Abstract
SCO4677 is one of a large number of similar genes in Streptomyces coelicolor that encode proteins with an HATPase_c domain resembling that of anti-sigma factors such as SpoIIAB of Bacillus subtilis. However, SCO4677 is not located close to genes likely to encode a cognate sigma or anti-anti-sigma factor. SCO4677 was found to regulate antibiotic production and morphological differentiation, both of which were significantly enhanced by the deletion of SCO4677. Through protein-protein interaction screening of candidate sigma factor partners using the yeast two-hybrid system, SCO4677 protein was found to interact with the developmentally specific σF, suggesting that it is an antagonistic regulator of σF. Two other proteins, encoded by SCO0781 and SCO0869, were found to interact with the SCO4677 anti-σF during a subsequent global yeast two-hybrid screen, and the SCO0869-SCO4677 protein-protein interaction was confirmed by coimmunoprecipitation. The SCO0781 and SCO0869 proteins resemble well-known anti-anti-sigma factors such as SpoIIAA of B. subtilis. It appears that streptomycetes may possess an extraordinary abundance of anti-sigma factors, some of which may influence diverse processes through interactions with multiple partners: a novel feature for such regulatory proteins.
Bacteria of the genus Streptomyces are highly adapted to living in soil environments and produce various valuable secondary metabolites. Their complex life cycle of morphological and physiological differentiation requires elaborate regulatory systems (6). The genome of the model species Streptomyces coelicolor A3(2) encodes 7,825 theoretical proteins, with more than 12% (≥900 gene products) encoding transcriptional regulators. The 65 sigma factors of S. coelicolor strikingly point to the complexity of its gene regulation (ScoDB at http://strepdb.streptomyces.org.uk) (2).
Sigma factors have been found to be important for differentiation. BldN and σN are required for formation of aerial hyphae (3, 8), while WhiG initiates the formation of spores from aerial hyphae (5): bldN, sigN, and whiG appear to be transcribed throughout aerial mycelium formation and development. On the other hand, σF is involved in the later stages of spore formation, and sigF transcription was detected only when sporulation started, and depended upon whiG and other early sporulation genes (whiA, whiB, whiH, whiI, and whiJ) (28). A sigF-null mutant formed smaller spores compared to the wild-type strain, with the spore wall being much thinner, and the chromosomal DNA of the sigF mutant was much less condensed (43). Among the seven other σF- and σN-like sigma factors of S. coelicolor (all members of the group 3 subfamily of sigma factors), σB and σH were identified as regulating both morphological differentiation and responses to osmotic stress (7, 30, 46, 50). σI and σJ, both particularly SigH-like, and σM were also reported to participate in the osmotic stress response (34, 50).
This sigma factor subfamily is exclusive to, but widespread among, gram-positive bacteria. Its members are often regulated by interaction with anti-sigma factors, which in low G+C gram-positive bacteria, all belong to a family of paralogous proteins that have in common the ability to phosphorylate other, alternative, partner proteins of a third class of paralogues (anti-anti-sigmas, or anti-sigma antagonists) through the serine protein kinase activity of their HATPase_c domain. The balance between the two alternative interactions of these anti-sigmas determines the extent to which the sigma factor is free to interact with RNA polymerase and promoters (11, 12, 13, 14).
Five anti-sigma factors have been identified experimentally in S. coelicolor, all encoded by genes adjacent to the genes for the target sigmas. These comprise three active against group 3 sigmas (UshX for σH [47], RsbA for σB [33], and RsmA for σM [17]) and two active against ECF sigma factors (RsrA for σR [27] and RsuA for σU [18, 19] possessing a conserved motif of the ZAS family). Overall, about 40 proteins encoded in the genome have an HATPase_c domain, giving rise to speculation that they may all be anti-sigmas active against group 3 sigmas (8, 17, 38), although they are generally rather highly diverged, with only a few being located close to genes for sigma factors or anti-anti-sigmas. One antagonist of such an anti-sigma factor has been experimentally identified (RsbV, which is antagonistic to RsbA [the σB-RsbA-RsbV interactions appear to be responsive to osmotic stress]) (33). RsbA and RsbV are both recognizably similar to their equivalents in other bacteria (33). A further 12 genes possibly encoding anti-anti-sigmas are annotated in the genome (ScoDB at http://strepdb.streptomyces.org.uk), including bldG, which is required for aerial growth, but whose target sigma factor has not been identified.
The SCO4677 product is one of the ca. 40 HATPase_c domain-containing putative anti-sigma factors. It is not located close to genes likely to encode either a sigma factor or an anti-anti-sigma. Here, the SCO4677 product has been shown through gene disruption to affect development, and yeast two-hybrid (Y2H) analysis has identified interactions of SCO4677 protein with a sigma factor (the development-specific σF) and two putative anti-anti-sigmas. This suggests that streptomycetes may have very complex regulation of sigma factor activity.
MATERIALS AND METHODS
Microorganisms used and maintenance of strains.
The microbial strains used in this study are listed in Table 1. S. coelicolor M600 was used for the construction of a cDNA library and null mutants. Conditions for Streptomyces plate culture on MS, R2YE, and SMM agar media were as previously described (32). E. coli DH5α was used for general DNA preparation. E. coli BW 25113/pIJ790 was the host for PCR-targeted mutagenesis and E. coli ET12567 and E. coli ET12567/pUZ8002 were used as nonmethylating hosts (20). Strains of E. coli were cultured on LB agar plates with appropriate antibiotics for the stable maintenance of plasmids or cosmids (45). Saccharomyces cerevisiae AH109 and Y187 strains were cultured on YPD(A) and synthetic dropout (SD) medium containing appropriate amino acid supplements for Y2H screening (Clontech).
TABLE 1.
Strains used in this study
| Strain | Descriptiona | Source or reference |
|---|---|---|
| Streptomyces coelicolor | ||
| M600 | Prototroph, SCP1−/SCP2− | JIC, United Kingdom |
| SMF5811 | M600 SCO4677::Aprr | This study |
| SMF5812 | pSET152K-SCO4677 integrated in SMF5811 | This study |
| SMF5813 | pSET152K integrated in SMF5811 | This study |
| SMF5814 | M600 SCO0781::Aprr | This study |
| SMF5815 | M600 SCO0869::Aprr | This study |
| Escherichia coli | ||
| DH5α | F− φdlacZΔM15 endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 gyrA96 Δ(lacZYA-argF)U169 λ−deoR phoA relA1 | 21 |
| ET12567 | dam13::Tn9 dcm6 hsdM hsdR recF143 16 zjj201::Tn10 galK2 galT22 ara14 lacY1 xyl5 leuB6 thi-1 tonA31 rspL136 hisG4 tsx78 mtl-1 glnV44 | 35 |
| Saccharomyces cerevisiae | ||
| AH109 | MATatrp1-901 leu2-3112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS-MEL1TATA-lacZ MEL1 | 25 |
| Y187 | MATα trp1-901 leu2-3112 ura3-52 his3-200 ade2-101 gal4Δ met− gal80Δ URA3::GAL1UAS-GAL1TATA-lacZ MEL1 | 22 |
| SMF5821 | Y187 harboring pGBKT7-SCO4677 | This study |
Aprr, apramycin resistant.
DNA manipulation.
The plasmids used in the present study are listed in Table S1 in the supplemental material. General procedures for in vitro DNA manipulation were performed as described previously (45). Genomic DNA was isolated from Streptomyces strains as described by Kieser et al. (32). Southern analysis was performed by the DIG DNA labeling system (Roche) (24).
Y2H analysis of interaction between SCO4677 and sigma factors.
Sigma factor genes (sigF, sigB, sigR, sigH, and hrdB) and SCO4677 were amplified by PCR using genomic DNA as a template. The primers are listed in Table S2 in the supplemental material. The amplified genes were cloned into pGADT7 encoding the GAL4 activation domain (AD), here termed pAD, and pGBKT7 encoding the GAL4 DNA binding domain (BD), here termed pBD. pAD derivatives with inserted sigma factor genes were used with pBD-SCO4677, and pBD derivatives with inserted sigma factor genes were used with pAD-SCO4677 to cotransform S. cerevisiae AH109. Interaction was examined by culturing cotransformants on quadruple dropout agar medium (QDO), that is, adenine, histidine, leucine, and tryptophan dropout SD medium [SD/Ade(−)/His(−)/Leu(−)/Trp(−)].
Construction of a cDNA library of S. coelicolor M600 and preparation of a mating strain for Y2H analysis.
Spores of S. coelicolor M600 were inoculated into 2xYT liquid medium and cultured for 7 h at 30°C. The pregerminated spores were then spread on a cellophane disc on R2YE agar and cultured for 3 and 6 days. Mycelium formed on the cellophane disc was harvested for RNA purification. Total RNA was purified as recommended by the manufacturer of the RNA isolation kits (Qiagen RNeasy Midi kit). The first-strand cDNA was synthesized by PCR by using 2 μg of purified total RNA and Moloney murine leukemia virus reverse transcriptase. Double-stranded cDNA was synthesized from the first-strand cDNA, in which PCR primers were designated to give a SMARTIII and CDSIII oligonucleotide sequence at the end of the DNA fragments. The cDNA linked with the SMARTIII/CDSIII oligonucleotide was introduced into S. cerevisiae AH109 together with linearized pGADT7-Rec (i.e., pAD), so that cDNA of S. coelicolor was inserted into pAD by natural recombination. After 3 to 4 days, clones grown on SD/Leu(−) plates were considered to represent a cDNA library (S. cerevisiae AH109/pAD-cDNA). The transformants were harvested and suspended in YPD medium supplemented with 25% (vol/vol) glycerol. The plasmids were extracted and amplified by retransformation of E. coli DH5α, and then the amplified plasmids were re-extracted and digested by NdeI and XhoI. Library strain aliquots containing more than 2 × 107 cells/ml were stored at −80°C until use.
Y2H screening of proteins interacting with SCO4677.
pBD-SCO4677 was introduced into S. cerevisiae Y187, creating the bait strain S. cerevisiae Y187/pBD-SCO4677 (SMF5821). The possible toxicity of SCO4677 protein to S. cerevisiae Y187 was evaluated by culturing SMF5821 in liquid medium SD/Trp(−)/Kan (20 μg/ml). To examine self-activation of pBD-SCO4677, pBD-SCO4677 was introduced into S. cerevisiae AH109 and cultured in adenine, histidine, and tryptophan dropout SD medium [SD/Ade(−)/His(−)/Trp(−)]. Mating between the bait strain, SMF5821 and the library strain was performed as recommended by the instructions for the Matchmaker Library Construction and Screening kit (Clontech). Cells (>109 cells/ml) of the bait strain SMF5821 (mating type α) were harvested from an actively growing culture in 50 ml of SD/Trp(−)/Kan (20 μg/ml) medium. These were mixed with cells (2 × 107) of the cDNA library strain AH109/pAD-cDNA (mating type a) and incubated in 2× YPDA medium containing kanamycin (50 μg/ml) at 30°C with mild agitation for 24 h. The mating cells were washed and harvested. The cells were suspended in 0.5× YPDA with kanamycin (50 μg/ml) and inoculated onto QDO medium, which selected colonies arising from interaction events. Secondary screening was done by comparing the intensity of the blue color developed on colony lifted filters soaked in buffer containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (4). Tertiary screening was carried out by quantitative determination of β-galactosidase activity, using o-nitrophenyl β-d-galactopyranoside (ONPG) as the color-developing substrate. The β-galactosidase unit was calculated as elsewhere (15). The candidates were finally confirmed by the determination of DNA sequences. The interaction between the baiting protein and baited protein was confirmed by cotransforming S. cerevisiae AH109 with pAD-X (full X gene) and pBD-SCO4677 and with pAD-SCO4677 and pBD-X (full X gene). pGADT7-T and pGBKT7-53 were used as a positive control, and pGADT7-T and pGBKT7-Lam were used as a negative control.
Expression and purification of proteins.
For the recombinant His-SCO0869, SCO0869 gene was amplified by PCR and cloned into pET19b to yield pET19b-SCO0869. For the recombinant GST-SCO4677, SCO4677 was amplified by PCR and cloned into pGEX4T-1. For expression, each of the plasmids was used to transform E. coli BL21(DE3)/pLysS. The resulting strains were inoculated into 20 ml of LB, cultured overnight, transferred to 5-liter fermentor jars (Kobiotech) containing 2 liters of LB medium, and cultured at 27°C and 170 rpm at 1 volume/volume/min. At an optical density at 600 nm of 0.6, the cultures were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubated for 3 h. Cells were harvested by centrifugation at 8,000 rpm for 20 min, and the pellets were resuspended in lysis buffer and sonicated. Cell lysates were centrifuged at 15,000 rpm for 20 min, and the supernatants were loaded onto Ni-NTA agarose (Qiagen) or GST-tag resin (Novagen), as appropriate. The eluates were dialyzed against storage buffer (20 mM Tris-Cl [pH 7.5], 60 mM KCl, 10 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 50% glycerol) and stored at −20°C. Purified proteins were confirmed by mass spectrometry analysis.
Coimmunoprecipitation.
Purified His-SCO0869 was incubated with GST-SCO4677 in binding buffer (50 mM Tris-Cl [pH 7.5], 50 mM KCl, 3 mM MgCl2, 0.1% NP-40) for 1 h at room temperature. The anti-GST antibody was added, and the mixture was incubated for 1 h at room temperature. nProtein A beads (Amersham Pharmacia) were added to the complex of proteins and antibodies and were further incubated for 1 h at room temperature. The beads were washed 10 times with binding buffer, and the proteins were eluted by boiling with sodium dodecyl sulfate sample buffer. Subsequently, the proteins were separated on two identical sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and transferred to membranes for Western blotting. One membrane was incubated with anti-His antibody, and the other was incubated with anti-GST antibody.
Gene disruption and complementation.
Selected genes were disrupted by the PCR targeting procedure (20). For the disruption of SCO4677, cosmid D31 containing SCO4677 was introduced into E. coli BW25113/pIJ790 by electroporation. An EcoRI/HindIII DNA fragment (1,384 bp) containing the apramycin resistance gene prepared from plasmid pIJ773 was used as the template for PCR to amplify an apramycin resistance cassette flanked by sequences upstream and downstream of SCO4677. The extended apramycin resistance cassette was introduced into E. coli BW25113/pIJ790/D31, and homologous recombination was induced between the extended apramycin resistance cassette and SCO4677 in cosmid D31. A strain, BW25113/pIJ790/D31ΔSCO4677, resistant to carbenicillin, kanamycin, and apramycin was selected as a candidate in which SCO4677 was disrupted. Cosmid D31ΔSCO4677 extracted from this strain was used to transform nonmethylating E. coli ET12567/pUZ8002. The transformants were mixed with preheated S. coelicolor M600 spores and then spread onto MS agar medium containing 10 mM MgCl2 for the conjugal transfer of the cosmid. Exconjugant strains in which the targeted gene had integrated into the chromosome were selected by their apramycin resistance. Double-crossover exconjugants were recognized by their sensitivity to kanamycin and resistance to apramycin. The deletion of SCO4677 in a selected mutant (SMF5811) was confirmed by Southern hybridization analysis. The disruption of SCO0781 and SCO0869 was accomplished and confirmed in the same manner.
For complementation, the 1,540-bp fragment containing the SCO4677 gene was amplified by PCR using com4677F and com4677R primers and then cloned into pSET152K, resulting in pSET152K-SCO4677. Nonmethylated pSET152K-SCO4677 was prepared from E. coli ET12567 and introduced into SMF5811 by protoplast transformation, creating SMF5812. As a control, pSET152K was introduced into SMF5811, creating SMF5813.
Fermentations.
The kinetics for mycelium growth, substrate utilization, and antibiotic production were analyzed during submerged culture at 30°C in jar fermentors (model KF-5L; KoBiotech, South Korea). Spores and/or mycelia formed freshly on MS agar plates were inoculated into 50 ml of GG1 medium (15 g of glucose, 15 g of glycerol, 15 g of Soytone (Difco), 3 g of NaCl, and 1 g of CaCO3 per liter [pH 7.2]) as the first seed culture and incubated for 48 h in a shaking incubator. The first seed culture was transferred to 50 ml of modified GYB medium (10 g of glucose and 15 g of yeast extract per liter [pH 7.2]) as the second seed culture and incubated for 36 h. The second seed culture was inoculated into 4 liters of SMM medium (32). The initial pH was adjusted to 7.2. The agitation and aeration conditions were 220 rpm and 1 volume/volume/min, respectively.
Analytical methods.
The first 5 ml of culture broth contained in the sampling line was discarded, and then about 20 ml of culture broth was taken for analysis. The concentration of ammonium ion and the dry cell weight (DCW) were measured immediately after sampling. For measurement of the DCW, the mycelium was collected by vacuum filtration (Whatman filter paper GF/C) and dried at 80°C for 24 h. The concentration of glucose in the cultures was measured with a glucose assay kit using glucose oxidase methods (BMI Korea) (31). Actinorhodin (Act) concentration was determined by a method described previously (44). Fermentation kinetic parameters were calculated as described previously (42).
RESULTS
In silico analysis of the protein encoded by SCO4677.
SCO4677 is annotated as encoding a putative regulatory protein of 144 amino acids, based on its resemblance to the product of orfA in the complex abaA locus previously shown to influence the level of antibiotic production (16) (SCO-DB genome annotation at http://streptomyces.org.uk). SCO4677 homologues exist mainly in actinomycetes such as Streptomyces spp. Nocardioides sp., and Frankia spp. The SCO4677 product shows 80% sequence identity with a protein encoded by SAV3140 in Streptomyces avermitilis. It includes a 97-amino-acid GHKL superfamily domain with the features of HATPases that are notably also present in anti-sigma factors such as SpoIIAB and RsbW from Bacillus stearothermophilus and Bacillus subtilis, respectively (Fig. 1). Recent bioinformatic analysis found that 27 SpoIIAB-like putative anti-sigma factors are encoded in the genome of S. coelicolor, all of them containing the GHKL domain (17). SCO4677 was not included in this list, but an earlier article did show SCO4677 (under the earlier name SCD31.02) in a phylogenetic tree of anti-sigma factors (38). It was thus possible that the SCO4677 product might be an anti-sigma factor. However, no obvious candidate genes encoding potential partner sigma or anti-anti-sigma factors are located near to SCO4677.
FIG. 1.
Amino acid alignment of SCO4677 protein, SpoIIAB, and RsbW. The protein encoded by SCO4677 was aligned with anti-sigma factors SpoIIAB of B. stearothermophilus and RsbW of B. subtilis. Motifs I to IV represent the conserved domains for ATP binding where the important residues for ATP binding are denoted by asterisks.
Functional analysis of SCO4677 null mutant.
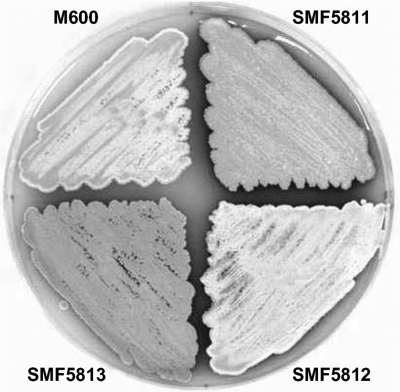
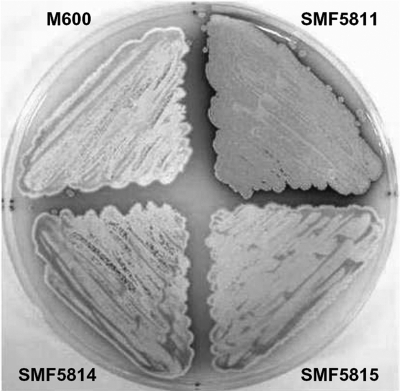
In order to characterize the function of the SCO4677 product, SCO4677 was deleted from S. coelicolor M600 by PCR targeting. To compare the phenotypes of S. coelicolor M600 (wild-type strain) and the ΔSCO4677 mutant (SMF5811), the strains were cultured on MS agar medium. Morphological differentiation and production of blue Act antibiotic were enhanced and accelerated in SMF5811 compared to the parent strain. To confirm the phenotype of SMF5811, we constructed SMF5812 containing pSET152K-SCO4677 and SMF5813 containing pSET152K. The phenotype of SMF5811 was complemented when pSET152K containing SCO4677 was introduced into SMF5811 (Fig. 2). Similar phenotypes were observed on R2YE and SMM agar media (not shown).
FIG. 2.
Phenotype of the SCO4677 null mutant. M600, SMF5811, SMF5812 and SMF5813 were cultured on MS agar medium for 4 days. The enhanced morphological differentiation of SCO4677 null mutant (SMF5811) was restored to the wild-type level by introducing pSET152K-SCO4677 into SMF5811. SMF5812 is SMF5811 containing pSET152K-SCO4677, and SMF5813 is SMF5811 containing pSET152K without an insert.
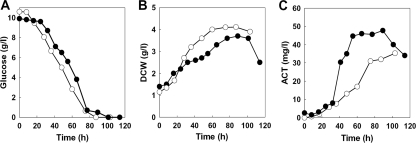
Growth and Act production in submerged batch cultures of the parent and SMF5811 were examined in SMM medium for 120 h by using a jar fermentor. Glucose utilization (Fig. 3A) and total cell mass (Fig. 3B) were not altered significantly in the mutant. The mutant initially grew at a rate comparable to that of the parent strain but showed an early growth pause, after which it continued growth at a reduced rate, eventually roughly further doubling its biomass (Fig. 3B), and produced Act abundantly (Fig. 3C). This could be interpreted as an early entry into the special transition phase preceding stationary phase seen in some culture conditions for streptomycetes (41).
FIG. 3.
Kinetic analysis of S. coelicolor strains M600 and SMF5811. M600 (○) and SMF5811 (•) were grown in SMM medium using jar fermentors. Glucose content (A), mycelial growth (DCW) (B), and Act level (C) were measured.
The SCO4677 gene encodes a protein that interacts with σF.
To further investigate whether the SCO4677 product is an anti-sigma factor, we looked at its ability to interact with certain sigma factors, using the Y2H system. We focused initially on four sigma factors, viz. σF (SCO4035), σB (SCO0600), σH (SCO5243), and σR (SCO5216), which have been reported to be involved in morphological differentiation and stress responses. Interaction with the housekeeping sigma factor HrdB (SCO5820) was also evaluated for comparison, since it belongs to a class of sigma factors that are not believed to interact with HATPase_c domain-containing anti-sigma factors. The genes, sigF, sigB, sigH, sigR, and hrdB for σF, σB, σH, σR, and HrdB were cloned into pGBKT7 (pBD) and pGADT7-Rec (pAD). The pBD-derived plasmids were used with pAD-SCO4677, and the pAD derivatives were used with pBD-SCO4677 to cotransform S. cerevisiae AH109.
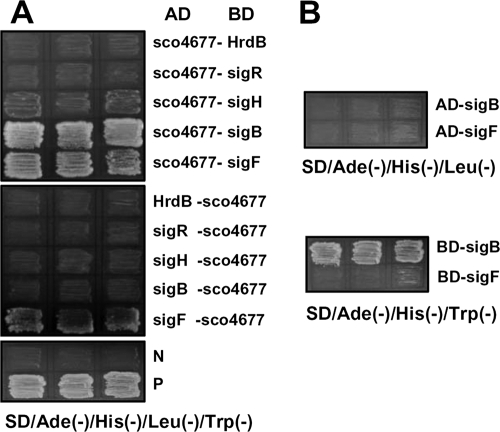
The transformants harboring pAD-SCO4677/pBD-sigF and pAD-SCO4677/pBD-sigB grew well on QDO agar medium, whereas those harboring pAD-SCO4677/pBD-sigH, pAD-SCO4677/pBD-sigR, and pAD-SCO4677/pBD-hrdB did not (upper panel in Fig. 4A). In experiments with the reciprocal arrangements of all potential interaction partners, only the transformant harboring pAD-sigF/pBD-SCO4677 grew well, while that harboring pAD-sigB/pBD-SCO4677 did not grow (middle panel in Fig. 4A). This anomaly was explained in experiments revealing that sigB constructs could activate the Y2H system even without an interaction partner—a property not shown by sigF, sigH, or sigR (Fig. 4B). Overall, the results indicate that the SCO4677 product interacts with σF, but not with σB, σH, σR, or HrdB, a finding consistent with the SCO4677 product acting as an anti-sigma factor for σF.
FIG. 4.
Screening for sigma factors interacting with SCO4677 protein. (A) Potential interaction of SCO4677 protein with HrdB, σR, σH, σB, or σF was tested by looking for strong growth on QDO agar plates, using the Y2H system. (B) Test of self-activation of σB and σF in S. cerevisiae AH109. The sigB and sigF genes were cloned into pAD and pBD and introduced into S. cerevisiae AH109, respectively. The strains containing pAD-sigB or pAD-sigF were streaked onto agar plates of SD/Ade(−)/His(−)/Leu(−). The strains containing pBD-sigB or pBD-sigF were streaked onto agar plates of SD/Ade(−)/His(−)/Trp(−). All strains were cultured for 5 days. P, positive control; N, negative control.
Screening for other gene products that interact with the SCO4677 protein.
If the interaction of SCO4677 protein with σF reflects an antagonism, precedent would suggest that there might be an alternative interacting partner for SCO4677 protein to serve as an anti-anti-sigma factor. To search for other gene products that interact with SCO4677 protein, we again applied the Y2H system. Mating between the baiting strain (S. cerevisiae Y187/pBD-SCO4677) and the cDNA pool strain (S. cerevisiae AH109/pAD-cDNA) was conducted. The mating efficiency was estimated to be 2.2% (the required mating efficiency is 2%). Of 68 clones growing well on QDO medium, 22 were subjected to colony-lift filter assay. A total of 16 of 22 clones were selected by quantitative β-galactosidase analysis and subjected to DNA sequence analysis. Excluding four plasmids with a gene inserted in the wrong orientation or reading frame, the DNA sequences of 10 clones matched with SCO0869, and those of the other 2 clones matched with SCO0781.
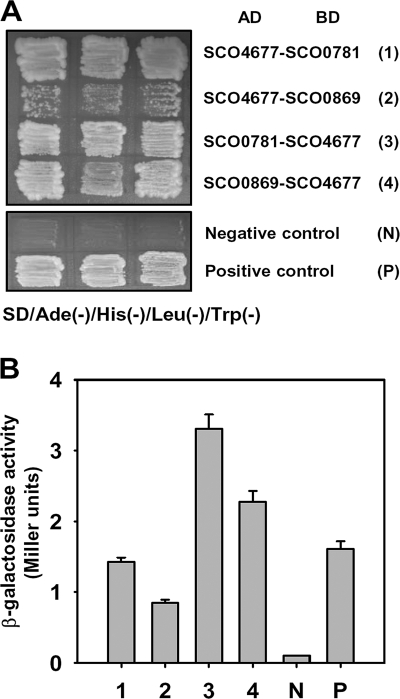
In order to confirm the interaction between the SCO4677 product and the products encoded by SCO0781 and SCO0869, these genes were cloned into pBD and pAD and used with the appropriate SCO4677 derivatives to cotransform S. cerevisiae AH109. All of the cotransformants except those containing pAD-SCO4677/pBD-SCO0869 grew well on QDO agar plates (Fig. 5A) and expressed β-galactosidase activity strongly (Fig. 5B), confirming the initial evidence of interactions of the SCO4677 product with the products of SCO0781 and SCO0869. To find out the role of SCO0781 and SCO0869 in morphological differentiation, SCO0781 and SCO0869 were disrupted in the chromosome of S. coelicolor M600. However, ΔSCO0781 (SMF5814) and ΔSCO0869 (SMF5815) mutants did not show obvious phenotypic differences from the wild-type strain (Fig. 6).
FIG. 5.
Confirmation of the interaction of SCO4677 protein with SCO0781 and SCO0869 proteins. The protein level interactions of SCO4677 with SCO0781 and of SCO4677 with SCO0869 were confirmed in the Y2H system by cotransforming the pairs into S. cerevisiae AH109. (A) Transformants were streaked onto QDO agar alongside positive and negative control strains and cultured for 5 days. (B) Quantification of the intensity of the interactions in panel A by β-galactosidase activity assays.
FIG. 6.
Phenotypic analysis of ΔSCO0781 and ΔSCO0869 mutants. The S. coelicolor wild-type strain and ΔSCO4677 (SMF5811), ΔSCO0781 (SMF5814), and ΔSCO0869 (SMF5815) mutants were cultured on MS agar medium for 3 days.
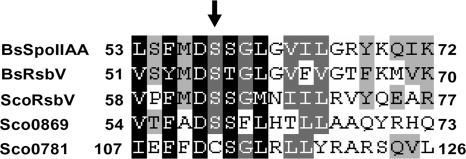
SCO0781 and SCO0869 are annotated as encoding anti-sigma factor antagonists (ScoDB at http://strepdb.streptomyces.org.uk), based on similarity with such anti-anti-sigma factors as SpoIIAA of B. subtilis. In alignments, SCO0781 showed 22% identities and 49% similarities, and SCO0869 showed 24% identities and 48% similarities with SpoIIAA. Anti-anti-sigma factors such as SpoIIAA contain a highly conserved serine residue that is phosphorylated by the serine kinase activity of their cognate anti-sigma factors. The SCO0869 product has this conserved serine residue, but it is replaced by a cysteine in the SCO0781 protein (Fig. 7). Therefore, it is likely that SCO0869 encodes a conventional anti-anti-sigma factor that antagonizes the sigma-binding function of the SCO4677 product in a manner responsive to phosphorylation by the anti-sigma factor's kinase activity, while the anti-anti-sigma factor activity of SCO0781 protein may be modulated in a hitherto unknown manner to provide sensory input.
FIG. 7.
Alignment of S. coelicolor SCO0781 and SCO0869 protein sequences with other anti-sigma factor antagonists. Internal portions from S. coelicolor SCO0781 and SCO0869 were compared to anti-σF antagonists from B. subtilis (BsSpoIIAA and BsRsbV) and S. coelicolor (ScoRsbV). The conserved serine residue phosphorylated by serine kinase anti-sigma factors is marked by an arrow but is replaced by a cysteine residue in SCO0781.
SCO4677 protein interacts with SCO0869 in vitro.
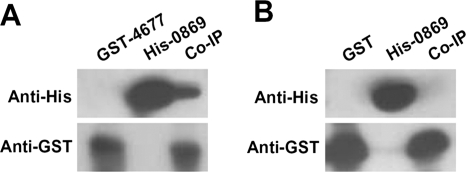
We confirmed the interaction of SCO4677 with SCO0869 in vitro by coimmunoprecipitation, using purified recombinant GST-SCO4677 and His-SCO0869 expressed in E. coli. The GST-SCO4677 and His-SCO0869 were incubated together and then further incubated with anti-GST antibody. GST antibodies were able to pull down not only GST-SCO4677 but also His-SCO0869, indicating the interaction of SCO4677 and SCO0869 (Fig. 8A). When His-SCO0869 and GST were incubated together and immunoprecipitated by anti-GST antibody, GST was detected but not SCO0869, confirming that the coimmunoprecipitation of His-SCO0869 with GST-SCO4677 was specific (Fig. 8B).
FIG. 8.
Interaction of SCO4677 with SCO0869 in vitro. Purified GST tagged SCO4677 (GST-4677) and His-tagged SCO0869 (His-0869) were used for coimmunoprecipitation. (A) GST-SCO4677 and His-SCO0869 were incubated together and immunoprecipitated with anti-GST antibodies prior to analysis by Western blotting. (B) For a negative control, GST and His-SCO0869 were incubated together and immunoprecipitated with anti-GST antibodies prior to analysis by Western blotting. The blots were detected with anti-GST or anti-His antibodies as indicated. Control tracks of the individual proteins (untreated with antibodies) were included as size standards.
DISCUSSION
A role of SCO4677 protein as an anti-sigma factor was experimentally supported by our evidence that it interacts directly with σF (encoded by SCO4035). It did not interact with σB or σH, both of which have previously been shown to interact with HATPase_c domain-containing anti-sigmas, so the interaction has considerable specificity. We therefore tentatively suggest that SCO4677 protein should be named RsfA (for regulator of sigma F antagonist). It is unusual for an anti-sigma to be encoded by a gene located far from the cognate sigma factor determinant. In the present study, we tested the interaction of SCO4677 with only four sigma factors, so it is quite possible that there might be other interacting partners of SCO4677.
The SCO4677 null mutant showed an accelerated sporulation and antibiotic production, perhaps associated with an early entry into transition phase, whereas sigF-null mutants were previously reported to have a white (43) or green (29) colony phenotype with delayed sporulation even after prolonged incubation. These somewhat complementary phenotypes support the hypothesis that the function of σF in morphological differentiation is inhibited by RsfA. The effect of SCO4677 disruption on Act antibiotic production was unexpected, since it is clear that σF is maximally abundant in developing spore chains, which do not produce Act, and Act production was not affected in a sigF-null mutant (28). One possible way to account for effects of SCO4677 at earlier stages of colony development would be to invoke interactions of RsfA with other partners (see below), though other less direct mechanisms, such as effects of the change in the growth curve, are also possible.
In the case, for example, of the SpoIIAB anti-sigma factor of B. subtilis, a further protein, SpoIIAA, interacts with SpoIIAB to alleviate the inhibition of the SpoIIAC sigma factor, and similar anti-sigma antagonists are often associated with SpoIIAB-like proteins (1, 9, 10, 13, 26, 36, 37, 48). It was therefore very striking that the SCO0869 and SCO0781 gene products, both of which weakly resemble SpoIIAA-like anti-anti-sigma factors, interacted with RsfA in the Y2H analysis, indicating that SCO0869 and SCO0781 encode anti-anti-sigmaF proteins. Thus, RsfA occupies the same place in a partner-switching relationship with a sigma factor and anti-anti-sigma factors as known anti-sigma factors.
Anti-anti-sigma factors such as SpoIIAA typically have a conserved serine residue that is phosphorylated by the serine kinase activity of the cognate anti-sigma (39). Interestingly, the serine residue is evident only in the SCO0869 protein but not in the SCO0781 protein (Fig. 7). Perhaps RsfA interacts in vivo with either anti-anti-sigma factor, with the SCO0869 protein interaction being controlled by RsfA-dependent phosphorylation and the SCO0781 interaction being responsive to a different input. The interaction of an anti-sigma factor with more than one anti-anti-sigma has not been observed before. Perhaps RsfA is relatively promiscuous in its partner choices. This might explain why SCO4677 is not located near either of the genes for its antagonists. Indeed, most SCO4677 homologues in S. coelicolor are located away from any recognizable candidate genes for such antagonists.
Although RsfA and SpoIIAB exhibited a low sequence similarity, the residues involved in ATP binding were conserved, as were those in many of the 27 SpoIIAB-like S. coelicolor proteins listed by Gaskell et al. (17) (which did not include the SCO4677 product, emphasizing the divergence of this protein family). Close homologues of RsfA are present in some other streptomycetes: SAV3140 of S. avermitilis showed 82%, and SGR4332 of S. griseus 66%, end-to-end similarity with RsfA (http://avermitilis.ls.kitasato-u.ac.jp).
Among the ca. 40 HATPase_c domain-containing genes in the S. coelicolor chromosome, 60% are similar in size to SCO4677 (which encodes 144 amino acids). The remaining 40% encode products with long N-terminal extensions, always including GAF and/or PAS domains, which are commonly associated with signal sensing and transduction. Thus, any anti-sigma activity associated with these proteins may be regulated by sensory input through these domains. In every case in which such domains are present, there is also a predicted serine phosphatase domain like that of SpoIIE in B. subtilis, which dephosphorylates SpoIIAA∼P to activate the forespore developmental program. This may indicate that all of these large proteins also interact with anti-anti-sigma partners in a manner controlled by the phosphorylation levels of those partners.
The abundant HATPase_c-encoding genes greatly outnumber both spoIIAA-like anti-anti-sigma factor genes and sigF-like sigma factor genes in the genome of S. coelicolor. An important question for the future will be to understand the interaction partners of all of these presumptive anti-sigma factors. From this point of view, the finding that RsfA interacts with more than one anti-anti-sigma may be significant: there could be further partners, including anti-sigmas for other sigma factors, for either or both of the SCO0781 and SCO0869 proteins. If, as seems possible, more of these interactions should prove to be somewhat promiscuous, the interpretation of mutant phenotypes will become difficult, since an absence of any one interacting partner may have knockon consequences for multiple signal transduction pathways.
Recently, in a combined transcriptome/proteome analysis of liquid cultures, it was reported that the expression of SCO4677 was strongly associated with the stationary phase and was reduced markedly in a bldA mutant (23). This indication that SCO4677 should function mainly to influence postgrowth processes is consistent with the phenotype of the ΔSCO4677 mutant and suggests that whatever sensory input is transmitted through bldA should potentially influence σF activity. Since there is no TTA codon in SCO4677 through which bldA might have a direct regulatory influence, it is possible that SCO4677 is itself dependent on a TTA-containing regulatory gene. A well-known and important TTA-containing developmental regulatory gene is adpA (bldH) (40, 49). Some features of the DNA sequence recognized by AdpA have been elucidated (51). However, we could not recognize such sequence features upstream of SCO4677.
Supplementary Material
Acknowledgments
This study was supported by the KICOS through a grant provided by the Korean Ministry of Science and Technology in 2007 (K20726000001-07B0100-00110) for collaborative research with the EU ActinoGEN project (IP005224).
E.S.K., J.Y.S., and D.W.K. were supported by the second stage of the Brain Korea 21 project.
Footnotes
Published ahead of print on 12 September 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Benson, A. K., and W. G. Haldenwang. 1993. Bacillus subtilis σB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc. Natl. Acad. Sci. USA 902330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417141-147. [DOI] [PubMed] [Google Scholar]
- 3.Bibb, M. J., V. Molle, and M. J. Buttner. 2000. σBldN, an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2). J. Bacteriol. 1824606-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breeden, L., and K. Nasmyth. 1985. Regulation of the yeast HO gene. Cold Spring Harbor Symp. Quant. Biol. 50643-650. [DOI] [PubMed] [Google Scholar]
- 5.Chater, K. F., C. J. Bruton, K. A. Plaskitt, M. J. Buttner, C. Méndez, and J. D. Helmann. 1989. The developmental fate of S. coelicolor hyphae depends upon a gene product homologous with the motility σ factor of B. subtilis. Cell 59133-143. [DOI] [PubMed] [Google Scholar]
- 6.Chater, K. F. 1993. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47685-713. [DOI] [PubMed] [Google Scholar]
- 7.Cho, Y. H., E. J. Lee, B. E. Ahn, and J. H. Roe. 2001. SigB, an RNA polymerase sigma factor required for osmoprotection and proper differentiation of Streptomyces coelicolor. Mol. Microbiol. 42205-214. [DOI] [PubMed] [Google Scholar]
- 8.Dalton, K. A., A. Thibessard, J. I. Hunter, and G. H. Kelemen. 2007. A novel compartment, the “subapical stem” of the aerial hyphae, is the location of a sigN-dependent, developmentally distinct transcription in Streptomyces coelicolor. Mol. Microbiol. 64719-737. [DOI] [PubMed] [Google Scholar]
- 9.Decatur, A. L., and R. Losick. 1996. Three sites of contact between the Bacillus subtilis transcription factor σF and its antisigma factor SpoIIAB. Genes Dev. 102348-2358. [DOI] [PubMed] [Google Scholar]
- 10.Delumeau, O., R. J. Lewis, and M. D. Yudkin. 2002. Protein-protein interactions that regulate the energy stress activation of σB in Bacillus subtilis. J. Bacteriol. 1845583-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diederich, B., J. F. Wilkinson, T. Magnin, M. Najafi, J. Errington, and M. D. Yudkin. 1994. Role of interactions between SpoIIAA and SpoIIAB in regulating cell-specific transcription factor σF of Bacillus subtilis. Genes Dev. 82653-2663. [DOI] [PubMed] [Google Scholar]
- 12.Dufour, A., and W. G. Haldenwang. 1994. Interaction between a Bacillus subtilis anti-σ factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 1761813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan, L., and R. Losick. 1993. SpoIIAB is an anti-σ factor that binds to and inhibits transcription by regulatory protein σF from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 902325-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan, L., S. Alper, and R. Losick. 1996. SpoIIAA governs the release of the cell-type specific transcription factor σF from its anti-sigma factor SpoIIAB. J. Mol. Biol. 260147-164. [DOI] [PubMed] [Google Scholar]
- 15.Estojak, J., R. Brent, and E. A. Golemis. 1995. Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell. Biol. 155820-5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernández-Moreno, M. A., A. J. Martín-Triana, E. Martínez, J. Niemi, H. M. Kieser, D. A. Hopwood, and F. Malpartida. 1992. abaA, a new pleiotropic regulatory locus for antibiotic production in Streptomyces coelicolor. J. Bacteriol. 1742958-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaskell, A. A., J. C. Crack, G. H. Kelemen, M. I. Hutchings, and N. E. Le Brun. 2007. RsmA is an anti-sigma factor that modulates its activity through a [2Fe-2S] cluster cofactor. J. Biol. Chem. 28231812-31820. [DOI] [PubMed] [Google Scholar]
- 18.Gehring, A. M., N. J. Yoo, and R. Losick. 2001. RNA polymerase sigma factor that blocks morphological differentiation by Streptomyces coelicolor. J. Bacteriol. 1835991-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon, N. D., G. L. Ottaviano, S. E. Connell, G. V. Tobkin, C. H. Son, S. Shterental, and A. M. Gehring. 2008. Secreted-protein response to σU activity in Streptomyces coelicolor. J. Bacteriol. 190894-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 1001541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 22.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75805-816. [DOI] [PubMed] [Google Scholar]
- 23.Hesketh, A., G. Bucca, E. Laing, F. Flett, G. Hotchkiss, C. P. Smith, and K. F. Chater. 2007. New pleiotropic effects of eliminating a rare tRNA from Streptomyces coelicolor, revealed by combined proteomic and transcriptomic analysis of liquid cultures. BMC Genomics 8261. http://www.biomedcentral.com/1471-2164/8/261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Höltke, H. J., W. Ankenbauer, K. Mühlegger, R. Rein, G. Sanger, R. Seibl, and T. Walter. 1995. The digoxigenin (DIG) system for non-radioactive labeling and detection of nucleic acids-an overview. Cell. Mol. Biol. 41883-905. [PubMed] [Google Scholar]
- 25.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1441425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang, C. M., M. S. Brody, S. Akbar, X. Yang, and C. W. Price. 1996. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor σB in response to environmental stress. J. Bacteriol. 1783846-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang, J. G., M. S. Paget, Y. J. Seok, M. Y. Hahn, J. B. Bae, J. S. Hahn, C. Kleanthous, M. J. Buttner, and J. H. Roe. 1999. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 184292-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelemen, G. H., G. L. Brown, J. Kormanec, L. Potúčková, K. F. Chater, and M. J. Buttner. 1996. The positions of the sigma-factor genes, whiG and sigF, in the hierarchy controlling the development of spore chains in the aerial hyphae of Streptomyces coelicolor A3(2). Mol. Microbiol. 21593-603. [DOI] [PubMed] [Google Scholar]
- 29.Kelemen, G. H., P. Brian, K. Flärdh, L. Chamberlin, K. F. Chater, and M. J. Buttner. 1998. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3 (2). J. Bacteriol. 1802515-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelemen, G. H., P. H. Viollier, J. Tenor, L. Marri, M. J. Buttner, and C. J. Thompson. 2001. A connection between stress and development in the multicellular prokaryote Streptomyces coelicolor A3(2). Mol. Microbiol. 40804-814. [DOI] [PubMed] [Google Scholar]
- 31.Keston, A. 1956. Specific colorimetric enzymatic reagents for glucose, abstr. 31c. Abstr. 129th Meet. Am. Chem. Soc. American Chemical Society, Washington, DC.
- 32.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom.
- 33.Lee, E. J., Y. H. Cho, H. S. Kim, B. E. Ahn, and J. H. Roe. 2004. Regulation of σB by an anti- and an anti-anti-sigma factor in Streptomyces coelicolor in response to osmotic stress. J. Bacteriol. 1868490-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, E. J., N. Karoonuthaisiri, H. S. Kim, J. H. Park, C. J. Cha, C. M. Kao, and J. H. Roe. 2005. A master regulator σB governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Mol. Microbiol. 571252-1264. [DOI] [PubMed] [Google Scholar]
- 35.MacNeil, D. J., J. L. Occi, K. M. Gewain, T. MacNeil, P. H. Gibbons, C. L. Ruby, and S. J. Danis. 1992. Complex organization of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene 115119-125. [DOI] [PubMed] [Google Scholar]
- 36.Magnin, T., M. Lord, J. Errington, and M. D. Yudkin. 1996. Establishing differential gene expression in sporulating Bacillus subtilis: phosphorylation of SpoIIAA (anti-anti-σF) alters its conformation and prevents formation of a SpoIIAA/SpoIIAB/ADP complex. Mol. Microbiol. 19901-907. [DOI] [PubMed] [Google Scholar]
- 37.Min, K. T., C. M. Hilditch, B. Diederich, J. Errington, and M. D. Yudkin. 1993. σF, the first compartment-specific transcription factor of Bacillus subtilis, is regulated by an anti-σ factor that is also a protein kinase. Cell 74735-742. [DOI] [PubMed] [Google Scholar]
- 38.Mittenhuber, G. 2002. A phylogenomic study of the general stress response sigma factor σB of Bacillus subtilis and its regulatory proteins. J. Mol. Microbiol. Biotechnol. 4427-452. [PubMed] [Google Scholar]
- 39.Najafi, S. M., A. C. Willis, and M. D. Yudkin. 1995. Site of phosphorylation of SpoIIAA, the anti-anti-sigma factor for sporulation-specific σF of Bacillus subtilis. J. Bacteriol. 1772912-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen, K. T., J. Tenor, H. Stettler, L. T. Nguyen, L. D. Nguyen, and C. J. Thompson. 2003. Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within the adpA gene. J. Bacteriol. 1857291-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novotna, J., J. Vohradsky, P. Berndt, H. Gramajo, H. Langen, X. M. Li, W. Minas, L. Orsaria, D. Roeder, and C. J. Thompson. 2003. Proteomic studies of diauxic lag in the differentiating prokaryote Streptomyces coelicolor reveal a regulatory network of stress-induced proteins and central metabolic enzymes. Mol. Microbiol. 481289-1303. [DOI] [PubMed] [Google Scholar]
- 42.Pirt, S. J. 1975. Principles of microbe and cell cultivation, p.29-41. Blackwell Scientific Publications, Oxford, United Kingdom.
- 43.Potúčková, L., G. H. Kelemen, K. C. Findlay, M. A. Lonetto, M. J. Buttner, and J. Kormanec. 1995. A new RNA polymerase sigma factor, σF, is required for the late stages of morphological differentiation in Streptomyces spp. Mol. Microbiol. 1737-48. [DOI] [PubMed] [Google Scholar]
- 44.Ryu, Y. G., M. J. Butler, K. F. Chater, and K. J. Lee. 2006. Engineering of primary carbohydrate metabolism for increased production of actinorhodin in Streptomyces coelicolor. Appl. Environ. Microbiol. 727132-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Sevciková, B., O. Benada, O. Kofronova, and J. Kormanec. 2001. Stress response sigma factor σH is essential for morphological differentiation of Streptomyces coelicolor A3(2). Arch. Microbiol. 17798-106. [DOI] [PubMed] [Google Scholar]
- 47.Sevciková, B., and J. Kormanec. 2002. Activity of the Streptomyces coelicolor stress-response sigma factor σH is regulated by an anti-sigma factor. FEMS Microbiol. Lett. 209229-235. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt, R., P. Margolis, L. Duncan, R. Coppolecchia, C. P. Moran Jr., and R. Losick. 1990. Control of developmental transcription factor σF by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 879221-9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takano, E., M. Tao, F. Long, M. J. Bibb, L. Wang, W. Li, M. J. Buttner, M. J. Bibb, Z. X. Deng, and K. F. Chater. 2003. A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Mol. Microbiol. 50475-486. [DOI] [PubMed] [Google Scholar]
- 50.Viollier, P. H., G. H. Kelemen, G. E. Dale, K. T. Nguyen, M. J. Buttner, and C. J. Thompson. 2003. Specialized osmotic stress response systems involve multiple SigB-like sigma factors in Streptomyces coelicolor. Mol. Microbiol. 47699-714. [DOI] [PubMed] [Google Scholar]
- 51.Yamazaki, H., A. Tomono, Y. Ohnishi, and S. Horinouchi. 2004. DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 53555-572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.