Abstract
Transcription-induced hypernegative supercoiling is a hallmark of Escherichia coli topoisomerase I (topA) mutants. However, its physiological significance has remained unclear. Temperature downshift of a mutant yielded transient growth arrest and a parallel increase in hypernegative supercoiling that was more severe with lower temperature. Both properties were alleviated by overexpression of RNase HI. While ribosomes in extracts showed normal activity when obtained during growth arrest, mRNA on ribosomes was reduced for fis and shorter for crp, polysomes were much less abundant relative to monosomes, and protein synthesis rate dropped, as did the ratio of large to small proteins. Altered processing and degradation of lacA and fis mRNA was also observed. These data are consistent with truncation of mRNA during growth arrest. These effects were not affected by a mutation in the gene encoding RNase E, indicating that this endonuclease is not involved in the abnormal mRNA processing. They were also unaffected by spectinomycin, an inhibitor of protein synthesis, which argued against induction of RNase activity. In vitro transcription revealed that R-loop formation is more extensive on hypernegatively supercoiled templates. These results allow us, for the first time, to present a model by which hypernegative supercoiling inhibits growth. In this model, the introduction of hypernegative supercoiling by gyrase facilitates degradation of nascent RNA; overproduction of RNase HI limits the accumulation of hypernegative supercoiling, thereby preventing extensive RNA degradation.
DNA supercoiling is tightly regulated by opposing enzymatic activities in bacterial cells. DNA gyrase, encoded by gyrA and gyrB, introduces (12, 13) and maintains (8) negative supercoiling, while topoisomerase I (topA) and topoisomerase IV (parC and parE) remove excess negative supercoiling (37, 41, 49). Too much negative supercoiling can be detrimental to cell growth, as initially revealed by genetic studies in which topA-null mutants of Escherichia coli were found to carry compensatory gyrA or gyrB mutations that reduced chromosomal supercoiling (7, 37). Plasmid DNA extracted from these topA-null mutants had very high levels of negative supercoiling (35). pBR322 topoisomers that could not be resolved by conventional agarose gel electrophoresis in the presence of chloroquine, a DNA-intercalating dye, were described by the term “hypernegative supercoiling” (35). Such topoisomers had a superhelical density that was more than twice the average seen with wild-type cells (35).
Hypernegative supercoiling was attributed to transcription of the tetA gene on pBR322 (36) and was explained by the twin-domain model in which a moving transcription complex generates negative supercoiling behind the complex and positive supercoiling in front of the complex (22). By removing the positive supercoils in the absence of topoisomerase I, DNA gyrase generates hypernegatively supercoiled DNA. Hypernegative supercoiling is particularly evident in plasmids carrying genes encoding membrane-bound proteins such as TetA (23, 25), presumably because coupled transcription-translation and membrane anchorage of nascent protein restrict DNA rotation. A requirement for membrane anchorage is bypassed if coupled transcription-translation originates from very strong promoters, such as the hybrid Tac promoter (40) or, as shown more recently, from a promoter recognized by T7 RNA polymerase (38). In each of these cases, the physiological significance of hypernegative supercoiling is unclear, since no phenotype is attributed to it. More importantly, hypernegative supercoiling is observed under conditions permissive for the growth of topA-null mutants.
In contrast, when cells are downshifted from 37 to 28°C and below, hypernegative supercoiling in topA-null mutants correlates with growth inhibition (28, 30). This is most evident when topA-null cells also carry a gyrB(Ts) allele that allows gyrase reactivation after temperature downshift (28, 30). In this situation, hypernegative supercoiling depends on transcription, but not translation. Moreover, it is not restricted to genes expressed from very strong promoters. Instead, downshift-based hypernegative supercoiling is related to R-loop formation coupled to gyrase activity (10, 29, 33). Overproduction of RNase HI, an enzyme that degrades the RNA of R-loops, compensates for the growth defects of topA-null mutants and reduces hypernegative supercoiling associated with temperature downshifts (11).
We recently used topA-null gyrB(Ts) strains to study the physiological consequences of temperature downshifts in the absence of topoisomerase I (2). Transient growth inhibition correlated with both a reduction in the rate of protein synthesis and with accumulation of truncated RNAs at the expense of normal full-length RNAs, phenotypes that were corrected by RNase HI overproduction (2). Since supercoiling measurements were not included in this work, whether and how these phenotypes are related to hypernegative supercoiling is unknown. The effect of RNase HI overproduction was explained in the context of local transcription-induced negative supercoiling without considering the global supercoiling level during the temperature downshift. We proposed that transcription-induced negative supercoiling promoted R-loop formation. The R-loops, when not removed rapidly by RNase HI, might act as roadblocks for subsequent transcribing RNA polymerases, thereby leading to the accumulation of shorter than full-length RNAs (2). According to this model, RNase HI overproduction, by continuously removing R-loops, would lead to substantial RNA degradation, which was not the case (2). Moreover, R-loops might turn over too rapidly to act as roadblocks for the majority of genes, since most E. coli genes are transcribed at low rates (45). Thus, an alternate explanation is required.
The present study showed that RNA degradation associated with hypernegative supercoiling was probably responsible for the growth inhibition of topA-null mutants. By limiting the accumulation of hypernegative supercoiling, RNase HI overproduction prevented RNA degradation. When the level of RNase HI was low, as in wild-type cells, hypernegative supercoiling accumulated, which led to extensive R-loop formation and RNA degradation. Thus, cells unable to keep supercoiling from reaching unacceptably high levels are compromised in the ability to grow due to extensive RNA degradation.
MATERIALS AND METHODS
E. coli strains and plasmids.
E. coli strains used in the present study are described in Table 1. The rneΔ14 derivatives of RFM445 and RFM475 were constructed by transduction with P1vir phage (32) grown on AC28 (MC1061, zce-726::Tn10 rneΔ14 [aaΔ636-845], obtained from A. J. Carpousis [19]). Tetracycline-resistant transductants were selected, and the presence of the rneΔ14 mutation was confirmed by PCR using the RNE1 d(CTGGAAATGTCCCGTCAG) and RNE2 d(CACTTCCGGTTGCGGTTC) oligonucleotides. The chromosomal lac fusion was obtained by using the system previously described by Simons et al. (39). pMD217 has been previously described (31). This pTrc99a derivatives contain a boxA antitermination regulatory sequence inserted immediately downstream of the IPTG-inducible Ptrc promoter. Oligonucleotides d(GCATAGATCTGCATTTACGTTGACACC) and d(CTGAAGATCTATCCGCCAAAACAGCC) were used in a PCR (Pfu polymerase from Stratagene) with pMD217 to amplify a DNA segment located between nucleotides 2982 and 327 of the pTrc99a sequence. This region also includes the lacIq gene. The PCR product was digested with BglII (BglII sites are present in both oligonucleotides) and cloned in pRS550 digested with BamHI to place the lac operon of pRS550 under the control of the Ptrc promoter. Sequencing was performed to confirm the integrity of the fusion. The fusion was transferred to the chromosome by using λRS45 phage as described previously (39). Single-copy integrations were confirmed by PCR as previously performed (34). pSK760 is a pBR322 derivative carrying the wild-type rnhA gene (11). pEM001 and pEM003 are pACYC184 derivatives that carry, respectively, the wild-type rnhA gene and a mutated and inactive version of this gene (31). pMD306 is a pMD217 derivative carrying the 567-bp HindIII fragment from the rrnB operon downstream of the Ptrc promoter (31).
TABLE 1.
E. coli strains used in this studya
| Strain | Genotype or description | Source or reference |
|---|---|---|
| MA249 | ilv metB his-29 trpA9605 pro thyA deoB (or deoC) gyrB221(Cour) gyrB203(Ts) zie-3163::Tn10kan | 14 |
| MA251 | ilv metB his-29 trpA9605 pro thyA deoB (or deoC) gyrB221(Cour) gyrB203(Ts) zie-3163::Tn10kan topA20::Tn10 | 14 |
| IB34 | MA251(pSK760) | This study |
| RFM445 | rpsL galK2 gyrB221(Cour) gyrB203(Ts) Δlac74 | 11 |
| RFM475 | rpsL galK2 gyrB221(Cour) gyrB203(Ts) Δ(topA cysB)204 Δlac74 | 11 |
| VU4 | RFM475(pMD306) | This study |
| VU7 | VU4(pEM001) | This study |
| VU8 | VU4(pEM003) | This study |
| PS63 | RFM475 φ(lacIq-Ptrc-lac) | This study |
| PS64 | RFM445 φ(lacIq-Ptrc-lac) | This study |
| PS66 | PS63(pEM001) | This study |
| PS68 | PS63(pEM003) | This study |
| PS70 | PS64(pEM001) | This study |
| PS72 | PS64(pEM003) | This study |
| PS123 | PS63 rneΔ\14 | This study |
| PS126 | PS64 rneΔ14 | This study |
Cour, coumermycin resistance.
Media and growth conditions.
Cells were grown in VB Casa (Vogel-Bonner) minimal medium (11) supplemented with Casamino Acids (0.1%), glucose (0.2%), required amino acids (0.5%), thiamine (5 μg/ml), and thymine (10 μg/ml). When needed, kanamycin was added to 25 μg/ml, chloramphenicol was added to 30 μg/ml, tetracycline was added to 10 μg/ml, and ampicillin was added to 50 μg/ml.
Preparation of S30 extracts and in vitro translation experiments.
S30 extracts from cells grown at 37°C and transferred or not to 28°C for 20 min were prepared as described previously (27), divided into aliquots, and stored at −80°C. In vitro translation experiments using the S30 extracts were performed essentially according to a protocol from Promega (technical bulletin TB219). The equivalent of an A260 of 0.6 of S30 extracts were used in the in vitro translation reactions with either 4 μg of phage MS2 RNA template (from Roche Applied Sciences) or 10 μg of poly(U) RNA template (from GE Healthcare). When MS2 mRNA template was used, the reaction contained 20 μl of the S30 premix solution without amino acids (Promega), 15 μCi (1 μl) of l-[35S]methionine (GE Healthcare), and 5 μl of the amino acids minus methionine mix (Promega). When poly(U) RNA template was used, the reaction contained 20 μl of the S30 premix solution without amino acids (Promega) and 0.25 μCi (5 μl) of l-[14C]phenylalanine (GE Healthcare). To measure the translation background activity in the S30 extracts, MS2 mRNA and poly(U) RNA were omitted in some reactions. After 1 h of incubation at either 37°C or 28°C, 3 ml of NaOH (1 M) was added, and the reactions were incubated on ice for 10 min. The reaction products were precipitated by adding 10 ml of an ice-cold solution containing 25% trichloroacetic acid (TCA) and 2% Casamino Acids and by incubating the tubes on ice for 30 min. The samples were deposited on 25-mm GF/B glass microfiber filters (Whatman), washed three times with 5% TCA and twice with 95% ethanol by using the Millipore 1225 sampling manifold system (Millipore). The filters were then dried at room temperature for 10 min. Filters were transferred to vials containing 10 ml of Cytoscint (MP Biomedicals), and the l-[35S]methionine or l-[14C]phenylalanine incorporation was measured in a scintillation counter (Beckman LS 6000 SC).
Sucrose density gradient fractionation of ribosomes and mRNA analysis.
Cells were grown to log phase (optical density at 600 nm [OD600] of ca. 0.5 to 0.6) at 37°C and transferred to 28°C for 20 min. After exposure to 400 μg of chloramphenicol for 2 min, the cells were chilled briefly on ice and pelleted by centrifugation at 4,000 rpm for 20 min at 4°C (Beckman J2-MI, JA-14 rotor). They were then resuspended in 10 ml of lysis buffer without lysozyme (25 mM Tris-HCl [pH 7.6], 60 mM KCl, 10 mM MgCl2, 20% sucrose), centrifuged at 4,000 rpm for 10 min at 4°C, resuspended in 500 μl of lysis buffer containing lysozyme (5 mg/ml), and stored at −80°C. The cells were lysed by three rounds of freeze-thawing. Then, 100 μl of 1% sodium deoxycholate, 50 μl of 6% Brij-58, and 20 U of RNase free DNase I (Roche Applied Sciences) were added to the lysates before they were incubated on ice for 10 min. The lysates were clarified by centrifugation at 13,000 rpm for 15 min at 4°C. The equivalent of 35 OD260 units were layered on 10 to 40% sucrose gradients made in 20 mM Tris-HCl (pH 7.8)-10 mM MgCl2-100 mM NH4Cl-2 mM dithiothreitol, and the gradients were centrifuged in a Beckman SW41 rotor at 35,000 rpm for 3 h at 4°C. After centrifugation the gradients were connected to a density gradient fractionation system (ISCO), fractionated, and recorded using an ISCO UA-6 absorbance detector.
The gradients were fractionated into 1-ml fractions from which RNAs were recovered by first adding 3 volumes of 100% ethanol. After 1 h at −80°C, RNAs were pelleted by centrifugation at 13,000 rpm for 15 min. The pellets were dried and resuspended in 50 μl of RNase-free deionized water. A portion (300 μl) of a solution composed of 1.2% sodium dodecyl sulfate (SDS), 2.3 mM EDTA, 46.6 mM sodium acetate (pH 5.5), and 100 μg of proteinase K (Roche Applied Sciences) was then added before the mixtures were incubated for 30 min at 65°C. RNAs from the mixtures were obtained by means of phenol chloroform (1:1) extraction and ethanol precipitation. Each RNA preparation was dissolved in 20 μl of RNase-free deionized water and stored at −80°C. Northern blots were performed as described previously (14). The membranes were probed with the appropriate random prime-labeled DNA fragments obtained by PCR by using the oligonucleotides d(GGACGTCACATTACCGTGC) and d(GCACGTACCCATGCGCTACG) for crp, the oligonucleotides d(GGCTGGCATTACAGACAG) and d(CTCTTCGCAGTTACGGTG) for yhdG, and the oligonucleotides d(GAGCTGACAGAACTATGT) and d(CGCAGCGTACCACGGTTG) for fis. Bands were detected by using a Phosphorimager Typhoon 9400 (GE Healthcare) with the ImageQuant image analysis software (Molecular Dynamics).
Time course of l-[35S]cysteine incorporation.
Cells were grown to log phase (OD600 of ca. 0.5 to 0.6) at 37°C and transferred to 28°C. After 20 min of incubation, 10 μCi of l-[35S]cysteine (GE Healthcare)/ml was added to the cells. Aliquots of the cells (500 μl) were recovered at various times as indicated and mixed with the same volume of cold nonradioactive cysteine (0.5%). After 10 min on ice the cells were recovered by centrifugation and resuspended in SDS loading buffer for lysis by boiling. The proteins were separated by SDS-polyacrylamide (10%) gel electrophoresis and transferred to a nitrocellulose membrane (Hybond-ECL; GE Healthcare). Bands were detected and quantified by using a Phosphorimager Typhoon 9400 with ImageQuant image analysis software.
Plasmid extraction for supercoiling analysis.
Cells were grown overnight at 37°C and diluted to an OD600 of 0.06 in prewarmed medium. They were grown to an OD600 of ∼0.5, at which time an aliquot of cells was recovered for plasmid extraction, while the remaining culture was transferred to the indicated temperature. Aliquots of cells were recovered for plasmid extractions at various times as indicated. Growth was arrested by transferring cells into a tube filled with ice, thus immediately lowering the temperature of the culture to 0°C. Plasmid DNAs were extracted by alkaline lysis as previously described (31).
Plasmid topoisomer analysis.
Agarose gel electrophoresis in the presence of chloroquine was performed in 0.5× Tris-borate-EDTA as described previously (31). After electrophoresis, the gels were dried and prepared for in situ hybridization (31) with a random prime-labeled probe corresponding to the bla gene of pMD306. Band densities of topoisomers were quantified by using a Phosphorimager Typhoon 9400 with ImageQuant image analysis software.
RNA extraction from cell cultures.
RNA extraction from growing cells was performed by using TRIzol reagent (Invitrogen) as described previously (2). Northern blots of RNA samples separated by electrophoresis in 1.2% agarose gels containing formaldehyde were performed as described previously (14). End-labeled FIS 3′ oligonucleotide (2) was used to detect fis mRNA (see Fig. 5). lacZ mRNA was detected by using a random prime-labeled probe obtained by PCR using the oligonucleotides LACPCR5 [d(CCGTCATAGCGATAACG)] and LACPCR3 [d(GCTGTTGACTGTAGCGG)]. lacA mRNA was detected by using a random prime-labeled probe obtained by PCR using the oligonucleotides LACA5′ [d(GATCGCATTGAACATGCC)] and LACA3′ [d(CCGGTCGTTTATTTCGCG)]. Bands were detected by using a Phosphorimager Typhoon 9400 with ImageQuant image analysis software.
FIG. 5.
Expression and processing of fis mRNAs after a temperature downshift. Cells were grown at 37°C to log phase as indicated in Materials and Methods before being transferred to 28°C. After 20 min, rifampin at 250 μg/ml (final concentration) was added to the cells, and the RNA was extracted at the indicated time (for time zero the RNA was extracted immediately before the addition of rifampin). Portions (10 μg) of the RNA samples were used for Northern blot analysis with an oligonucleotide probe hybridizing to fis as described in Materials and Methods. The strains used were MA249 [gyrB(Ts)], MA251 [gyrB(Ts) topA], and IB34 [gyrB(Ts) topA/pSK760]. + RNase HI indicates that RNase HI was overproduced (pSK760).
R-loop formation in vitro.
pJP461 (33) and pTW120 (48) were purified from DH5α cells by using a Plasmid Midi Kit (Qiagen). Hypernegatively supercoiled templates were prepared by transcription in the presence of DNA gyrase (John Innes Enterprises, Ltd.) as described previously (33). After electrophoresis in agarose gel containing 7.5 μg of chloroquine/ml, we were able to estimate that more than 90% of the topoisomers were hypernegatively supercoiled (migration at the bottom of the gel as unresolved topoisomers). Transcription reactions (25 μl) in 40 mM Tris-HCl (pH 8.0), 15 mM MgCl2, 50 mM dithiothreitol, and 0.5 mg of bovine serum albumin/ml were performed in the presence of 20 U of T3 or T7 RNA polymerase (USB), 0.5 μg of template DNA, a 0.4 mM concentration of each NTP, and 2 μCi of both [32P]ATP and [32P]UTP (3,000 Ci/mmol; GE Healthcare). When indicated, RNase HI (20 ng) was added. Transcription reactions were incubated at 37°C for 15 min. They were stopped by adding EDTA to a 20 mM final concentration. Samples were treated with 250 ng of RNase A for 1 h at 37°C. After phenol extraction, the samples were loaded on agarose gels (1%) in Tris-borate-EDTA buffer for electrophoresis. After staining with ethidium bromide and photography under UV light, gels were dried and exposed to a storage phosphor screen, and the RNA bands were revealed by using a Phosphorimager Typhoon 9400 with ImageQuant image analysis software.
RESULTS
The growth rate of a topA-null gyrB(Ts) mutant at low temperature correlates with restricted accumulation of hypernegatively supercoiled DNA.
To better understand the physiology of topA-null cells after temperature downshifts, supercoiling levels must be measured at regular intervals during and after the transient growth inhibition. Indeed, still unknown is whether the eventual growth resumption of topA-null gyrB(Ts) cells accompanied by the accumulation of full-length RNAs (2) is related to changes in supercoiling levels. This question is also worth addressing considering the fact that GyrB proteins should remain active throughout incubation at low temperatures after the downshift. It is therefore possible that hypernegative supercoiling persists following the downshift, thus suggesting that it is not directly related to growth inhibition. On the other hand, if hypernegative supercoiling is directly involved in growth inhibition, it should be relaxed when growth resumes.
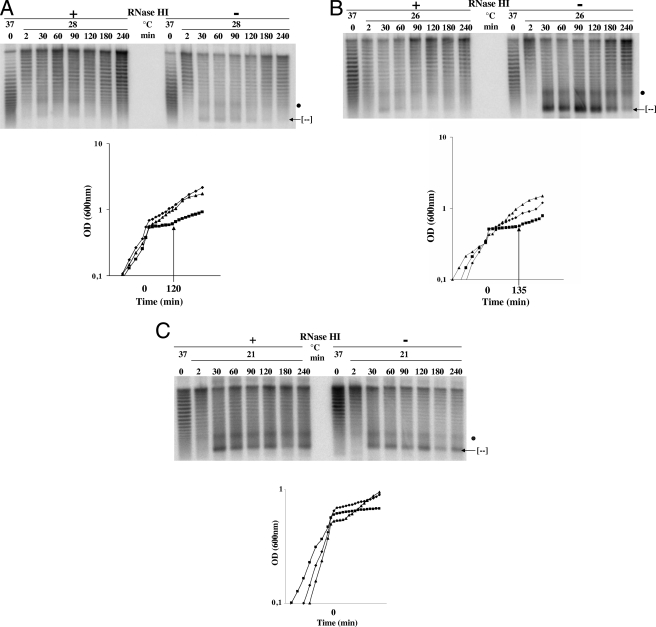
We sought to determine whether transient growth inhibition is related to hypernegative supercoiling by comparing two ΔtopA gyrB (Ts) strains, VU7 and VU8, after a shift from the permissive temperature of 37 to 28°C. Strain VU7 expresses RNase HI from plasmid pEM001; VU8 carries the vector pEM003 that has a mutated, inactive rnhA gene. As a supercoil reporter, both strains carry pMD306 (pMD306 contains the lacIq gene, which stimulates R-loop-dependent hypernegative supercoiling [4]). Cell growth was monitored, and plasmid DNA was extracted at various times before and after temperature downshift. To observe hypernegative supercoiling, DNA samples were subjected to agarose gel electrophoresis in the presence of the DNA intercalator chloroquine. Topoisomers of pMD306 were then detected by hybridization with a specific probe. Temperature downshift caused a transient growth arrest (Fig. 1A) and transient accumulation of hypernegative supercoils when RNase HI was not overproduced (strain VU8). Growth resumed 2 h after the shift, and the level of hypernegatively supercoiled DNA dropped. Overproduction of RNase HI (strain VU7) prevented significant accumulation of hypernegatively supercoiled DNA and growth inhibition (Fig. 1A).
FIG. 1.
Correlation between hypernegative supercoiling and growth inhibition after temperature downshifts. Cells were grown at 37°C to log phase as indicated in Materials and Methods before being transferred to 28°C (A), 26°C (B), or 21°C (C). Plasmid DNA from the topA-null strains was extracted just before the temperature downshift (time zero) and at the indicated times after the downshift. DNA was loaded on an agarose gel with 7.5 μg of chloroquine/ml and probed to reveal hypernegatively supercoiled pMD306 (top panels). Note that at this chloroquine concentration, the more relaxed topoisomers migrate faster, except hypernegatively supercoiled DNA, which also migrates rapidly (indicated by [-]). The dot points to a signal reflecting a negative supercoiling-dependent structural transition that was previously observed for this plasmid following two-dimensional gel analysis (4, 31). On the top of the gels, “+” and “-” indicate, respectively, that RNase HI was overproduced (pEM001) or not overproduced (pEM003). The arrow on the growth curves (bottom) indicates the time when growth resumed following the downshifts. Symbols: ⧫, VU7 [gyrB(Ts) topA/pMD306 and pEM001]; ▪, VU8 [gyrB(Ts) topA/pMD306 and pEM003]; ▴, RFM445 [gyrB(Ts)].
A more severe shift to 26°C led to a greater accumulation of hypernegative supercoiling. The growth inhibition was similar to that observed at 28°C but with a slightly longer lag (Fig. 1B, RNase HI not overproduced). As seen with the shift to 28°C, resumption of growth was accompanied by relaxation of hypernegatively supercoiled DNA. At 26°C, overproduction of RNase HI failed to completely eliminate excess negative supercoiling, and the growth rate of the topA-null strain (diamonds) was slightly lower than that of strain RFM445 (triangles), the isogenic topA+ gyrB(Ts) derivative.
In an even more severe shift to 21°C, growth of the topA-null strain not overproducing RNase HI (VU8) failed to resume, and the high level of hypernegatively supercoiled DNA persisted (Fig. 1C). In this case, overproduction of RNase HI (VU7) slightly reduced the proportion of hypernegatively supercoiled topoisomers, and cells grew, albeit at a significantly lower rate than seen with the isogenic topA+ gyrB(Ts) strain (Fig. 1C). Thus, the transient inability of downshifted topA-null gyrB(Ts) cells to grow and accumulate full-length RNAs was related to hypernegative supercoiling. The ability of RNase HI to correct these problems was linked to its capacity to restrain hypernegative supercoiling.
Failure to accumulate full-length RNAs during growth inhibition of topA-null gyrB(Ts) mutants at low temperature.
We previously showed that rRNA synthesis can be defective in topA-null gyrB(Ts) mutants (14). Since rRNA transcription and ribosome biogenesis are linked (5, 20), defective ribosomes could render RNA susceptible to RNases, thus leading to RNA degradation (17). We examined the possibility that ribosomes of topA-null gyrB(Ts) mutants were defective by measuring protein synthesis in S30 extracts from both topA-null gyrB(Ts) and isogenic topA+ gyrB(Ts) cells. Cells were grown to log phase at 37°C, and half of each culture was transferred to 28°C. Translation initiation and elongation in S30 extracts were measured by monitoring either l-[35S]methionine or l-[14C]phenylalanine incorporation with phage MS2 RNA or poly(U) RNA, respectively, as templates. No significant difference was seen between the strains irrespective of temperature (data not shown). We conclude that ribosomes of topA-null gyrB(Ts) cells are fully functional.
To further examine RNA features associated with growth inhibition, we examined yhdG-fis and crp mRNA (2). Previous work with whole-cell extracts showed that the yhdG-fis operon was overexpressed in the topA-null gyrB(Ts) mutant at the low, nonpermissive temperature and that RNAs corresponding to the proximal yhdG gene accumulated, whereas RNAs from the distal fis gene were barely detectable (2). In the topA+ strain or in the topA-null mutant overproducing RNase HI, yhdG-fis operon expression was lower, but RNA corresponding to the distal fis gene was more abundant than RNA from the proximal yhdG gene (2).
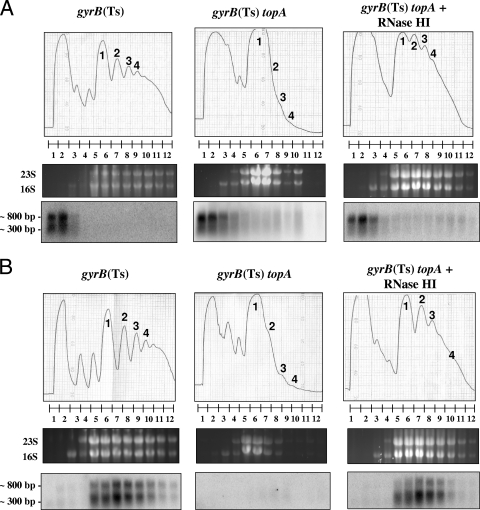
To assess translatable RNA, we used probes to detect RNA corresponding to the proximal yhdG gene and the distal fis gene in various ribosome fractions displayed by sucrose density-gradient centrifugation. The top panels of Fig. 2A show sedimentation profiles with the numbers indicating monosomes (1, one ribosome per mRNA) and polysomes (2, 3, or 4; two or more ribosomes per mRNA). The relative distribution of yhdG RNA in the fractions was similar in all strains. Figure 2B shows that fis RNA was easily detected in the ribosome fractions of both the topA+ strain and the topA-null strain overproducing RNase HI, but such RNA was absent in fractions from the topA-null strain not overproducing RNase HI. When present, fis RNA (Fig. 2B) but not yhdG RNA (Fig. 2A) was mostly found associated with the ribosomes (monosomes and polysomes), which probably reflects a stronger ribosome-binding site for fis RNA (a strong ribosome-binding site is expected for a gene encoding a very abundant protein such as Fis). Thus, less fis RNA was ribosome associated in cells undergoing growth inhibition.
FIG. 2.
Association of yhdG and fis mRNAs with ribosomes after a temperature downshift. Cells were grown at 37°C to log phase as indicated in Materials and Methods before being transferred to 28°C. After 20 min, cells were recovered for sucrose gradient fractionation of ribosomes and mRNA analysis as described in Materials and Methods. Top panels show the ribosome profiles (A260) with the numbers pointing to the peaks corresponding to monosomes (1, one ribosome per RNA) and to polysomes (2, 3, and 4; two or more ribosomes per RNA). The numbers below the ribosome profiles correspond to the different fractions from which the RNA was extracted. The middle panels show the ethidium bromide-stained gels with 23S and 16S rRNA. These gels were used for Northern blot analysis as described in Materials and Methods with probes corresponding to yhdG (a) or to fis (b). The strains used were MA249 [gyrB(Ts)], MA251 [gyrB(Ts) topA], and IB34 [gyrB(Ts) topA/pSK760]. + RNase HI indicates that RNase HI was overproduced (pSK760).
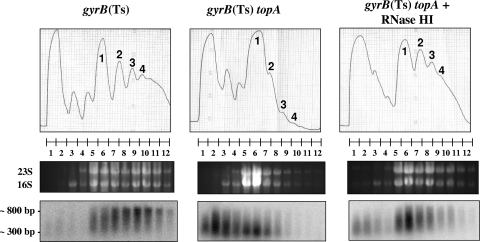
The crp gene was previously found to be expressed at the same level in all strains at the nonpermissive temperature (2). However, in the topA-null strain not overproducing RNase HI, full-length crp mRNA was barely detected, while short stable RNAs corresponding to the 5′ portion of the gene accumulated. Figure 3 shows the presence of crp RNA in the ribosome fractions of both gyrB(Ts) and topA gyrB(Ts) strains, but with much shorter RNA species detected in the ribosome fractions from the topA-null strain not overproducing RNase HI (middle panel, lanes 5 to 12). When RNase HI was overproduced, longer RNAs were detected (right panel, lanes 5 to 12).
FIG. 3.
Association of crp mRNAs with ribosomes after a temperature downshift. Cells were grown and recovered for sucrose gradient fractionation of ribosomes and mRNA analysis as described in the legend to Fig. 2. The top, middle, and bottom panels are as described in the legend to Fig. 2. The probe used for the Northern blot analysis corresponds to crp.
Figures 2 and 3 also show that the ribosome profiles of the topA-null gyrB(Ts) strain not overproducing RNase HI were very different from those of the other strains. Indeed, a much larger proportion of monosomes were observed; polysomes were barely visible. This result indicated the presence of shorter translation-competent RNAs in the topA-null gyrB(Ts) cells at the low, nonpermissive temperature. In agreement with this interpretation, the preferential accumulation of monosomes was also observed when cellular RNA was extensively degraded due to overproduction of the endoribonuclease MazF in E. coli cells (50).
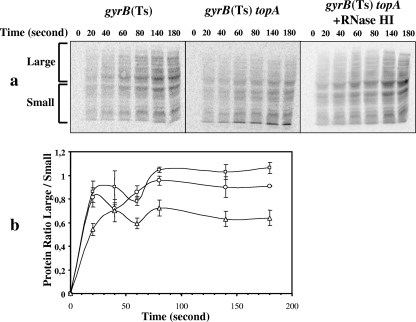
The failure to accumulate full-length RNAs suggested that protein synthesis would be inhibited in downshifted topA-null gyrB(Ts) cells. In addition, the accumulation of short translatable RNAs at the expense of longer ones would favor the synthesis of small proteins over larger ones, as previously observed in cells overproducing the MazF (50) or ChpBK (51) endoribonucleases. To examine protein synthesis, proteins were labeled by adding l-[35S]cysteine to the cell cultures at 28°C. Proteins were analyzed by polyacrylamide gel electrophoresis. We confirmed the previous finding (2) that the protein synthesis rate is reduced in downshifted topA-null gyrB(Ts) cells (data not shown). Figure 4a shows one set of gels that was representative of three independent experiments. Ratios of large to small proteins were obtained by quantifying band intensities in all lanes in the two areas delimited by the brackets labeled large and small (Fig. 4a, left). Figure 4b shows the results of the three independent experiments and demonstrates that the absence of topoisomerase I reduced the large/small ratio of proteins by ca. 40% at a nonpermissive temperature [compare topA+ gyrB(Ts) (squares) to topA gyrB(Ts) (triangles) for times 60 to 180 s].
FIG. 4.
Protein synthesis after a temperature downshift. Cells were grown at 37°C to log phase before being transferred to 28°C. After 20 min, l-[35S]cysteine was added, and aliquots were recovered at different times to monitor protein synthesis as described in Materials and Methods. Samples were analyzed by polyacrylamide gel electrophoresis as described in Materials and Methods. Shown in panel a is the result of one experiment. The two areas delimited by the brackets, respectively, labeled large and small were chosen to calculate the large/small ratios graphically represented in panel b (three independent experiments). Symbols: □, MA249 [gyrB(Ts)]; ▵, MA251 [gyrB(Ts) topA]; ○, IB34 [gyrB(Ts) topA/pSK760].
Collectively, the data indicate that a failure to accumulate full-length RNAs caused a shortage of appropriate templates in topA-null gyrB(Ts) cells, thereby leading to protein synthesis impairment and growth inhibition.
Incorrect processing and degradation of fis and lac RNAs in topA-null gyrB(Ts) mutants at a low temperature.
An inability to accumulate full-length RNAs might be related to either premature termination of transcription or to RNA degradation. In previous work, 5′-proximal probes were used to address the possibility that topA-null gyrB(Ts) cells synthesize truncated RNAs at the nonpermissive temperature (2). Here, we used probes corresponding to internal regions of yhdG-fis and lac operons to address the possibility that RNA is synthesized but degraded in topA-null gyrB(Ts) cells at the nonpermissive temperature.
yhdG-fis RNA is normally processed by an endonucleolytic cleavage to generate two RNA species in which the longer one corresponds to fis and the shorter one to yhdG (3). To monitor the processing of yhdG-fis RNA at the nonpermissive temperature, rifampin was added to cell cultures previously transferred to 28°C, and RNA was extracted at various times. RNA was then probed with an oligonucleotide corresponding to the 3′ end of the fis gene. As expected, yhdG-fis RNA was processed to give the RNA carrying fis both in topA+ (Fig. 5, lanes 1 to 6) and in topA mutant cells overproducing RNase HI (Fig. 5, lanes 13 to 18). However, in topA mutant cells not overproducing RNase HI, yhdG-fis RNA was more slowly processed, a smear was clearly visible, and the RNA corresponding to fis was barely detected (Fig. 5, lanes 7 to 12). Thus, fis RNA was synthesized in topA mutant cells not overproducing RNase HI, but it was incorrectly processed.
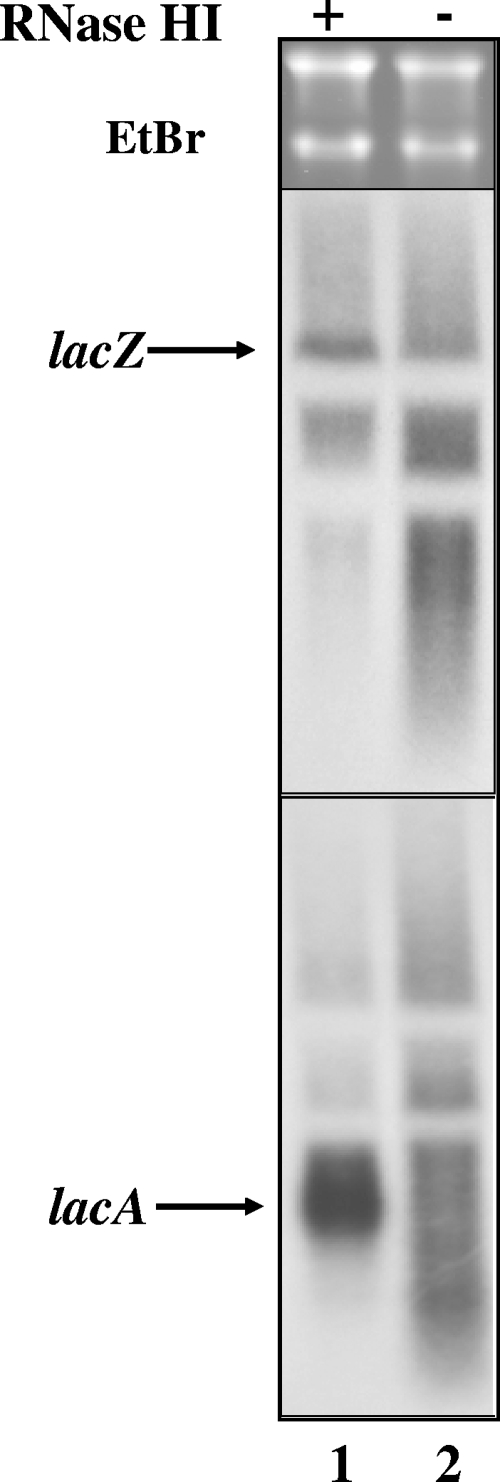
We previously used 5′ probes to demonstrate that topA-null gyrB(Ts) cells accumulate truncated lacZ RNAs at the nonpermissive temperature (2). In agreement with this finding, β-galactosidase synthesis was dramatically reduced in these cells. Here, we used a random-primed probe corresponding to the 3′ region of the lacZ gene to detect lacZ RNA degradation. To facilitate detection of lac RNAs by Northern blot analysis, we placed the lac operon under the control of the strong IPTG-inducible Ptrc promoter on the chromosome of isogenic strains. Figure 6 (top panel) shows that lacZ RNA was synthesized but extensively degraded in topA-null gyrB(Ts) cells when RNase HI is not overproduced (lane 2). We next followed the synthesis of lacA, the third and last member of the lac operon. lacA RNA is normally produced following successive endonucleolytic cleavages of lac RNA by RNase P and RNase E in the intergenic region between lacY and lacA (21). Figure 6 (bottom panel) shows accumulation of correctly processed lacA RNA in topA mutant cells overproducing RNase HI (lane 1). However, lacA RNA was detected as a smear in topA mutant cells not overproducing RNase HI (lane 2). Thus, although lac RNA was synthesized in topA mutant cells not overproducing RNase HI, it was not correctly processed and was degraded, as was the case for fis RNA. These data indicate that the failure to accumulate full-length RNAs in topA-null gyrB(Ts) cells is due largely to altered processing and degradation. Extensive R-loop formation could inhibit normal RNA processing and instead lead to RNA degradation by RNase HI (see below and Discussion).
FIG. 6.
Expression and processing of lac mRNAs after a temperature downshift. Cells were grown at 37°C to log phase as indicated in Materials and Methods before being transferred to 28°C. At 5 min after this transfer, IPTG was added to 1 mM to induce lac expression from the chromosomal Ptrc-lac fusion, and 30 min later aliquots of the cell cultures were obtained for RNA extraction. The strains used were PS66 [gyrB(Ts) topA φ(lacIq-Ptrc-lac)/pEM001] and PS68 [gyrB(Ts) topA φ(lacIq-Ptrc-lac)/pEM003]. + and -, respectively, indicate that RNase HI was overproduced (pEM001) or not overproduced (pEM003). Fifteen μg of RNA were used for Northern blot analysis with random prime-labeled probes hybridizing to lacZ (top panel) or lacA (bottom panel) as described in Materials and Methods.
RNase E is not responsible for the extensive RNA degradation in topA-null gyrB(Ts) mutants at low temperature.
RNase E (rne) is the major endoribonuclease involved in RNA degradation in E. coli (18). To examine RNase E involvement in RNA degradation of topA-null gyrB(Ts) mutants at nonpermissive temperature, we introduced the rneΔ14 mutation into isogenic topA+ and topA mutant strains carrying the lac operon under the control of the Ptrc promoter (among viable rne mutations, rneΔ14 is best able to stabilize the otherwise very unstable lacZ mRNA synthesized by T7 RNA polymerase (19). The rneΔ14 slightly promoted the accumulation of RNA products hybridizing to the lacA probe in topA+ cells (Fig. 7, compare lane 1, rne+ with lane 3, rneΔ14). In topA mutant cells, the rneΔ14 mutation promoted accumulation of lacA degradation products but not normal lacA RNA (Fig. 7, compare lane 5, rne+ with lane 7, rneΔ14). Thus, RNase E does not initiate lacA RNA degradation in topA-null gyrB(Ts) mutants at the nonpermissive temperature. Similar results were obtained for fis RNA (not shown).
FIG. 7.
Effects of rneΔ14 and spectinomycin on the processing of lacA mRNAs after a temperature downshift. Cells were grown at 37°C to log phase as indicated in Materials and Methods. Spectinomycin (final concentration, 400 μg/ml) was added to one-half of the culture and 5 min later the cells were transferred to 28°C. Five minutes after this transfer, IPTG was added to 1 mM to induce lac expression from the chromosomal Ptrc-lac fusion, and thirty minutes later aliquots of the cell cultures were taken for RNA extraction. The strains used are PS63 [gyrB(Ts) topA φ(lacIq-Ptrc-lac)], PS64 [gyrB(Ts) φ(lacIq-Ptrc-lac)], PS123 [gyrB(Ts) topA φ(lacIq-Ptrc-lac rneΔ14], and PS126 [gyrB(Ts) φ(lacIq-Ptrc-lac rneΔ14]. Spc is spectinomycin.
To examine whether extensive RNA degradation requires de novo synthesis of a RNase following the temperature downshift, cells were treated with the translation inhibitor, spectinomycin, 5 min before transfer from 37 to 28°C. Truncated lacA RNAs accumulated whether or not spectinomycin was added to cultures of both rne+ and rneΔ14-derivatives of topA-null mutants (Fig. 7, compare lanes 5 and 7, minus spectinomycin with, respectively, lanes 6 and 8, plus spectinomycin). However, the addition of spectinomycin caused a significant reduction in the amount of RNA detected with the lacA probe in all strains (Fig. 7, compare lanes 1, 3, 5, and 7, minus spectinomycin to, respectively, lanes 2, 4, 6, and 8, plus spectinomycin). We attribute this reduction to polarity effects (1, 46), which are expected to more severely affect lacA, the last gene of the operon. Moreover, in topA+ cells, the addition of spectinomycin strongly promoted the accumulation of larger RNA species (Fig. 7, compare lane 1 to lane 2, respectively, minus and plus spectinomycin). This is in agreement with the previously described stabilizing effects of translation inhibitors on mRNAs due to the titration of RNase E, presumably by unstable rRNAs that are overproduced when protein synthesis is inhibited (24). Thus, extensive RNA degradation in topA mutant cells does not require the de novo synthesis of a RNase following the temperature downshift. Rather, the temperature downshift may lead to extensive accumulation of RNA substrates for a preexisting RNase, other than RNase E.
In vitro transcription of hypernegatively supercoiled templates leads to extensive R-loop formation.
The observation that growth resumption coincided with DNA relaxation suggested that hypernegative supercoiling is tightly linked to RNA degradation. Furthermore, DNA supercoiling can directly affect RNA features only by acting during transcription. Based on experimental evidence, we recently proposed a self-promoting cycle of R-loop formation involving gyrase and RNase HI that ultimately leads to hypernegative supercoiling. In this model, transcription of hypernegatively supercoiled templates leads to extensive and non-sequence-specific R-loop formation (9).
Two plasmids were examined that allowed transcription of a cloned DNA insert from either the physiological or the reverse orientation by the use of T7 or T3 RNA polymerase, respectively. Plasmid pJP461, which carries the 567-bp HindIII fragment of the E. coli rrnB operon, exhibits a small amount of R-loop formation in its physiological orientation (31, 33). A second plasmid, pTW120 (48), carries 41 highly G-rich repeats of the mouse Sγ3 class switch region that allow efficient formation of stable R-loops (6, 15, 47, 48). Stable R-loops significantly formed on templates having wild-type supercoiling levels only when the 41 repeats of the mouse Sγ3 class switch region were transcribed in their physiological, G-rich orientation, as shown by the RNase HI-sensitive gel retardation of pTW120 during electrophoresis (Fig. 8, top panel: compare lane 11 [- RNase HI] to lane 12 [+ RNase HI]). However, these R-loops were short since only a very faint plasmid-associated radioactive RNA signal was detected (Fig. 8, lane 11, bottom panel).
FIG. 8.
R-loop formation on hypernegatively supercoiled templates in vitro. In vitro transcription reactions were performed as described in Materials and Methods in the presence of both [32P]UTP and [32P]ATP. Samples were loaded on an agarose gel without chloroquine. The bottom panel is the autoradiography of the gel to reveal RNA in R-loops. [-] indicates hypernegatively supercoiled templates.
When hypernegatively supercoiled templates were transcribed, extensive R-loop formation occurred with each transcribed sequence (Fig. 8, lanes 5, 7, 13, and 15). This R-loop formation was accompanied by a strong radioactive RNA signal, reflecting the presence of long R-loops. A loss of preference for G-rich regions was indicated by the fact that R-loop formation was detected irrespective of the transcription orientation of the cloned DNA fragments and the RNA polymerase used (T3 or T7). Consequently, transcription of hypernegatively supercoiled DNA templates promotes extensive, sequence-independent R-loop formation. In this context, the presence of RNase HI could lead to extensive RNA degradation and could explain the failure of topA-null gyrB(Ts) cells to accumulate full-length RNAs at the nonpermissive temperature (see Discussion).
DISCUSSION
Mechanism of growth inhibition by hypernegative supercoiling.
Two pathways exist by which topA mutants acquire hypernegative supercoiling. One involves an imbalance between gyrase and the relaxing enzymes, topoisomerases I and IV that allows negative supercoils to accumulate due to restricted DNA rotation associated with transcription (twin-domain model). The other involves formation of RNase HI-sensitive R-loops behind transcription complexes. The latter appears to be involved in the transient accumulation of hypernegative supercoils and growth inhibition that occurs when a topA-null mutant is shifted to low temperature. Overproduction of RNase HI alleviates both (Fig. 1). Accumulation of truncated RNAs at the expense of full-length functional RNAs then impaired protein synthesis and inhibited growth. When hypernegative supercoiling was relaxed, full-length functional RNAs accumulated, and growth resumed. As expected, restoration of the ribosome profile, i.e., the emergence of polysomes at the expense of monosomes, accompanied growth resumption (data not shown). Thus, the known sensitivity of topA-null mutants to low temperature (30, 42) is due to accumulation of hypernegatively supercoiled DNA that then triggers RNA degradation.
Inhibition of topoisomerase IV significantly stimulated the accumulation of hypernegatively supercoiled DNA in topA-null mutants, suggesting that this topoisomerase is involved in removal of hypernegative supercoiling (44). We found that hypernegative supercoiling becomes more stable as temperature decreases (E. Massé et al., unpublished results). It may be that topoisomerase IV preferentially loses activity under these conditions. With various topA-null strains containing or lacking compensatory mutations, we observed a correlation between growth inhibition and hypernegative supercoiling at low temperature (28, 30). The dissimilar extent of RNA degradation, determined by the level of hypernegative supercoiling present in these various strains, may explain their different growth rates.
The data presented above also explain the paradoxical observation that overproduction of RNase HI, an enzyme that degrades RNA, allows the accumulation of full-length RNAs. Overproduction of RNase HI limits the accumulation of hypernegatively supercoiled DNA, thereby preventing extensive RNA degradation. We have found that hypernegatively supercoiled DNA accumulates during a very short time interval following the downshift, after which it persists due to inefficient relaxation (V. Usongo et al., unpublished results). Therefore, RNase HI activity must be high enough in the cell to efficiently inhibit hypernegative supercoiling within a narrow time window. Although our in vivo and in vitro data suggest that RNase HI overproduction acts by preventing the formation of hypernegatively supercoiled DNA (10, 29, 31, 33), it may also promote the rapid removal of these topoisomers. For example, by limiting the formation of hypernegatively supercoiled DNA, RNase HI overproduction may prevent the saturation of topoisomerase IV activity, thereby facilitating relaxation of hypernegative supercoils.
Origin of RNA degradation.
Although the exact mechanism that links hypernegative supercoiling to RNA degradation is not fully characterized, our data allow us to discard several possibilities. Experiments with the rneΔ14 mutation indicated that RNase E is not responsible for the extensive RNA degradation occurring in the topA-null gyrB(Ts) mutant at low temperatures. As expected, we found that the rneΔ14 mutation did not improve the growth of this topA mutant (not shown).
In vitro transcription of hypernegatively supercoiled templates generated extensive, sequence-independent R-loop formation. Therefore, hypernegative supercoiling in the topA mutant may also lead to extensive sequence-independent R-loop formation. RNA in the R-loops can then be degraded by RNase HI, thus explaining the failure to accumulate full-length RNAs and subsequent growth inhibition. Interestingly, fragments on the 3′ side of RNase HI cleavage carry a monophosphorylated 5′ terminus. Such RNAs are preferred substrates for the 5′-end-dependent RNase E, as opposed to mRNAs that normally have a triphosphorylated 5′ terminus (26). This could explain the rapid degradation in the topA mutant of RNAs from fis and lacA that correspond to the distal part of polycistronic RNAs. This would also be compatible with the observation that the rneΔ14 mutation significantly promoted the accumulation of truncated lacA RNAs in the topA mutant.
Fragments from the 5′ side of RNase HI cleavage can be degraded from their 3′ end by exoribonucleases such as PNPase and RNase II (18). Interestingly, we found that RNAs such as yhdG that correspond to the proximal part of operons tend to accumulate in topA-null mutants at the nonpermissive temperature (2; I. Baaklini et al., unpublished observation). The activity of the cellular exoribonucleases may become saturated due to the rapid degradation of RNAs by RNase HI. In agreement with this interpretation, we found that overproducing PNPase or RNase II reduced the accumulation of RNAs that corresponded to the proximal part of both monocistronic and polycistronic RNAs (C. Hraiky et al., unpublished observation). Thus, the apparent 3′-to-5′ RNA decay in the topA-null mutant, as opposed to the normal 5′-to-3′ pathway initiated by the binding of RNase E to the 5′ end of mRNAs (18), would be compatible with RNase HI, not RNase E, initiating RNA decay in topA-null cells after temperature downshifts.
Therefore, according to our model, R-loops inhibit normal RNA processing since RNase E does not have access to RNA in RNA-DNA hybrids. However, such R-loops are targets for RNase HI, thus leading to RNA degradation. When there is no hypernegatively supercoiled DNA (at 37°C or when RNase HI is sufficiently overproduced) extensive R-loop formation is prevented, and there is no significant RNA degradation. This explains why the RNA pattern in topA mutant cells overproducing RNase HI is normal. The level of RNase HI activity required to efficiently inhibit gyrase-mediated hypernegative supercoiling is much higher than the level required to degrade the RNA in R-loops because RNase HI must compete with the strong supercoiling activity of gyrase following downshifts. RNase HI activity becomes toxic only in the presence of hypernegatively supercoiled DNA, because it then leads to extensive RNA degradation. In topA+ cells, where the supercoiling level is normal, the wild-type level of RNase HI activity is sufficient to rapidly eliminate the few R-loops that form during transcription.
A more direct test of RNase HI involvement in RNA degradation in topA mutants would involve inactivating the rnhA gene: deleting rnhA should allow the accumulation of full-length functional RNAs, and it should correct the growth problems of topA mutants at nonpermissive temperatures. Although double topA rnhA-null mutants cannot be constructed (11, 30), we have shown that depletion of RNase HI activity in topA-null cells eventually leads to extensive DNA relaxation, segregation defects, and growth inhibition (44). Extensive DNA relaxation was related to a cellular response leading to gyrase inhibition, possibly to protect cells from the deleterious effect of R-loop-dependent hypernegative supercoiling by gyrase. We found that topA-null mutants depleted in RNase HI activity accumulated normal full-length RNAs following temperature downshifts (P. Sanscartier et al., unpublished observation). Segregation problems leading to growth inhibition were likely due to replication defects related to the lack of RNase HI activity (44).
Although our data are compatible with a direct involvement of RNase HI in RNA degradation in topA mutants, we cannot exclude the contribution of a yet-unknown mechanism triggered by hypernegative supercoiling. Such a mechanism, however, must be consistent with the finding that de novo protein synthesis is not required for RNA degradation following temperature downshifts. As stated in Results, several of the phenotypes seen for the topA-null mutants following temperature downshifts are reminiscent to toxin-antitoxin systems such as MazEF (50) or ChpBIK (51). In these systems, the stable toxin (an endoribonuclease) is produced from a polycistronic RNA that also encodes the unstable antitoxin. Conditions that inhibit the expression of the polycistronic RNA, such as the addition of protein synthesis inhibitors including spectinomycin, lead to the rapid elimination of the antitoxin, thus allowing the endoribonuclease to degrade RNA. However, our observation that the RNA degradation phenotype of topA-null cells exposed to spectinomycin at 37°C is still only observed when such cells are transferred to 28°C (2; I. Baaklini and M. Drolet, unpublished results) is not easily explained by the induction of a toxin-antitoxin system.
Transcription arrest may also be a mechanism by which R-loops inhibit gene expression (2, 9, 14). With yeast, such a mechanism has been proposed (16) and recently supported by the results of in vitro experiments (43). With E. coli, pulse-labeling experiments showed that the rate of rRNA synthesis is reduced in topA-null gyrB(Ts) cells not overproducing RNase HI at the nonpermissive temperature (14). Therefore, in E. coli, R-loops could act as roadblocks for RNA polymerases transcribing highly expressed genes, such as those encoding rRNA (rrn operons).
Recovery of growth.
The transient growth inhibition shown in Fig. 1 is also observed with partially defective topoisomerase I: strain RS2 (41), when shifted from 37°C to 17°C, undergoes a 7-h growth lag (K. Drlica, unpublished observation). This lag is alleviated by a compensatory gyrB mutation (strain SD7 [7]), which suggests that gradual reduction of gyrase activity may restore growth in topA mutants. It will now be interesting to determine whether the cellular response leading to gyrase inhibition when RNase HI activity is depleted in topA mutants (44) contributes to the recovery of growth at low temperature.
Acknowledgments
This study was supported by grant FNR 12667 from the CIHR to M.D. and NIH grants AI35257, AI63431, and AI73491 to K.D. M.D. was a Chercheur-Boursier Senior from the FRSQ.
We thank A. J. Carpousis for the strain carrying the rneΔ14 mutation and Robert Crouch for the RNase HI used in the in vitro transcription experiments. We also thank Patrick Hallenbeck for editing of the manuscript.
Footnotes
Published ahead of print on 12 September 2008.
REFERENCES
- 1.Adhya, S., and M. Gottesman. 1978. Control of transcription termination. Annu. Rev. Biochem. 47967-996. [DOI] [PubMed] [Google Scholar]
- 2.Baaklini, I., C. Hraiky, F. Rallu, Y.-C. Tse-Dinh, and M. Drolet. 2004. RNase HI is required for efficient full-length RNA synthesis in the absence of topoisomerase I in Escherichia coli. Mol. Microbiol. 54198-211. [DOI] [PubMed] [Google Scholar]
- 3.Ball, C. A., R. Osuna, K. C. Ferguson, and R. C. Johnson. 1992. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J. Bacteriol. 1748043-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broccoli, S., F. Rallu, P. Sanscartier, S. M. Cerritelli, R. J. Crouch, and M. Drolet. 2004. Effects of RNA polymerase modifications on transcription-induced supercoiling and associated R-loop formation. Mol. Microbiol. 521769-1779. [DOI] [PubMed] [Google Scholar]
- 5.Condon, C., C. Squires, and C. L. Squires. 1995. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 59623-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniels, G. A., and M. Lieber. 1995. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 235006-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiNardo, S., K. A. Voelkel, R. Sternglanz, A. E. Reynolds, and A. Wright. 1982. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell 3143-51. [DOI] [PubMed] [Google Scholar]
- 8.Drlica, K., and M. Snyder. 1978. Superhelical Escherichia coli DNA: relaxation by coumermycin. J. Mol. Biol. 120145-154. [DOI] [PubMed] [Google Scholar]
- 9.Drolet, M. 2006. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol. Microbiol. 59723-730. [DOI] [PubMed] [Google Scholar]
- 10.Drolet, M., X. Bi, and L. F. Liu. 1994. Hypernegative supercoiling of the DNA template during transcription elongation in vitro. J. Biol. Chem. 2692068-2074. [PubMed] [Google Scholar]
- 11.Drolet, M., P. Phoenix, R. Menzel, E. Massé, L. F. Liu, and R. J. Crouch. 1995. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc. Natl. Acad. Sci. USA 923526-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gellert, M., K. Mizuuchi, M. H. O'Dea, and H. A. Nash. 1976. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. US 733872-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gellert, M., M. H. O'Dea, T. Itoh, and J. Tomizawa. 1976. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc. Natl. Acad. Sci. USA 734474-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hraiky, C., M. A. Raymond, and M. Drolet. 2000. RNase H overproduction corrects a defect at the level of transcription elongation during rRNA synthesis in the absence of DNA topoisomerase I in Escherichia coli. J. Biol. Chem. 27511257-11263. [DOI] [PubMed] [Google Scholar]
- 15.Huang, F. T., K. Yu, C. L. Hsieh, and M. R. Lieber. 2006. Downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc. Natl. Acad. Sci. USA 1035030-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huertas, P., and A. Aguilera. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12711-721. [DOI] [PubMed] [Google Scholar]
- 17.Iost, I., and M. Dreyfus. 1995. The stability of Escherichia coli lacZ mRNA depends upon the simultaneity of its synthesis and translation. EMBO J. 143252-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kushner, S. R. 2005. mRNA decay and processing, p. 327-345. In N. P. Higgins (ed.), The bacterial chromosome. ASM Press, Washington, DC.
- 19.Leroy, A., N. F. Vanzo, S. Sousa, M. Dreyfus, and A. J. Carpousis. 2002. Function in Escherichia coli of the non-catalytic part of RNase E: role in the degradation of ribosome-free mRNA. Mol. Microbiol. 451231-1243. [DOI] [PubMed] [Google Scholar]
- 20.Lewicki, B. T., T. Margus, J. Remme, and K. H. Nierhaus. 1993. Coupling of rRNA transcription and ribosomal assembly in vivo. Formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage T7 RNA polymerase. J. Mol. Biol. 231581-593. [DOI] [PubMed] [Google Scholar]
- 21.Li, Y., and S. Altman. 2004. Polarity effects in the lactose operon of Escherichia coli. J. Mol. Biol. 33931-39. [DOI] [PubMed] [Google Scholar]
- 22.Liu, L. F., and J. C. Wang. 1987. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 847024-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lodge, J. K., T. Kazic, and D. E. Berg. 1989. Formation of supercoiling domains in plasmid pBR322. J. Bacteriol. 1712181-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez, P. J., I. Marchand, O. Yarchuk, and M. Dreyfus. 1998. Translation inhibitors stabilize Escherichia coli mRNAs independently of ribosome protection. Proc. Natl. Acad. Sci. USA 956067-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch, A. S., and J. C. Wang. 1993. Anchoring of DNA to the bacterial cytoplasmic membrane through cotranscriptional synthesis of polypeptides encoding membrane proteins or proteins for export: a mechanism of plasmid hypernegative supercoiling in mutants deficient in DNA topoisomerase I. J. Bacteriol. 1751645-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackie, G. A. 1998. Ribonuclease E is a 5′-end-dependent endonuclease. Nature 395720-723. [DOI] [PubMed] [Google Scholar]
- 27.Mackie, G. A., B. C. Donly, and P. C. Wong. 1990. Coupled transcription-translation of ribosomal proteins, p. 191-211. In G. Spedding (ed.), Ribosomes and protein synthesis: a practical approach. IRL Press, Oxford, United Kingdom.
- 28.Massé, E., and M. Drolet. 1999. Relaxation of transcription-induced negative supercoiling is an essential function of Escherichia coli DNA topoisomerase I. J. Biol. Chem. 27416654-16658. [DOI] [PubMed] [Google Scholar]
- 29.Massé, E., and M. Drolet. 1999. Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J. Biol. Chem. 27416659-16664. [DOI] [PubMed] [Google Scholar]
- 30.Massé, E., and M. Drolet. 1999. R-loop-dependent hypernegative supercoiling in Escherichia coli topA mutants preferentially occurs at low temperatures and correlates with growth inhibition. J. Mol. Biol. 294321-332. [DOI] [PubMed] [Google Scholar]
- 31.Massé, E., P. Phoenix, and M. Drolet. 1997. DNA topoisomerases regulate R-loop formation during transcription of the rrnB operon in Escherichia coli. J. Biol. Chem. 27212816-12823. [DOI] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 33.Phoenix, P., M. A. Raymond, E. Massé, and M. Drolet. 1997. Roles of DNA topoisomerases in the regulation of R-loop formation in vitro. J. Biol. Chem. 2721473-1479. [DOI] [PubMed] [Google Scholar]
- 34.Powell, B. S., M. P. Rivas, D. L. Court, Y. Nakamura, and C. L. Turnbough, Jr. 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 231278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruss, G. J. 1985. DNA topoisomerase I mutants. Increased heterogeneity in linking number and other replicon-dependent changes in DNA supercoiling. J. Mol. Biol. 18551-63. [DOI] [PubMed] [Google Scholar]
- 36.Pruss, G. J., and K. Drlica. 1986. Topoisomerase I mutants: the gene on pBR322 that encodes resistance to tetracycline affects plasmid DNA supercoiling. Proc. Natl. Acad. Sci. USA 838952-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pruss, G. J., S. H. Manes, and K. Drlica. 1982. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell 3135-42. [DOI] [PubMed] [Google Scholar]
- 38.Samul, R., and F. Leng. 2007. Transcription-coupled hypernegative supercoiling of plasmid DNA by T7 RNA polymerase in Escherichia coli topoisomerase I-deficient strains. J. Mol. Biol. 374925-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 5385-96. [DOI] [PubMed] [Google Scholar]
- 40.Spirito, F., and L. Bossi. 1996. Long-distance effect of downstream transcription on activity of the supercoiling-sensitive leu-500 promoter in a topA mutant of Salmonella typhimurium. J. Bacteriol. 1787129-7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sternglanz, R. S. DiNardo, K. A. Voelkel, Y. Nishimura, Y. Hirota, K. Becherer, L. Zumstein, and J. C. Wang. 1981. Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and transposition. Proc. Natl. Acad. Sci. USA 782747-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stupina, V. A., and J. C. Wang. 2005. Viability of Escherichia coli topA mutants lacking DNA topoisomerase I. J. Biol. Chem. 280355-360. [DOI] [PubMed] [Google Scholar]
- 43.Tous, C., and A. Aguilera. 2007. Impairment of transcription elongation by R-loops in vitro. Biochem. Biophys. Res. Commun. 360428-432. [DOI] [PubMed] [Google Scholar]
- 44.Usongo, V., F. Nolent, P. Sanscartier, C. Tanguay, S. Broccoli, I. Baaklini, K. Drlica, and M. Drolet. 2008. Depletion of RNase HI activity in Escherichia coli lacking DNA topoisomerase I leads to defects in DNA supercoiling and segregation. Mol. Microbiol. 69968-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei, Y., J. M. Lee, C. Richmond, F. R. Blattner, J. A. Rafalski, and R. A. LaRossa. 2001. High-density microarray-mediated gene expression profiling of Escherichia coli. J. Bacteriol. 183545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yarchuk, O., N. Jacques, J. Guillerez, and M. Dreyfus. 1992. Interdependence of translation, transcription and mRNA degradation in the lacZ gene. J. Mol. Biol. 226581-596. [DOI] [PubMed] [Google Scholar]
- 47.Yu, K., F. Chedin, C. L. Hsieh, T. E. Wilson, and M. R. Lieber. 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4442-451. [DOI] [PubMed] [Google Scholar]
- 48.Yu, K., D. Roy, M. Bayramyan, I. S. Haworth, and M. R. Lieber. 2005. Fine-structure analysis of activation-induced deaminase accessibility to class switch region R-loops. Mol. Cell. Biol. 251730-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zechiedrich, E. L., A. B. Khodursky, S. Bachellier, R. Schneider, D. Chen, D. M. Lilley, and N. R. Cozzarelli. 2000. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 2758103-8113. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, Y., J. Zhang, K. P. Hoeflich, M. Ikura, G. Qing, and M. Inouye. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12913-923. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, Y., L. Zhu, J. Zhang, and M. Inouye. 2005. Characterization of ChpBK, an mRNA interferase from Escherichia coli. J. Biol. Chem. 28026080-26088. [DOI] [PubMed] [Google Scholar]