoint at 12 h following induction in which up- and downregulated genes were transiently down- or upregulated, respectively. The transient changes in these 1,043 genes appeared to reflect a switch back to a vegetative growth state. Such a checkpoint was not seen in HIF hormogonia. Genes uniquely upregulated in HIF hormogonia included those encoding proteins hypothesized to synthesize a metabolite repressor of hormogonium differentiation. Approximately 34 to 42% of the 6,893 printed genes were differentially transcribed during hormogonium differentiation; about half of those genes were upregulated, and 1,034 genes responded within 0.5 h after induction. These collective results indicate extensive and rapid global changes in the transcription of specific genes during the differentiation of these specialized filaments.
The price of motility in Nostoc species is a requirement for the transient differentiation of nongrowing filaments called hormogonia. Hormogonia differentiate from vegetative filaments in a variety of filamentous cyanobacteria; in most of these species, hormogonia are the only motile form (23). Restriction of motility to hormogonia is a phenotypic characteristic that distinguishes members of the heterocyst-forming genera Nostoc and Anabaena, the latter of which does not differentiate hormogonia (23, 24). Essentially all laboratory studies of hormogonium differentiation have utilized heterocyst-forming species (15, 20, 29). Hormogonia are motile through a gliding mechanism, which requires association with the substratum and may involve type IV pili (11) and/or directional secretion of a polysaccharide slime (17). In heterocyst-forming genera, such as Nostoc, Calothrix, Tolypothrix, and Fischerella, the cells of hormogonium filaments are distinctly smaller and of different shape than cells of vegetative filaments (29). Hormogonium filaments lack heterocysts and are incapable of nitrogen fixation in air.
A variety of environmental changes, either positive or negative for growth, can induce the differentiation of hormogonia, thereby allowing active motility in order for the organisms to exploit a favorable, or to escape an inhospitable, microhabitat. Examples of positive signals include changes in the quantity (3) or quality (9, 25) of light, addition of iron to iron-starved cultures (10), and transfer from aged to fresh medium (15, 29). Transfer to fresh medium is most effective when it involves a large dilution factor, presumably to attenuate an autogenic differentiation-repression activity (15). Conversely, starvation for phosphate (32) or for combined nitrogen (3) and transition from suspension to surface growth (16) also induce hormogonium differentiation. All Nostoc strains that are symbiotically competent with plants differentiate hormogonia; the hormogonia function as infection units of the plant partners, and their differentiation is induced by a factor(s) produced by the plants (1, 4, 19).
Hormogonia reflect a transient nongrowth state, and their differentiation occurs as a cycle, following induction, back to a growing state (5). Upon induction of differentiation and entrance into the cycle, there is an immediate cessation of net biomass increase, including DNA replication (15), protein accumulation (5), and chlorophyll and phycobiliprotein synthesis (9) in all cells of a filament. However, cell division continues for one or two rounds, resulting in the smaller-sized cells in the hormogonium filaments. Cyanobacteria, in general, contain multiple copies of their chromosomes (14); thus, even in the case of septation in the absence of DNA replication, hormogonium cells are likely to receive at least one genome equivalent. If the filaments contain heterocysts, the filaments fragment at the junctions between the heterocysts and vegetative cells, releasing a short small-celled filament (5). Vegetative filaments lacking heterocysts appear to fragment randomly, yielding the somewhat shorter hormogonium filaments. Following filament fragmentation, hormogonia initiate gliding and remain motile for about 48 to 60 h, after which they become sessile, the end cells of the filaments begin to differentiate into heterocysts if the cultures lack combined nitrogen (N), and macromolecules and pigments start to accumulate (5, 21).
The general decline in pigment accumulation (9) and induced synthesis of gas vacuole proteins (3, 9) imply that differential gene expression occurs in hormogonia. Klint et al. (18) identified 10 proteins that were uniquely present in hormogonia. We recently defined the global transcription profiles of Nostoc punctiforme in single-time-point assays of heterocyst-containing, dinitrogen-grown cultures of akinetes 3 days following induction and hormogonia at 24 h after induction by starvation for combined N, all in reference to ammonium-grown cultures (6). The 24-h hormogonium snapshot assay remarkably revealed that 944 and 883 genes were up- and downregulated, respectively. In this report, we have defined the time courses (0.5 to 24 h) of global differential gene expression following induction of hormogonia in N. punctiforme by starvation for ammonium (N stress-induction [NSI]) and by exposure to the exudate of a symbiotic plant partner, the bryophyte hornwort Anthoceros punctatus, which contains a hormogonium-inducing factor (HIF) (5). The results establish that the signals for induction of differentiation lead to two distinctly different temporal patterns of gene expression but that a set of genes common to both induction signals can be defined. Moreover, in the NSI hormogonia only, a transcription checkpoint was apparent at the 12-h time point, at which point upregulated genes were transiently downregulated and downregulated or nondifferentially transcribed genes were transiently upregulated.
MATERIALS AND METHODS
Cultures and culture conditions.
N. punctiforme strain ATCC 29133 (PCC 73102) and its derivatives were grown under standard culture conditions (6) in the minimal salts medium of Allen and Arnon, diluted fourfold (AA/4) (2). Strain UCD 153 (capable of both hormogonium and heterocyst differentiation) was used for both of the hormogonium time course experiments. The N. punctiforme culture medium was supplemented with 2.5 mM NH4Cl and 5.0 mM morpholinepropanesulfonic acid (MOPS), pH 7.8, prior to the beginning of the experiments.
The hornwort plant partner, A. punctatus, was cultured in Hutner's medium containing ammonium nitrate as a nitrogen source under standard conditions (12). To obtain HIF, A. punctatus cultures, at a density of 168.34 g of fresh weight (± 2.44 g; n = 3), were starved for N by transfer to 700 ml of AA/4 medium without N supplementation but buffered with 5.0 mM MOPS. These cultures were incubated under standard culture conditions for 1 week. To harvest HIF, large pieces of plant tissue were manually removed under sterile conditions, and the remaining liquid was filtered through four layers of sterile cheesecloth to remove any other remaining pieces of plant tissue. N. punctiforme filaments were suspended in the HIF solution immediately upon removal of plant tissue to begin the HIF hormogonium time course experiments.
For NSI hormogonia, N. punctiforme cultures were grown under conditions identical to those described above, harvested by centrifugation at 1,000 × g for 5 min, and then suspended in fresh N-free AA/4 medium. Time points were collected, and the time course was used in these analyses only if at 24 h the entire culture differentiated motile hormogonia. Any time course in which the filaments did not differentiate into hormogonia within 24 h (delayed differentiation) or in which only a fraction of the filaments differentiated hormogonia (incomplete differentiation in which some filaments differentiate heterocysts) was not used in these analyses. Since hormogonium filaments tend to clump together during differentiation in suspension cultures, samples were suspended in separate flasks and incubated under identical conditions, with volumes relevant to the amount harvested at each time point, thus ensuring that the same amount of biomass was harvested at each time point. For consistency, both hormogonium time courses were performed with cells of the same age after inoculation and the same incubation conditions and inoculated at the same density from parental cultures of similar age and culture conditions.
Sample preparation and hybridizations.
Cells in two 50-ml volumes at a density of 2 to 3 μg of chlorophyll a per ml were harvested by centrifugation at 1,000 × g for 5 min, excess medium was removed, and the pellets were quickly frozen in liquid nitrogen, followed by long-term storage at −80°C. Cell density, RNA isolations from the frozen samples, cDNA synthesis, and dye labeling were performed exactly as in previous microarray studies (6). The N. punctiforme DNA microarray was constructed in-house using internal PCR-generated products of 6,893 open reading frames computationally predicted from the genome sequence (22) (GenBank accession numbers CP001037 to CP001042) and printed in duplicate. All microarray hybridizations were done with an automated Tecan HS4800 hybridization station at 50°C. Slides were scanned using a GenePix 3000A scanner (MDS Analytical Technologies). Northern blot hybridizations, using 20 μg of total RNA per lane, were performed as previously described (7, 28) but with RNA from a biological replicate different from that analyzed in the microarray studies.
Data analysis.
Raw data were generated by GenePix Pro, version 3.0, in laser scanning of microarray slides. Three biological replicates of each time course with reciprocal dye swaps were used in all of the analyses, thereby yielding 12 technical replicates. Data were normalized, and M values (log2 of the ratio of experimental to reference) were calculated, with T0 as a reference, using the statistical programming language R and the free source software package LIMMA GUI (linear model for microarray analysis graphical user interface) as previously described (6). Statistically significant differentially expressed genes are defined as those with calculated B values greater than 0, as previously described (6). In R, B is defined as the log-odds that a gene is differentially expressed; i.e., eB, with the percent probability calculated as (eB/1 + eB) × 100. B takes into account multiple testing, such as t and P values. Functional gene assignments were determined also as previously described (6). Compilation of data and variance statistics analysis were performed in Excel. Time course transcription patterns were analyzed using the K-means clustering algorithm from the program Genesis (27). The input file for K-means analysis was the normalized M values from each of the time points as generated by LIMMA. The raw data for the experiments in this paper, in the form of .gpr files, are accessible at http://microbiology.ucdavis.edu/faculty/jcMeeks/ under the paper title.
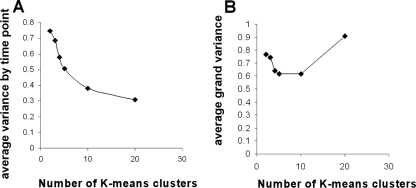
To determine how many clusters to use for further analysis, Genesis was applied to cluster the time course array data into 2, 3, 4, 5, 10, and 20 clusters. The gene list for these cluster divisions containing M values at all the time points analyzed was used for variance analysis. Average variance per time point was calculated as follows: within each cluster, the variance per time point was calculated using Excel; then an average calculated from those values for that cluster and an overall average for all of the clusters were calculated using the averages for each cluster. Average grand variance was calculated also within each cluster, but all of the datum points from all of the time points were used in the calculation for that cluster. The average variance per time point better reflects how well the data cluster at each of the time points in the analysis, whereas the grand variance is a reflection of how variable the data are over the entire time course. Figure 1A shows the average variance per time point as a function of the number of K-means clusters for the HIF-induced hormogonium time course. Analogously, Fig. 1B shows the results of grand variance as a function of the number of K-means clusters. Because each cluster has the same weight in average calculations, regardless of the number of genes within the cluster, the grand variance increase at larger numbers of clusters reflects the further delineations of clusters of genes with large changes during the time course. For example, a cluster with four gene members that vary from an M value of 1 to an M of 6 will have a higher variance than a cluster with 100 gene members that vary only from an M value of 1 to an M of 2. We chose to use 10 clusters for further analysis because, as shown in Fig. 1A, the maximum decrease in variance occurs up to that number of clusters. Variance analysis of the NSI-grown hormogonium time course showed very similar results (data not shown).
FIG. 1.
Average and grand variance of the HIF-induced cluster data. Panel A shows the calculated average variance by time point as a function of the number of clusters specified in K-means analysis. Panel B shows the average of the grand variance as a function of the number of K-means clusters.
To further determine the identity of genes that are different between the two time courses, the distribution of genes in each cluster was sorted relative to their clustering in the other time course (Table 1). Datum cells that contained 10 genes or less, or outliers, were examined in more detail on an individual basis. A gene was not considered significantly different between the two time courses if the following applied: (i) it clustered within one of the 12-h hour checkpoint clusters in the NSI-grown hormogonium time course (clusters 2, 4, 5, 6, and 7; these genes and clusters are described separately below); (ii) if the overall pattern of expression was the same between the two time courses and the 24-h time points for the two time courses are the same; (iii) if the entire time course for both is below an M value of 0; and (iv) if the largest M value is <2 in the time course.
TABLE 1.
Matrix depiction of the distribution in reciprocal clusters of differentially expressed genes from hormogonia induced by NSI and HIF
| HIF cluster no. | Total no. of genes in HIF (NSI) cluster | No. of differentially regulated genes expressed in hormogonia induced by HIF (NSI)a:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| NSI cluster no.
| |||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| 1 | 265 (86) | 27 (27) | 117 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (3) | 75 (2) | 1 (25) | 44 (9) | 0 (20) |
| 2 | 329 (380) | 0 (117) | 1 (1) | 162 (0) | 0 (0) | 90 (169) | 3 (8) | 0 (7) | 10 (67) | 1 (11) | 62 (0) |
| 3 | 270 (592) | 0 (0) | 0 (162) | 2 (2) | 49 (1) | 188 (26) | 1 (340) | 0 (0) | 4 (4) | 0 (55) | 26 (2) |
| 4 | 90 (135) | 0 (0) | 0 (0) | 1 (49) | 79 (79) | 10 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (7) |
| 5 | 244 (290) | 0 (0) | 169 (90) | 26 (188) | 0 (10) | 0 (0) | 0 (2) | 21 (0) | 16 (0) | 12 (0) | 0 (0) |
| 6 | 840 (67) | 3 (1) | 8 (3) | 340 (1) | 0 (0) | 2 (0) | 11 (11) | 1 (0) | 347 (10) | 10 (41) | 118 (0) |
| 7 | 80 (171) | 2 (75) | 7 (0) | 0 (0) | 0 (0) | 0 (21) | 0 (1) | 70 (70) | 0 (3) | 1 (1) | 0 (0) |
| 8 | 693 (3,852) | 25 (1) | 67 (10) | 4 (4) | 0 (0) | 0 (16) | 10 (347) | 3 (0) | 128 (128) | 442 (3,324) | 14 (22) |
| 9 | 3817 (738) | 9 (44) | 10 (1) | 55 (0) | 0 (0) | 0 (12) | 41 (10) | 1 (1) | 3,325 (422) | 210 (210) | 166 (18) |
| 10 | 270 (587) | 20 (0) | 0 (62) | 2 (26) | 7 (0) | 0 (0) | 0 (118) | 0 (0) | 22 (14) | 18 (166) | 201 (201) |
Boldface indicates the best correspondence. NSI and HIF clusters 8 and 9, respectively, contain genes that show no significant differential transcription; therefore, other than the reciprocal correspondence, any other apparent correspondence to these clusters is ignored. In several cases, one cluster shows correspondence to two reciprocal clusters; for examples, HIF cluster 1 corresponds to NSI clusters 2 and 7, HIF cluster 2 corresponds to NSI clusters 3 and 5, NSI cluster 2 corresponds to HIF clusters 2 and 5, NSI cluster 3 corresponds to HIF clusters 2 and 6, NSI cluster 4 corresponds to HIF clusters 3 and 4, and NSI cluster 7 corresponds to HIF clusters 1 and 7. NSI clusters 1 and 6 appear to be unique, having no correspondence to HIF clusters.
RESULTS AND DISCUSSION
Basic parameters of time course analyses.
These experiments were designed to look specifically at the expression of genes following induction of hormogonium differentiation by two different environmental signals. Both time courses begin with the stress of N deprivation; thus, heterocyst differentiation is also a possibility (20, 31). In a filament of N. punctiforme following NSI, a few vegetative cells can differentiate into heterocysts, or all can differentiate into a hormogonium filament; but the two processes are mutually exclusive for a single filament (20). Therefore, a mixed population of hormogonia or heterocyst-containing filaments can occur. However, we specifically chose to examine only cultures that differentiated 100% of their vegetative filaments into hormogonia within 24 h. In the HIF hormogonium time course, the filaments were also exposed to compounds released into the medium by N-starved A. punctatus tissue that have previously been shown to induce high levels of hormogonia in Nostoc species (5).
The K-means algorithm is used to divide the entire set of microarray data into subsets or clusters of genes that show similarities in their expression patterns over time (13). Figure 1B shows that the grand variance of HIF hormogonia increases with the number of clusters from 10 to 20. This is an indication that the cluster increases in this range involve expression patterns from genes that are highly variable over the time course. The ideal number of clusters would be achieved when, statistically, the entire data set is separated into groups of genes that have the most similar patterns of expression. As groupings of genes become more similar, variance decreases, as shown by the average variance analysis by time point. This complex data set of hormogonia differentiation, however, shows that by greatly increasing the number of clusters, there is not a corresponding gain in variance decrease. This is not the case with other, less complex data sets. For example, a similar comparison of an NSI hormogonium time course leading to heterocyst differentiation shows that grand variance continues to decrease with a greater number of clusters because the gain of clusters only divides similar sets of genes that do not change over time into further subsets of clusters (data not shown). That is, it takes fewer clusters to statistically describe the expression patterns of the heterocyst induction data set than the hormogonium induction data (data not shown). This observation is consistent with previous results that show a 3.7-fold greater number of genes that are statistically significantly differentially transcribed at the 24-h time point for hormogonia than for heterocysts (6). More powerfully, however, the clustering analyses show not only that a greater number of genes are differentially expressed during hormogonium differentiation but also that the differences in the expression patterns are more complex, with more types of patterns of expression than in heterocyst differentiation. It is clear from this analysis that the differentiation of nongrowing hormogonia, whose physiological role is in dispersion to more favorable growth habitats, is substantially more complex than the differentiation of nitrogen-fixing heterocysts, a developmental and metabolic adaptation that allows for continued growth of vegetative filaments in an N-poor habitat.
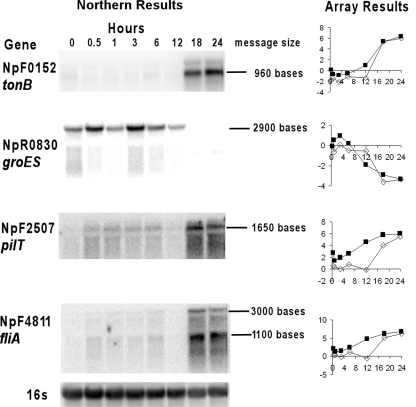
Correlation between Northern hybridization and microarray data.
Figure 2 shows the expression of genes randomly chosen from the list of statistically significant differentially transcribed genes at the 24-h time point for the NSI hormogonia. The Northern blots on the left in Fig. 2 are from a different biological replicate than the three used in the microarray results shown in the plots on the right. The results for all of the up-and downregulated genes are consistent between the Northern blot and the microarray analyses, qualitatively confirming the validity of the microarray analysis. Specifically note that the differential expression level in the Northern blots and the array results with NSI hormogonia both transiently decrease at 12 h following induction in the cases of upregulated genes NpF2507 (pilT) and NpF4811 (fliA, encoding an alternative sigma factor). Such a transition is not apparent in the HIF hormogonia. This transient change will be analyzed in a subsection below.
FIG. 2.
Comparison between Northern blot and microarray analyses of selected genes. Northern blots of RNA from NSI hormogonium cultures for the genes indicated are shown on the left side, with the corresponding transcription profiles from the microarray experiments on the right. The array results are plotted as normalized M values on the y axes and time in hours on the x axes. Open symbols are for the NSI hormogonium time course, and filled symbols are for the HIF hormogonium time course. The same blot was successively stripped and reprobed. A probe of 16S rDNA, showing relative RNA loads per lane for the blot, is given in the very bottom left panel.
A common set of genes encoding proteins for hormogonium differentiation and function and unique groupings of genes dependent on the method of induction.
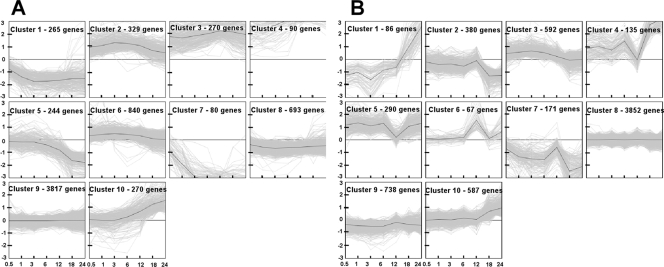
The microarray time course data for both methods of hormogonium induction that were subjected to the K-means clustering analysis with a 10-cluster limit are shown in Fig. 3. (The complete data set of M values with gene identities is given in Table S1 in the supplemental material. Cluster numbering by the Genesis algorithm is arbitrary and was not changed.) The two time courses have a cluster of many genes that show no significant change throughout the experiment: cluster 9 for the HIF hormogonium time course with 3,817 genes and cluster 8 for the NSI hormogonium time course with 3,852 genes. HIF cluster 8 and NSI cluster 9, both with members having less than a twofold change, may also be included in the list of genes whose expression does not change in a statistically robust manner. These are genes whose proteins are most likely not to be uniquely involved in hormogonium differentiation or function; they account for only 65 to 66% of the printed genome, implying that a substantial fraction (∼34%) of the genome is differentially expressed during hormogonium formation.
FIG. 3.
K-Means clustering results with a 10-cluster limit. (A) The HIF hormogonium time course. (B) The NSI hormogonium time course. The data are plotted as normalized M values on the y axes and time in hours on a nonlinear scale on the x axes. The dark line in each plot represents the average of all the genes within that cluster. The cluster numbers are assigned by the Genesis program and were not reassigned.
Somewhat similar clusters of genes are up- and downregulated following induction by both methods (Fig. 3). Genes in clusters 2, 3, and 4 are immediately, or ultimately (cluster 10), upregulated following exposure to HIF, whereas those in clusters 1 and 7 are immediately, or ultimately (cluster 5), downregulated. An analogous pattern can be seen in the NSI hormogonium time course; genes in clusters 4 and 5 are immediately, or ultimately (cluster 10), upregulated, and those in cluster 7 are immediately, or ultimately (cluster 2), downregulated. The numbers of differentially transcribed genes in the respective clusters are tabulated in correspondence to the two methods of induction in Table 1. This depiction elaborates both the similarity and differences that can be visualized in the two time courses and emphasizes the observation that NSI clusters 1 and 6 have expression patterns with no direct correspondence in the HIF clusters. The differentially upregulated HIF clusters 2, 3, 4, and 10 most strongly correspond to NSI clusters 3, 5, 4, and 10, in the order stated. However, HIF clusters 2 and 3 also show less correspondence to NSI clusters 5 and 4, respectively. The downregulated clusters show split correspondence, where HIF cluster 1 has correspondence with members of both NSI clusters 2 and 7, while HIF clusters 5 and 7 singly correspond with NSI clusters 2 and 7, respectively. Moreover, members of NSI cluster 4 show a split correspondence with HIF clusters 3 and 4, while NSI cluster 3 splits with HIF clusters 2 and 6; the temporal patterns of the latter three clusters are similar (Fig. 3), with the variation dependent on the higher average degree of upregulation in HIF cluster 2. The different numbers of differentially expressed genes in corresponding clusters, the splitting of cluster correspondence, and the occurrence of unique clusters, all imply differences in the route of hormogonium formation, dependent on the method of induction.
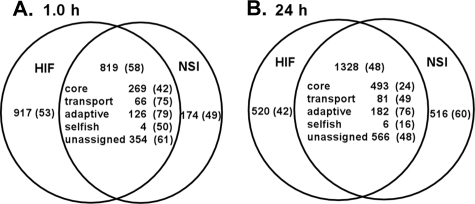
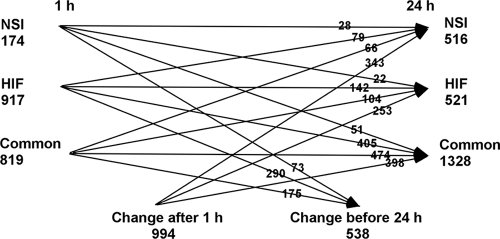
To more precisely determine the extent of the similarity and differences in differential gene expression following NSI and HIF hormogonium induction, the results of genes with LIMMA B values of >0, identifying significantly differentially expressed genes (6, 26), were extracted, compiled, and sorted in Excel. The sum total of results from this analysis are given in Fig. 4A for 1.0 h and in Fig. 4B for 24 h following induction (a complete list of functional gene assignments and the statistical parameters of their up- or downregulation at 1 and 24 h, dependent on the method of induction, is given in Tables S2 and S3 in the supplemental material). By 24 h after induction, the sum total of genes sort into three categories; there is a set of 1,328 members common to both methods of induction and two groups unique to the method of induction that are about 2.5-fold smaller than the common set. The distribution of genes in the three categories at 24 h dynamically changes throughout the time courses (Fig. 5). A common set of 819 genes can be defined by 1.0 h after induction (Fig. 4A); however, only 474 of those genes are also present in the common set by 24 h (Fig. 5). The remainder of the 24-h common set arises from three different groupings: in the first and second groups, 405 of the 917 genes unique to HIF hormogonia and 51 of the 174 genes unique to NSI hormogonia at 1.0 h transition to the 24-h common set (Fig. 5); in the third group, genes differentially transcribed late following induction (HIF clusters 5 and 10 and NSI clusters 1, 2 and 10) contribute the remaining 398 members of the 24-h common set (Fig. 5). Genes transcribed early and assigned to a unique induction category also transition to the same or the alternative induction category by 24 h (a complete list of genes that transition from the 1- to 24-h groupings is given in Table S4 in the supplemental material). The total number of statistically significant differentially transcribed genes at 24 h is 2,365. Inclusion of the 538 genes that were differentially transcribed early in a burst pattern but whose differential transcription was not apparent at 24 h (Fig. 5) yields 2,903 genes, or 42% of the printed genome, a value somewhat higher than that calculated from the cluster plots above.
FIG. 4.
Venn diagrams showing the numbers of genes differentially transcribed, uniquely and in common, in differentiating hormogonia at 1.0 h (A) and 24 h (B) following induction by HIF or NSI. Functional categories are given for the common set of genes at each time point, with the values in parentheses showing the percentages of genes upregulated.
FIG. 5.
Schematic diagram of the global changes in gene expression with time between 1 and 24 h. At the sides of the diagram the numbers of genes in the common set and of those unique to induction by HIF or NSI after 1 and 24 h of incubation, as indicated, are given. The lines with arrows show where the 1-h genes transition and the sources of the 24-h genes, with the respective numbers of genes indicated Change after 1 h refers to the differentially transcribed genes listed as present at 24 h that were not present at 1 h. Change before 24 h refers to the differentially transcribed genes listed as present at 1 h that were not present at 24 h.
All three of these 24-h groupings are of interest. We will first focus on genes of the common set. The unique NSI and HIF groupings are analyzed in the subsections below.
The common set of genes defines the potential physiology of hormogonia, irrespective of the method of induction. In this analysis, we will not describe these genes in the detailed context of their putative functional roles in metabolic pathways leading to hormogonium differentiation and function. Rather, we will address the extent and timing of change in overall functional categories. The largest common set category is of genes encoding proteins of unassigned function (Fig. 4B), and the majority of these are conserved hypothetical proteins (see Table S3 in the supplemental material). In the context of core metabolism, 76% of the 493 differentially transcribed genes are downregulated by 24 h, including those encoding proteins of the photosynthetic units and ATP synthesis. This downregulation is consistent with the nongrowth physiology of hormogonia and associated repressed metabolism. The majority of the downregulation occurs within 1.0 h following induction; this observation applies to 30% of the downregulated core metabolic genes of the common set and to 37% of the 1.0-h unique metabolic genes of the HIF- and NSI-grown hormogonia that transition into the 24-h common set. Overall, transcription of 66% of the downregulated core metabolic genes is altered within 1.0 h of induction. The most notable exception in downregulation is of genes encoding the protein synthetic machinery, which show a notable delay; the majority of protein synthesis genes is found in clusters 2 and 5 of NSI and HIF grown hormogonia, respectively (Fig. 3; see also Tables S2 and S3 in the supplemental material). This delay is consistent with the required synthesis of hormogonium-specific proteins.
A similar overall temporal pattern occurs with genes encoding transport proteins, although the numbers of transport genes up- and downregulated by 24 h are about equal. In this case, 65% and 73% of the 40 downregulated and 41 upregulated 24-h common set of transport genes, respectively, transition from the 1.0-h groupings. With respect to adaptive metabolic genes, 76% of the differentially transcribed genes are upregulated by 24 h (Fig. 4B), and 46% of those genes encode signal transduction proteins (see Table S3 in the supplemental material). In this case, 47% of the upregulated 1.0-h common set of adaptive genes and 60% of the signal transduction-encoding genes transition into the 24-h common set of adaptive metabolism (see Tables S2, S3, and S4 in the supplemental material). Including the unique 1.0-h HIF and NSI hormogonium genes that transition to the 24-h common adaptive set, transcription of 70% of the upregulated adaptive genes occurs within 1.0 h following induction. We previously noted the transcriptional enrichment of genes encoding signal transduction proteins in hormogonia (6). However, we now see that about 80% of those genes in the 24-h common set are enhanced from an NH4+-grown constitutive level (data not tabulated). This observation implies that transcription of these genes is regulated by at least two different transcriptional factors or cascades.
These data reflect a rapid transition and commitment to the hormogonium state (1 h is from 4 to 2.8% of the 24- to 36-h vegetative cell generation time). Nevertheless, altered transcription of roughly 30% of the differentially expressed 24-h common set of genes is delayed beyond 1.0 h, which we suggest is related to the regulated, sequential, transition from vegetative cell to hormogonium physiology.
Differentially transcribed genes unique to induction by NSI: the occurrence of a transcriptional checkpoint.
Induction by NSI compared to HIF yields three differences in both the patterns of differential gene expression and of specific metabolic systems: (i) there are unique transcription patterns in NSI clusters 1 and 6; (ii) there is a change in transcription patterns at 12 h following induction in NSI clusters 2, 4, 5, 6, and 7; and (iii) there is an unusual upregulation of genes for heterocyst structures and nitrogen fixation transiently at both 12 h and at 24 h (this latter observation will be elaborated in a subsequent publication).
The 86 genes in NSI cluster 1 are immediately downregulated and then strongly upregulated by 18 and 24 h after induction; a comparable pattern is not seen in HIF clusters. Slightly more than 50% (46 genes) of the genes in this cluster are present in the 24-h common set of genes. The 67 genes in NSI cluster 6 uniquely show no significant differential transcription except at the 12-h, and less so the 24-h, time points. This cluster of genes is enriched in genes that encode adaptive proteins (48%), including 18 putatively involved in synthesis of the unique heterocyst envelope, as well as three genes encoding a cytochrome oxidase specific to heterocysts (30).
The most dramatic difference in the NSI hormogonium time course from that of HIF involves clusters that show a strong transcriptional change at the 12-h time point; the specific NSI clusters are 2, 4, 5, and 7 in addition to 6 (Fig. 3). The M value of the up- (clusters 4 and 5) or downregulated (clusters 2 and 7) gene expression returns to essentially that of the reference (i.e., that of time zero) at 12 h after induction. The abrupt return to the time zero level of expression does not persist to the next time point; the results at 18 h show that the previously up- or downregulated genes return to, or exceed, their prior 6-h M values. This abrupt change in differential gene expression has the characteristics of a transcriptional checkpoint, and we will provisionally refer to it as such. The checkpoint is highly reproducible in these experiments where cultures were selected that differentiate only into hormogonia, both in the array and Northern hybridization assays (Fig. 2). There was no specific collocation on the chromosome for either the up- or downregulated genes at the 12-h checkpoint (data not shown).
Randomly chosen genes in each of the 12-h checkpoint clusters are presented in more detail in Fig. 6. In each case, except for the deviations at 12 h, the NSI and the HIF hormogonium time courses show similar patterns of transcription with comparable levels of differential expression, especially at 24 h. The chosen genes encode a variety of functions ranging from transiently downregulated pilin synthesis (NpF0069) and chemotaxis-like proteins (NpF5640) characteristic of hormogonia to a transiently upregulated secondary polyketide synthesis protein (NpR3452) with an unknown role and a photosynthetic reaction center (NpF3818) plus a signal transduction serine/threonine protein kinase (NpR1081), the latter two of which are characteristic of vegetative cell growth in limiting or excess N. The HIF patterns of transcription (Fig. 3 and 6) indicate that in these genes, the choice to differentiate into hormogonia was made early in the time course; thus, they encountered no 12-h checkpoint. We suggest that HIF is a much stronger inducing signal for hormogonium differentiation than N starvation and that, in spite of N stress, the checkpoint is overridden by an early and unequivocal commitment. This interpretation is supported by two observations: (i) the number of genes showing delayed transcriptional regulation in NSI clusters 1, 2, and 10 (1,153 genes) is twofold greater than the number in HIF clusters 5 and 10 (514 genes) (Fig. 3); (ii) the number of genes unique to HIF hormogonia at the 1.0-h time point that transition into the 24-h common set of hormogonium genes is eightfold greater than the number of genes unique to NSI hormogonia that transition (Fig. 5).
FIG. 6.
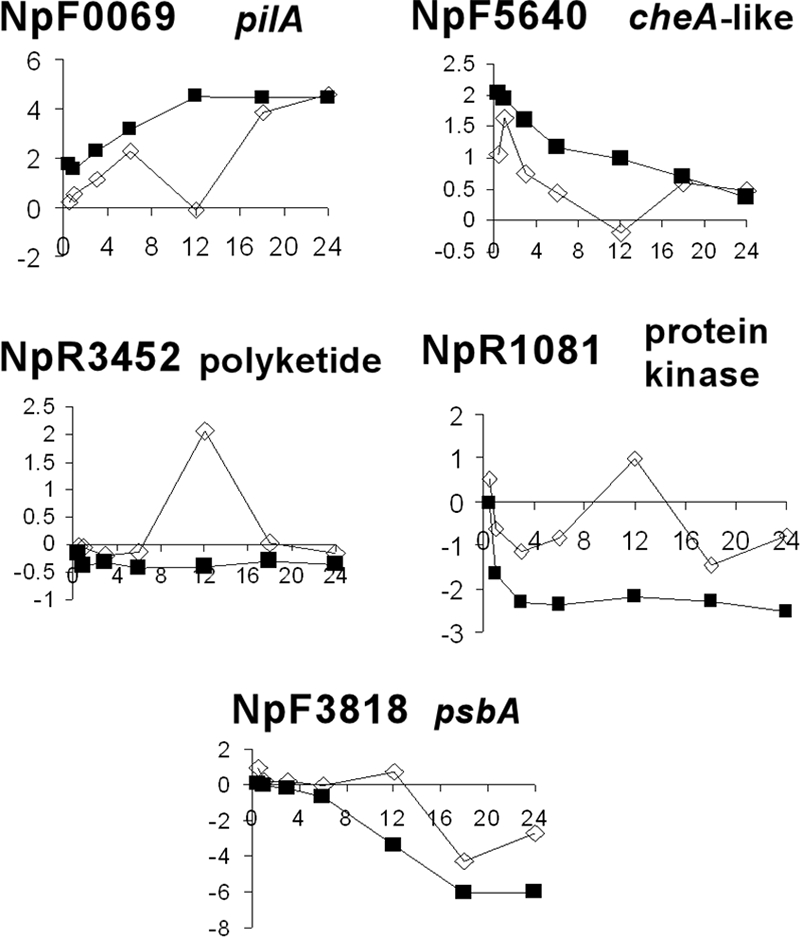
Microarray results for representative genes in 12-h checkpoint clusters for the NSI hormogonium time course compared to the HIF hormogonium time course. Array results are plotted as normalized M values on the y axes, with time in hours on the x axes. Open symbols are data from the NSI hormogonium time course; filled symbols represent data from the HIF hormogonium time course.
Table 2 summarizes the functional gene categories for the members of the 12-h checkpoint clusters. In the case of combined NSI clusters 4 and 5, which display elevated levels of expression at 0.5 and 24 h, 26% of the total downregulated checkpoint gene members encode adaptive proteins. The majority of these 112 adaptive proteins are involved in signal transduction including 16 histidine kinases, 5 serine/threonine protein kinases, 9 response regulators, 9 combined histidine and serine/threonine protein kinases, and 9 combined histidine kinase and response regulator proteins (see Table S5 in the supplemental material). The 88 core metabolic genes represent a breadth of function, with a slight enrichment in synthesis of cell envelope components. A substantial number (48% of the total) of genes that encode proteins with unassigned function are also present in these checkpoint clusters, and their physiological roles remain to be defined. For the genes downregulated during hormogonium differentiation (clusters 2 and 7), 59% of the transiently upregulated checkpoint gene products are involved in core metabolism, reflecting a return to vegetative cell growth, but there is also a marked fraction (29%) of genes encoding proteins with unassigned function and a dearth of genes encoding adaptive proteins (27% as many as upregulated genes in clusters 4 and 5).
TABLE 2.
Functional categories of genes in the 12-h checkpoint of NSI hormogonia
| Functional category | No. of genes in NSI cluster(s):
|
||
|---|---|---|---|
| 4 and 5 | 6 | 2 and 7 | |
| Core metabolism | 88 | 11 | 327 |
| Transport metabolism | 21 | 6 | 27 |
| Adaptive metabolism | 111 | 31 | 31 |
| Selfish metabolism | 0 | 1 | 4 |
| Unassigned metabolism | 205 | 18 | 162 |
| Sum of all genes | 425 | 67 | 551 |
The 12-h checkpoint in NSI hormogonia appears to mark when a “decision” is instigated on whether to transiently switch away from the sensory, but nongrowth, hormogonium state (5, 6) back to a vegetative growth state, or to differentiate into a heterocyst, or to proceed along a pattern of expression that will lead to a transcription level consistent for hormogonium formation. In the experiments reported here, the last choice was made. It is possible that the cultures that did not commit to hormogonium differentiation by 24 h and were not analyzed (see Materials and Methods) would display a different post-12-h gene expression pattern, one related to heterocyst differentiation or no differentiation. The most intriguing question derived from these results is the unknown mechanism(s) that might modulate such an extensive (1,046 genes) and nearly equivalent reciprocal up- and downregulation of transcription.
Genes uniquely induced by exposure to HIF.
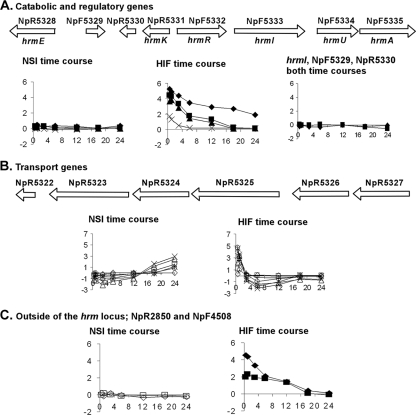
In a specific comparison of the two hormogonium induction time courses using outliers from the cluster analysis (Table 1), genes in the hrm locus are immediately apparent (7) (Fig. 7). The genes in this locus were first identified by a random transposition into hrmA, in which the mutant was more highly infective of A. punctatus than the wild type (8). Three genes (hrmIUA) encode proteins with significant similarity to those of hexuronic acid catabolism; other genes encode a transcriptional repressor (hrmR), a carbohydrate kinase (hrmK), an aldehyde dehydrogenase (hrmE), and proteins of an ABC sugar transporter (NpR5323 to NpR5327). The gene products from this locus are hypothesized to synthesize a metabolite repressor of hormogonium differentiation (7, 8). This is clearly an early response locus for HIF. All of the genes with M values of >2 show an early increase in expression upon exposure to HIF; hrmI is not expressed in either the HIF or NSI hormogonium time course. Only three of these genes, NpR5323, NpR5324, and NpR5325, encoding components of the ABC transporter, show an increase in the NSI hormogonium time course, with an M of >2, and this occurs only late during the time course. Two other genes outside of the hrm locus that also had a similar early response to HIF were found during the outlier analysis: NpR2850 (unassigned, with some similarity to an epimerase involved in antibiotic synthesis) and NpF4508 (encoding a conserved hypothetical protein). The upstream region of these genes contains sequences with similarity to the previously identified binding site of HrmR (7). However, it is not known whether HrmR also controls their transcription. Previous studies of gene expression in the hrm locus suggested that an aqueous extract of A. punctatus was required for induction of transcription (8). Based on the gene expression patterns presented here, it is now clear that the hormogonium-repressing factors are also present in the exudate containing HIF.
FIG. 7.
Transcription profiles from microarray analysis of genes in the hrm locus. Above each set of graphs in panels A and B is a schematic of the genes in the region of the chromosome, as indicated by the gene location numbers above the arrows. In panel A, where previously published, the gene designations of the catalytic and regulatory proteins are given below the gene schematic; NpF5329 and NpR5330 are conserved hypothetical genes. The transport protein-encoding genes shown in panel B have the following computational identities: sugar transporter (NpR5323), ABC permease (NpR5324), ABC ATP-binding protein (NpR5325), and sugar-binding periplasmic protein (NpR5326 and NpR5327). (C) With respect to the genes outside the hrm locus, NpR2850 (⧫) encodes a putative epimerase, and NpF4508 (▪) encodes a conserved hypothetical protein. In each plot, the y axes are normalized M values, and the x axes show time in hours.
The fact that hormogonia are induced by exposure to HIF implies that either synthesis of the repressor metabolite lags behind the induction response and the metabolite is not effective in repressing the differentiation of hormogonia once initiated or that the hrmI hexuronate isomerase gene product is essential.
Additional genes uniquely upregulated in NSI or HIF hormogonia.
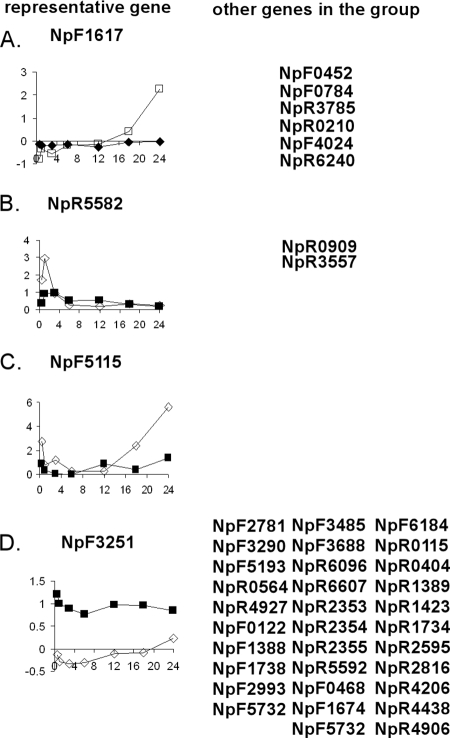
We have identified four other groups of transcriptional differences in which NSI- or HIF-dependent genes are uniquely upregulated compared to the reciprocal method of induction. They are grouped by their transcription patterns. There are 10 genes uniquely elevated during the NSI hormogonium time course, seven of which are only elevated at 24 h, represented by NpF1617, which encodes a conserved hypothetical protein (Fig. 8A), and three of which are elevated early in a burst pattern in the time course, represented by NpR5582, encoding an unassigned protein with some similarity to protochlorophyllide reductases (Fig. 8B). Only three of the late upregulated genes encode known proteins, all of which are members of gene families: a heat shock 20 protein (NpF0784), a photosystem I reaction center (NpR0210), and a transaldolase (NpF4024). The known early upregulated gene, NpR0909, encodes a putative asparaginase involved in glycoprotein cleavage. One unique gene, NpF5115, encoding a putative cell envelope adhesion protein, responds early and late in the NSI hormogonium time course (Fig. 8C). Thirty-two genes show a moderately higher (ca. twofold) overall expression during the entire HIF hormogonium time course (Fig. 8D). The members of this group fall into the functional categories of unassigned (13 members), core metabolism (8 members, mostly cell envelope), transport (4 members), and adaptive (8 members, all sensory and signal transduction). This group may represent genes uniquely involved in sensing and/or responding to plant factors.
FIG. 8.
Examples of genes with different transcription patterns in the two time courses of hormogonium differentiation. A representative gene from each group is plotted on the left in each panel, and additional members of the group are listed on the right side. Open symbols are data from the NSI hormogonium time course; filled symbols are from the HIF hormogonium time course. On the y axes are normalized M values; x axes show time in hours.
Conclusions.
The results of this study confirm that, while hormogonia do not grow, they reflect a highly dynamic transcriptional state from within less than 0.5 h after their induction through to their maturation as motile filaments. Between 34 and 42% of the 6,893 printed genes of the N. punctiforme genome are differentially transcribed during hormogonium differentiation, and about half of them are upregulated. These roughly 2,900 genes encompass at least as many genes as are present in many unicellular cyanobacteria (see http://bacteria.kazusa.or.jp/cyanobase) and are reflective of the extreme breadth of physiological potential in the cyanobacteria as a group.
We assume, but have no specific experimental evidence, that translation is largely coupled to the transcriptional patterns in hormogonia; certainly, downregulation of pigment synthesis and upregulation of gas vacuole synthesis can be seen in fluorescence and bright-field microscopy (3, 9, 21, 29). If these processes are bellwethers of the physiological state of the cells, hormogonia are much more active in protein degradation and synthesis than anticipated from time-dependent measurements of static protein content (5). Based on transcription profiles of ribosomal proteins (Fig. 3A, cluster 5), the bulk of the new protein synthesis may occur within the first 12 h following induction before declining to a new, lower level although transcription of new genes remains high at 24 h. Similarly, in the absence of an external source of nitrogen, there must be turnover of nucleic acids to fuel the synthesis of new mRNAs. The hormogonium developmental state, therefore, may be one of extensive cellular reorganization using recycled monomers. However, hormogonia have the capacity to transport specific compounds that are apparently not required in the vegetative state, as evidenced by the differential transcription of genes encoding transport proteins.
In the presence of a strong inducer, such as HIF, commitment to the hormogonium developmental state is rapid and unequivocal; within 0.5 h after induction 345 genes are downregulated, and 689 are upregulated. These results imply that the transcriptional regulatory factors are either present in the vegetative cells prior to induction of differentiation or very rapidly synthesized. Overall, in HIF and NSI grown hormogonia, 855 and 993 genes, respectively, are upregulated, and 946 and 898 genes, respectively, are downregulated by 24 h. In NSI hormogonia, the checkpoint phenomenon results in the transient and reciprocal up- and downregulation of 618 and 425 genes, respectively. The fact that the checkpoint genes are not collocated in the genome implies that large-scale, regional changes in chromosome or plasmid morphology are not likely to play a major role in their specific differential transcription. All of these results reflect extensive global changes in gene transcription, the mechanisms of which have yet to be investigated.
Supplementary Material
Acknowledgments
This work was supported by grant EF-0317104 from the NSF and page charges by IOS-0822008.
We thank Sydney Kustu for advice and encouragement in cluster analysis.
Footnotes
Published ahead of print on 12 September 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adams, D. G. 2000. Symbiotic interactions, p. 523-561. In B. A. Whitton and M. Potts (ed.), The ecology of cyanobacteria, their diversity in time and space. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 2.Allen, M. B., and D. I. Arnon. 1955. Studies on nitrogen-fixing blue-green algae. I. Growth and nitrogen fixation by Anabaena cylindrica Lemm. Plant Physiol. 30366-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong, R. E., P. K. Hayes, and A. E. Walsby. 1983. Gas vacuole formation in hormogonia of Nostoc muscorum. J. Gen. Microbiol. 128263-270. [Google Scholar]
- 4.Bergman, B. 2002. The Nostoc-Gunnera symbiosis, p. 207-232. In A. Rai, B. Bergman, and U. Rasmussen (ed.), Cyanobacteria in symbiosis. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 5.Campbell, E. L., and J. C. Meeks. 1989. Characteristics of hormogonia formation by symbiotic Nostoc spp. in response to the presence of Anthoceros punctatus or its extracellular products. Appl. Environ. Microbiol. 55125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, E. L., M. L. Summers, H. Christman, M. E. Martin, and J. C. Meeks. 2007. Global gene expression patterns of Nostoc punctiforme in steady-state dinitrogen-grown heterocyst-containing cultures and at single time points during the differentiation of akinetes and hormogonia. J. Bacteriol. 1895247-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, E. L., F. C. Y. Wong, and J. C. Meeks. 2003. DNA binding properties of the HrmR protein of Nostoc punctiforme responsible for transcriptional regulation of genes involved in differentiation of hormogonia. Mol. Microbiol. 47573-582. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, M. F., and J. C. Meeks. 1997. A hormogonium regulating locus, hrmUA, of the cyanobacterium Nostoc punctiforme strain ATCC 29133 and its response to an extract of a symbiotic plant partner Anthoceros punctatus. Mol. Plant-Microbe Interact. 10280-289. [DOI] [PubMed] [Google Scholar]
- 9.Damerval, T., G. Guglielmi, J. Houmard, and N. Tandeau de Marsac. 1991. Hormogonium differentiation in the cyanobacterium Calothrix: a photoregulated developmental process. Plant Cell 3191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglas, D., A. Peat, B. A. Whitton, and P. Wood. 1987. Influence of iron status on structure of the cyanobacterium (blue-green alga) Calothrix parientina. Cytobiosis 47155-165. [Google Scholar]
- 11.Duggan, P. S., P. Gottardello, and D. G. Adams. 2007. Molecular analysis of genes in Nostoc punctiforme involved in pilus biogenesis and plant infection. J. Bacteriol. 1984547-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enderlin, C. S., and J. C. Meeks. 1983. Pure culture and reconstitution of the Anthoceros-Nostoc symbiotic association. Planta 158157-165. [DOI] [PubMed] [Google Scholar]
- 13.Gyaneshwar, P., O. Paliy, J. McAuliffe, D. L. Popham, M. I. Jordan, and S. Kustu. 2005. Sulfur and nitrogen limitation in Escherichia coli K-12: specific homeostatic responses. J. Bacteriol. 1871074-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herdmann, M., M. Janvier, R. Rippka, and R. Y. Stanier. 1979. Genome size in cyanobacteria. J. Gen. Microbiol. 11173-85. [Google Scholar]
- 15.Herdmann, M., and R. Rippka. 1988. Cellular differentiation: hormogonia and baeocytes. Methods Enzymol. 167232-242. [Google Scholar]
- 16.Hernandez-Muniz, W., and S. E. Stevens, Jr. 1987. Characteristics of the motile hormogonia of Mastigocladus laminosus. J. Bacteriol. 169218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoiczyk, E., and W. Baumeister. 1998. The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria. Curr. Biol. 81161-1168. [DOI] [PubMed] [Google Scholar]
- 18.Klint, J., L. Ran, U. Rasmussen, and B. Bergman. 2006. Identification of developmentally regulated proteins in cyanobacterial hormogonia using a proteomic approach. Symbiosis 4187-95. [Google Scholar]
- 19.Meeks, J. C. 1998. Symbiosis between nitrogen-fixing cyanobacteria and plants. BioScience 48266-276. [Google Scholar]
- 20.Meeks, J. C., E. L. Campbell, M. L. Summers, and F. C. Wong. 2002. Cellular differentiation in the cyanobacterium Nostoc punctiforme. Arch. Microbiol. 178395-403. [DOI] [PubMed] [Google Scholar]
- 21.Meeks, J. C., and J. Elhai. 2002. Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associated symbiotic growth states. Microbiol. Mol. Biol. Rev. 6694-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meeks, J. C., J. Elhai, T. Thiel, M. Potts, F. Larimer, J. Lamerdin, P. Predki, and R. Atlas. 2001. An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium. Photosyn. Res. 7085-106. [DOI] [PubMed] [Google Scholar]
- 23.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdmann, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbial. 1111-61. [Google Scholar]
- 24.Rippka, R., and M. Herdman. 1992. Pasteur culture collection of cyanobacteria in axenic culture. Institute Pasteur, Paris, France.
- 25.Robinson, B. L., and J. H. Miller. 1970. Photomorphogenesis in the blue-green alga Nostoc commune 584. Physiol. Plant 23461-472. [Google Scholar]
- 26.Smyth, G. K. 2005. Limma: linear models for microarray data, p. 397-420. In R. Gentleman, V. Carry, S. Dudoit, R. Irizarry, and W. Huber (ed.), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY.
- 27.Sturn, A., J. Quackenbush, and Z. Trajanoski. 2002. Genesis: cluster analysis of microarray data. Bioinformatics 18207-208. [DOI] [PubMed] [Google Scholar]
- 28.Summers, M. L., J. G. Wallis, E. L. Campbell, and J. C. Meeks. 1995. Genetic evidence of a major role for glucose-6-phosphate dehydrogenase in nitrogen fixation and dark growth of the cyanobacterium Nostoc sp. strain ATCC 29133. J. Bacteriol. 1776184-6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tandeau de Marsac, N. 1994. Differentiation of hormogonia and relationships with other biological processes, p. 825-842. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Boston, MA.
- 30.Valladares, A., A. Herrero, D. Pils, G. Schmetterer, and E. Flores. 2003. Cytochrome c oxidase genes required for nitrogenase activity and diazotrophic growth in Anabaena sp. PCC 7120. Mol. Microbiol. 471239-1249. [DOI] [PubMed] [Google Scholar]
- 31.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Boston, MA.
- 32.Wood, P., A. Peat, and B. A. Whitton. 1986. Influence of phosphorus status on fine structure of the cyanobacterium (blue-green alga) Calothrix parientina. Cytobiosis 4789-99. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.