Abstract
Vibrio cholerae, the causative agent of the disease cholera, can generate rugose variants that have an increased capacity to form biofilms. Rugosity and biofilm formation are critical for the environmental survival and transmission of the pathogen, and these processes are controlled by cyclic diguanylate (c-di-GMP) signaling systems. c-di-GMP is produced by diguanylate cyclases (DGCs) and degraded by phosphodiesterases (PDEs). Proteins that contain GGDEF domains act as DGCs, whereas proteins that contain EAL or HD-GYP domains act as PDEs. In the V. cholerae genome there are 62 genes that are predicted to encode proteins capable of modulating the cellular c-di-GMP concentration. We previously identified two DGCs, VpvC and CdgA, that can control the switch between smooth and rugose. To identify other c-di-GMP signaling proteins involved in rugosity, we generated in-frame deletion mutants of all genes predicted to encode proteins with GGDEF and EAL domains and then searched for mutants with altered rugosity. In this study, we identified two new genes, cdgG and cdgH, involved in rugosity control. We determined that CdgH acts as a DGC and positively regulates rugosity, whereas CdgG does not have DGC activity and negatively regulates rugosity. In addition, epistasis analysis with CdgG, CdgH, and other DGCs and PDEs controlling rugosity revealed that CdgG and CdgH act in parallel with previously identified c-di-GMP signaling proteins to control rugosity in V. cholerae. We also determined that PilZ domain-containing c-di-GMP binding proteins contribute minimally to rugosity, indicating that there are additional c-di-GMP binding proteins controlling rugosity in V. cholerae.
Vibrio cholerae causes cholera and is a natural inhabitant of aquatic environments (14, 34). Seasonal cholera outbreaks occur where the disease is endemic and can spread worldwide. V. cholerae's ability to cause epidemics is tied to its ability to survive in aquatic habitats (14, 34). One key factor that is important for environmental survival and transmission of V. cholerae is the microbe's ability to form biofilms, which are matrix-enclosed surface-associated communities (1, 14, 34).
V. cholerae can generate colony variants, termed smooth and rugose, that differ significantly in their biofilm-forming capacities (58). Smooth-to-rugose conversion occurs spontaneously under a variety of conditions, including carbon limitation, growth in biofilms, and treatment with bactericidal agents (39, 54, 58). Rugose variants have been isolated from environmental biofilm samples collected in Bangladesh, indicating that the smooth-to-rugose switch can also occur in natural environments (24). In addition, Morris et al. have shown that rugose variants can cause cholera when given orally to human volunteers, thus demonstrating that rugose variants can infect humans (39). Several molecular mechanisms controlling the smooth-to-rugose switch have been found, but each of these mechanisms does not function in all strains. The identified molecular alterations to create rugosity include the loss of HapR (the master regulator of quorum sensing) (19, 57, 59), FlaA (a major flagellin subunit) (29, 55), or CytR (a regulator of nucleoside uptake and catabolism) (20). In our prototype strain (V. cholerae O1 El Tor, A1552), hapR mutants form rugose colonies, but flaA and cytR mutants form smooth colonies. These results demonstrate that there are multiple ways by which the smooth-to-rugose switch can take place.
Rugosity and formation of mature biofilms require extracellular matrix components. A major component of the V. cholerae biofilm matrix is the VPS (named for Vibrio polysaccharide) exopolysaccharide. VPS production is essential for the development of three-dimensional biofilm structures (58) and is mediated by proteins encoded by the vps genes, which are organized into vps-I and vps-II clusters on the large chromosome (58). Protein components of the V. cholerae biofilm matrix are also required for rugosity and the formation of a wild-type biofilm (15, 16). Biofilm matrix production is positively controlled by transcriptional regulators VpsR and VpsT (8, 56) and negatively regulated by the quorum-sensing transcriptional regulator HapR (19, 57, 59), as well as the cyclic AMP (cAMP) and cyclic AMP receptor protein regulatory complex (31).
Cyclic diguanylate (c-di-GMP) has emerged as a ubiquitous second messenger in bacteria that controls the transition from a free-living, motile lifestyle to a biofilm lifestyle (42). c-di-GMP production and degradation is controlled by diguanylate cyclases (DGCs) and phosphodiesterases (PDEs), respectively. Proteins that contain GGDEF domains act as DGCs, whereas proteins that contain EAL or HD-GYP domains act as PDEs (44, 46, 47). In addition, proteins carrying PilZ domains, which are shown to bind c-di-GMP, are one type of downstream protein that relays signals to cellular processes (11, 36, 40, 45). V. cholerae has 31 genes that encode proteins with a GGDEF domain, 12 genes that encode proteins with an EAL domain, 10 genes that encodes proteins with both GGDEF and EAL domains, 9 genes that encode proteins with a HD-GYP domain, and 4 genes that encode proteins with a conserved PilZ domain (2, 18). Studies to date have shown that c-di-GMP regulates biofilm formation, motility, virulence, and smooth-to-rugose phase variation in V. cholerae (5-7, 27, 32, 40, 41, 50, 52, 53). We recently demonstrated that rugose variants have increased c-di-GMP levels compared to smooth variants, leading to elevated biofilm formation in the rugose forms (4, 6, 32). The rugosity-associated increase in c-di-GMP in our prototype rugose strain is caused by a single amino acid change in a DGC protein, which we called VpvC (6). However, other genetic variations can also cause rugosity. For example, we found that increased transcription of cdgA, encoding another DGC, causes rugosity in hapR mutants (4). Furthermore, an increase in c-di-GMP due to loss of a key PDE, CdgC, MbaA, or RocS, in the rugose genetic background leads to formation of super-rugose colonies that are more opaque and corrugated than rugose (7, 32, 41).
In the present study, we investigated whether other genes encoding DGCs and PDEs contribute to rugosity and, in turn, biofilm formation in V. cholerae. We identified and characterized two such genes, cdgG and cdgH, encoding GGDEF domain proteins. Through epistasis analysis, we determined that many c-di-GMP signaling proteins act in parallel pathways to control rugosity. We also determined that PilZ domain-containing c-di-GMP receptors contribute minimally to rugosity, indicating that there are additional c-di-GMP receptors controlling rugosity.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in the present study are listed in Table 1. Escherichia coli DH10B and CC118 λpir strains were used for DNA manipulation, and E. coli S17-1 λpir strains were used for conjugation with V. cholerae. Knockout mutants of V. cholerae strains and V. cholerae strains carrying plasmids with lacZ transcriptional fusions and multicopy vectors were generated as described earlier (32). V. cholerae cultures were grown in Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, 1% NaCl [pH 7.5]) with aeration at 30°C. E. coli cultures were grown in the same medium with aeration at 37°C. Antibiotics (rifampin and ampicillin) were added at 100 μg/ml unless otherwise noted. For the induction of gene expression in strains carrying arabinose-inducible vectors, l-arabinose was added to the growth medium at a final concentration of 0.2% (wt/vol) for c-di-GMP quantification experiments and 0.02% (wt/vol) for complementation analysis via colony morphology and biofilm assays.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| CC118λpir | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1 λpir | 21 |
| S17-1λpir | recA thi pro rK− mK+ RP4:2-Tc::MuKm Tn7 Tpr Smr λpir | 13 |
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 endA1 araΔ139 Δ(ara leu)7697 galU galK λ−rpsL (Strr) nupG | Invitrogen |
| V. cholerae | ||
| Fy_Vc_1 | V. cholerae O1 El Tor A1552, smooth variant, Rifr | 57 |
| Fy_Vc_2 | V. cholerae O1 El Tor A1552, rugose variant, Rifr | 57 |
| Fy_Vc_3 | Fy_Vc_1 ΔlacZ, Rifr | 8 |
| Fy_Vc_4 | Fy_Vc_2 ΔlacZ, Rifr | 8 |
| Fy_Vc_237 | Fy_Vc_1 mTn7-GFP, Rifr Gmr | 5 |
| Fy_Vc_240 | Fy_Vc_2 mTn7-GFP, Rifr Gmr | 32 |
| Fy_Vc_231 | Fy_Vc_1 Δvps-I operon, Rifr | 5 |
| Fy_Vc_234 | Fy_Vc_2 Δvps-I operon, Rifr | 6 |
| Fy_Vc_337 | Fy_Vc_1 ΔflaA, Rifr | This study |
| Fy_Vc_339 | Fy_Vc_2 ΔflaA, Rifr | This study |
| Fy_Vc_970 | Fy_Vc_1 ΔVC0900, Rifr | This study |
| Fy_Vc_3074 | Fy_Vc_1 ΔVC0900 ΔlacZ, Rifr | This study |
| Fy_Vc_3083 | Fy_Vc_1 ΔVC0900 mTn7-GFP, Rifr Gmr | This study |
| Fy_Vc_917 | Fy_Vc_2 ΔVC0900, Rifr | This study |
| Fy_Vc_3071 | Fy_Vc_2 ΔVC0900 ΔlacZ, Rifr | This study |
| Fy_Vc_3080 | Fy_Vc_2 ΔVC0900 mTn7-GFP, Rifr Gmr | This study |
| Fy_Vc_1592 | Fy_Vc_1 ΔVC1067, Rifr | This study |
| Fy_Vc_3077 | Fy_Vc_1 ΔVC1067 ΔlacZ, Rifr | This study |
| Fy_Vc_2089 | Fy_Vc_1 ΔVC1067 mTn7-GFP, Rifr Gmr | This study |
| Fy_Vc_861 | Fy_Vc_2 ΔVC1067, Rifr | This study |
| Fy_Vc_1139 | Fy_Vc_2 ΔVC1067 ΔlacZ, Rifr | This study |
| Fy_Vc_1232 | Fy_Vc_2 ΔVC1067 mTn7-GFP, Rifr Gmr | This study |
| Fy_Vc_3211 | Fy_Vc_1 ΔVC0900 ΔVC1067, Rifr | This study |
| Fy_Vc_3217 | Fy_Vc_1 ΔVC0900 ΔVC1067 ΔlacZ, Rifr | This study |
| Fy_Vc_3208 | Fy_Vc_2 ΔVC0900 ΔVC1067, Rifr | This study |
| Fy_Vc_3214 | Fy_Vc_2 ΔVC0900 ΔVC1067 ΔlacZ, Rifr | This study |
| Fy_Vc_1745 | Fy_Vc_2 ΔvpvC, Rifr | 6 |
| Fy_Vc_3584 | Fy_Vc_2 ΔvpvC ΔVC1067, Rifr | This study |
| Fy_Vc_183 | Fy_Vc_1 ΔhapR, Rifr | 4 |
| Fy_Vc_344 | Fy_Vc_1 ΔcdgA, Rifr | 32 |
| Fy_Vc_1675 | Fy_Vc_1 ΔhapR ΔcdgA, Rifr | 4 |
| Fy_Vc_3635 | Fy_Vc_1 ΔhapR ΔVC1067, Rifr | This study |
| Fy_Vc_3638 | Fy_Vc_1 ΔcdgA ΔVC1067, Rifr | This study |
| Fy_Vc_3643 | Fy_Vc_1 ΔhapR ΔcdgA ΔVC1067, Rifr | This study |
| Fy_Vc_346 | Fy_Vc_1 ΔcdgC, Rifr | 32 |
| Fy_Vc_354 | Fy_Vc_1 ΔrocS, Rifr | 32 |
| Fy_Vc_356 | Fy_Vc_1 ΔmbaA, Rifr | 32 |
| Fy_Vc_3599 | Fy_Vc_1 ΔrocS ΔcdgC, Rifr | This study |
| Fy_Vc_3611 | Fy_Vc_1 ΔrocS ΔmbaA, Rifr | This study |
| Fy_Vc_3602 | Fy_Vc_1 ΔmbaA ΔcdgC, Rifr | This study |
| Fy_Vc_3614 | Fy_Vc_1 ΔrocS ΔmbaA ΔcdgC, Rifr | This study |
| Fy_Vc_3590 | Fy_Vc_1 ΔrocS ΔVC0900, Rifr | This study |
| Fy_Vc_3593 | Fy_Vc_1 ΔmbaA ΔVC0900, Rifr | This study |
| Fy_Vc_3587 | Fy_Vc_1 ΔcdgC ΔVC0900, Rifr | This study |
| Fy_Vc_3617 | Fy_Vc_1 ΔrocS ΔmbaA ΔcdgC ΔVC0900, Rifr | This study |
| Fy_Vc_1643 | Fy_Vc_2 ΔplzA, Rifr | This study |
| Fy_Vc_1647 | Fy_Vc_2 ΔplzB, Rifr | This study |
| Fy_Vc_1651 | Fy_Vc_2 ΔplzC, Rifr | This study |
| Fy_Vc_1636 | Fy_Vc_2 ΔplzD, Rifr | This study |
| Fy_Vc_1640 | Fy_Vc_2 ΔplzE, Rifr | This study |
| Fy_Vc_2504 | Fy_Vc_2 ΔplzA ΔplzC ΔplzD ΔplzE, Rifr | This study |
| Fy_Vc_3655 | Fy_Vc_2 ΔplzA ΔplzB ΔplzC ΔplzD ΔplzE, Rifr | This study |
| Fy_Vc_3661 | Fy_Vc_2 ΔplzA ΔplzC ΔplzD ΔplzE mTn7-GFP, Rifr | This study |
| Fy_Vc_3781 | Fy_Vc_2 ΔvpvC ΔplzC, Rifr | This study |
| Fy_Vc_3784 | Fy_Vc_2 ΔVC1067 ΔplzC, Rifr | This study |
| Plasmids | ||
| pGP704-sac28 | pGP704 derivative; sacB, Apr | 8 |
| pFY-449 | pGP704-sac28:: ΔVC0900, Apr | This study |
| pFY-399 | pGP704-sac28:: ΔVC1067, Apr | This study |
| pCC2 | pGP704-sac28:: ΔlacZ, Apr | 8 |
| pSM1 | pWM91:: ΔflaA(VC2188) | 38 |
| pAJH5 | pWM91:: Δvps-I operon | 20 |
| pFY-367 | pGP704-sac28:: ΔvpvC, Apr | 6 |
| pFY-9 | pGP704-sac28:: ΔhapR, Apr | Gary Schoolnik |
| pFY-149 | pGP704-sac28:: ΔcdgA, Apr | 32 |
| pFY-151 | pGP704-sac28:: ΔcdgC, Apr | 32 |
| pFY-161 | pGP704-sac28:: ΔrocS, Apr | 32 |
| pFY-163 | pGP704-sac28:: ΔmbaA, Apr | 32 |
| pFY-640 | pGP704-sac28:: ΔplzA, Apr | This study |
| pFY-641 | pGP704-sac28:: ΔplzB, Apr | This study |
| pFY-642 | pGP704-sac28:: ΔplzC, Apr | This study |
| pFY-643 | pGP704-sac28:: ΔplzD, Apr | This study |
| pFY-644 | pGP704-sac28:: ΔplzE, Apr | This study |
| pMCM11 | pGP704::mTn7-GFP, Gmr Apr | 32 |
| pUX-BF13 | oriR6K helper plasmid, provides Tn7 transposition function in trans, Apr | 3 |
| pBAD/myc-His-B | Arabinose-inducible expression vector with C-terminal myc epitope and His6 tags | Invitrogen |
| pFY-573 | pBAD/myc-His-B::VC0900, Apr | This study |
| pFY-572 | pBAD/myc-His-B::VC1067, Apr | This study |
| pFY-590 | pBAD/myc-His-B::VC0900, E445A and E446A mutations, Apr | This study |
| pFY-645 | pBAD/myc-His-B::VC0900, R434A and D437A mutations, Apr | This study |
| pFY-589 | pBAD/myc-His-B::VC1067, G605A mutation, Apr | This study |
| pCC12 | pRS415 vpsL promoter, Apr | 8 |
| pCC10 | pRS415 vpsR promoter, Apr | 8 |
| pCC25 | pRS415 vpsT promoter, Apr | 8 |
Rifr, rifampin resistance; Apr, ampicillin resistance; Gmr, gentamicin resistance.
DNA manipulations.
The DNA oligonucleotides used in the present study were purchased from Operon Technologies (Alameda, CA) and are listed in Table S1 in the supplemental material. A PCR method was used to generate in-frame deletions of the cdgG and cdgH genes utilizing previously published methods (32). Deletion vectors were constructed by using pGP704sac28 suicide plasmid as described previously (32). For overexpression studies, cdgG and cdgH were cloned into pBAD/myc-His-B plasmid using the primers listed (see Table S1 in the supplemental material). Construction of cdgG and cdgH vectors harboring point mutations was done utilizing a previously published method (6). The deletion and overexpression constructs were sequenced (UC Berkeley DNA Sequencing Facility), and only the clones without any undesired mutations were utilized.
Generation of V. cholerae deletion mutants and green fluorescent protein tagging of V. cholerae strains.
V. cholerae deletion mutants and green fluorescent protein-tagged V. cholerae strains were generated as described previously (17, 32).
β-Galactosidase assays.
β-Galactosidase assays were performed and Miller units were calculated as previously described (32, 37).
c-di-GMP quantification.
The amount of c-di-GMP was quantified by two-dimensional thin-layer chromatography (2D-TLC) as previously described (32, 52) with the following modifications. Briefly, bacteria were grown in morpholinepropanesulfonic acid (MOPS) minimal medium containing 0.75 mM KH2PO4 at 30°C for overnight. The cells were then diluted 1:50 into fresh MOPS minimal medium containing 0.25 mM KH2PO4 and grown to an optical density at 600 nm of 0.6 ± 0.05 at 30°C. Then, 50 μCi of [32P]orthophosphate was added to 0.5 ml of cell suspension, followed by further growth for 1 h. Labeled nucleotides were extracted by using previously published methods (52). Portions (10 μl) of total nucleotides were separated on TLC plates as previously described (52).
Colony morphology.
For colony morphology assays, V. cholerae colonies were grown on LB agar plates for 1 to 2 days at 30°C. Colonies were photographed by using a Nikon CoolPix 4500 digital camera.
Biofilm assays.
The biofilm-forming capacities of the V. cholerae strains were determined using cover glass chambers (Lab-Tek). Dilutions (1:100, 2 ml) in LB medium from overnight-grown cultures were placed into chambers. Biofilms were formed under static conditions at 30°C for 8 h, washed twice with 1 ml of LB medium, and then visualized by using confocal laser scanning microscopy (CLSM). Acquired images were analyzed with the COMSTAT program (22). For flow cell experiments, biofilms were grown at room temperature in flow chambers (individual channel dimensions of 1 by 4 by 40 mm) supplied with 2% LB medium supplemented with 0.9% NaCl (0.02% peptone, 0.01% yeast extract, 0.9% NaCl) at a flow rate of 4.5 ml/h. Assembly of the flow cell system and image acquisitions were done as previously described (57).
Motility assays.
LB soft agar plates (0.3% agar) were used to determine the motility of bacterial strains (57). The diameter of the migration zone was measured after 18 h of incubation at 30°C.
RESULTS
CdgH positively regulates rugosity-associated phenotypes.
In V. cholerae, there are a total of 62 genes that are predicted to encode proteins capable of producing or degrading c-di-GMP (18). Fifty-three of these genes encode proteins with GGDEF and/or EAL domains. We previously studied 10 of them and had shown that VpvC, CdgA, CdgC, MbaA, and RocS regulate rugosity in V. cholerae O1 El Tor A1552 strain (4, 6, 32). In the present study, we analyzed the contribution of the remaining genes encoding proteins with GGDEF and/or EAL domains to rugosity. To this end, we created in-frame deletions of all GGDEF/EAL genes (except VC0515, since the genomic region appears to be different between our prototype strain and the sequenced N16961 strain) in the rugose genetic background; we then screened the mutants for alterations in colony corrugation when grown on LB agar plates. Through this screen of 42 mutants, we identified two additional mutants exhibiting a significant change in colony corrugation.
The first mutant had a deletion in VC1067, which encodes a protein with a GGDEF domain, now termed cdgH for cyclic diguanylate H. The second mutant had a deletion in VC0900, which encodes a protein with a GGDEF domain, now termed cdgG for cyclic diguanylate G.
We first investigated the contribution of CdgH to rugosity-associated phenotypes by analyzing colony corrugation and biofilm formation. Colonies formed by the rugose cdgH mutant (RΔcdgH) on LB agar plates appeared less corrugated and flatter compared to those formed by rugose, a finding consistent with producing less VPS (Fig. 1A). Similarly, RΔcdgH formed thinner and less-structured biofilms compared to rugose after 8 h of biofilm development at 30°C under static conditions (Fig. 1B). To quantify the differences in biofilm architecture, we used the COMSTAT program and quantified various biofilm parameters: biomass, average thickness, maximum thickness, substratum coverage, roughness, and the surface area/volume ratio. Although, substratum coverage, roughness, and the surface area/volume ratio remained the same in RΔcdgH compared to rugose, a significant reduction in the biomass and average and maximum thicknesses was observed in RΔcdgH biofilms compared to rugose biofilms (Table 2). Rugosity and biofilm formation are directly linked to VPS production (58). Thus, we evaluated the transcription of vps genes in RΔcdgH using a vpsL-lacZ transcriptional fusion (vpsL is the first gene of the vps-II cluster). We also measured the transcription of genes encoding the positive regulators of VPS biosynthesis, VpsR and VpsT, using vpsR-lacZ and vpsT-lacZ transcriptional fusions. We observed a decrease in transcription of vpsL, vpsR, and vpsT genes in RΔcdgH compared to that in rugose (Fig. 1C). Taken together, we found CdgH is required for wild-type levels of vps expression and biofilm formation.
FIG. 1.
Phenotypic characterization of CdgH in the rugose genetic background. (A) Colony morphologies of rugose and RΔcdgH strains that were grown for 24 and 48 h on LB agar plates at 30°C. (B) Three-dimensional biofilm structures of rugose and RΔcdgH strains that are formed 8 h postinoculation under static conditions at 30°C. Images were acquired with CLSM, with top-down (large panes) and orthogonal (side panels) views of biofilms shown. Scale bar, 30 μm. (C) Transcription of vpsL-lacZ, vpsR-lacZ, and vpsT-lacZ fusions determined in rugose and RΔcdgH strains by measuring β-galactosidase activity in cells that were grown to mid-exponential phase in LB medium at 30°C. The result shown is representative of three independent experiments. Error bars represent the standard deviations. (D) Diameter of migration zone of rugose, RΔcdgH, RΔvps-I, and RΔflaA strains measured in LB soft agar plates (0.3%) after 18 h of incubation at 30°C. The data shown are representative of three independent experiments. Error bars represent the standard deviations.
TABLE 2.
COMSTAT analysis of biofilmsb
| Straina | Biomass (μm3/μm2) | Thickness (μm)
|
Substratum coverage | Roughness | Surface area to vol (μm2/μm3) | |
|---|---|---|---|---|---|---|
| Avg | Maximum | |||||
| Rugose | ||||||
| Rugose (WT) | 19.61 ± 3.01 | 29.35 ± 3.11 | 80.69 ± 14.60 | 0.95 ± 0.07 | 0.57 ± 0.10 | 1.43 ± 0.16 |
| RΔcdgG | 14.17 ± 3.87 | 21.00 ± 4.70 | 65.69 ± 19.72 | 0.93 ± 0.08 | 0.67 ± 0.01 | 1.57 ± 0.29 |
| RΔcdgH | 11.54 ± 2.66 | 15.91 ± 5.18 | 50.68 ± 9.98 | 0.95 ± 0.03 | 0.63 ± 0.07 | 1.48 ± 0.11 |
| Smooth | ||||||
| Smooth (WT) | 15.17 ± 2.39 | 16.48 ± 3.55 | 26.87 ± 3.22 | 0.98 ± 0.03 | 0.18 ± 0.01 | 1.37 ± 0.52 |
| SΔcdgG | 16.41 ± 3.14 | 18.31 ± 4.70 | 50.41 ± 9.01 | 0.85 ± 0.09 | 0.34 ± 0.06 | 1.38 ± 0.70 |
| SΔcdgH | 14.58 ± 3.99 | 15.24 ± 5.06 | 23.91 ± 4.49 | 0.98 ± 0.02 | 0.17 ± 0.04 | 1.08 ± 0.49 |
WT, wild type.
All values are means ± standard deviations derived from at least four z-series image stacks.
VPS biosynthesis and flagellar motility are inversely regulated by the c-di-GMP signaling system in V. cholerae (5, 6, 27, 32, 52, 58). Therefore, we sought to understand the effect of CdgH on motility. To this end, we measured the migration zone formed by RΔcdgH and rugose strains when grown on LB soft agar plates. RΔcdgH exhibited an increase in motility compared to rugose (Fig. 1D). We also used RΔflaA (a rugose strain harboring flaA deletion) and RΔvps-I (a rugose strain harboring vps-I cluster deletion) as controls in our assay. Since enhanced VPS production can negatively impact the motility of the rugose variant, increased motility behavior of the RΔcdgH could be due to decreased vps expression. Thus, we compared the motility of the RΔcdgH mutant to that of the RΔvps-I mutant. Motility of RΔcdgH was higher than that of RΔvps-I, indicating that the CdgH affect on motility is not due simply to a decrease in VPS production.
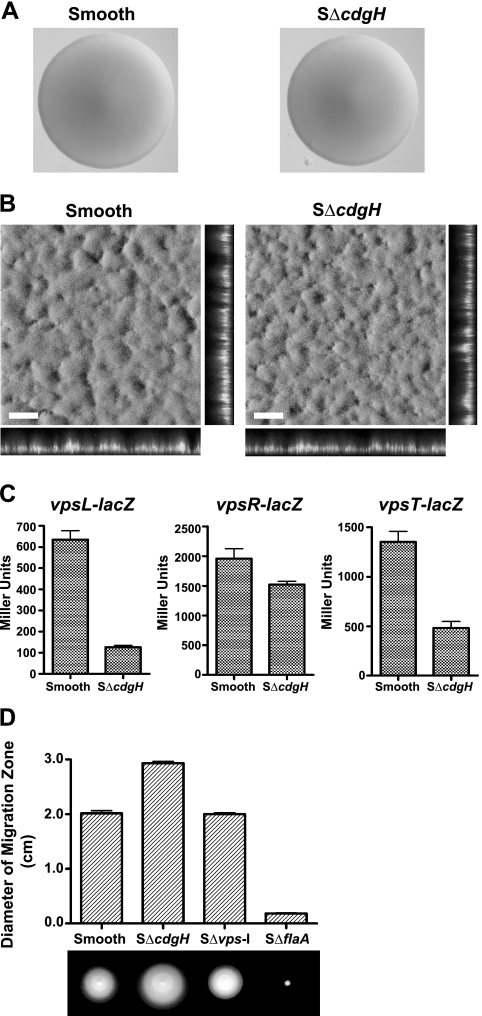
To gain further insight into the role of CdgH in rugosity, we also analyzed a cdgH mutant in the smooth genetic background. This approach is useful because smooth strains have lower cellular c-di-GMP levels, vps transcription, and VPS production, and thus one can assess alterations in these processes more readily. In the smooth genetic background, deletion of cdgH (SΔcdgH) did not affect colony morphology (Fig. 2A). SΔcdgH did not have a significant defect in biofilm formation as analyzed using the COMSTAT program (Fig. 2B and Table 2). Interestingly, the transcription of vpsL and vpsT was markedly decreased in SΔcdgH compared to smooth (Fig. 2C), indicating that while a mutation in ΔcdgH decreases vps gene expression, it is not sufficient to eliminate biofilm formation. We determined that SΔcdgH exhibited increased motility compared to smooth (Fig. 2D), further indicating that CdgH controls motility.
FIG. 2.
Phenotypic characterization of CdgH in the smooth genetic background. (A) Colony morphologies of smooth and SΔcdgH strains that were grown for 24 h on LB agar plates at 30°C. (B) Three-dimensional biofilm structures of smooth and SΔcdgH strains that are formed 24 h postinoculation in a once-through flow cell system. Images were acquired with CLSM, with top-down (large panes) and orthogonal (side panels) views of biofilms shown. Scale bar, 30 μm. (C) Transcription of vpsL-lacZ, vpsR-lacZ, and vpsT-lacZ fusions determined in smooth and SΔcdgH strains by measuring β-galactosidase activity in cells that were grown to mid-exponential phase in LB medium at 30°C. The result shown is representative of three independent experiments. Error bars represent the standard deviations. (D) Diameter of migration zone of smooth, SΔcdgH, SΔvps-I, and SΔflaA strains measured in LB soft agar plates (0.3%) after 18 h of incubation at 30°C. The data shown are representative of three independent experiments. Error bars represent the standard deviations.
CdgH acts as a DGC.
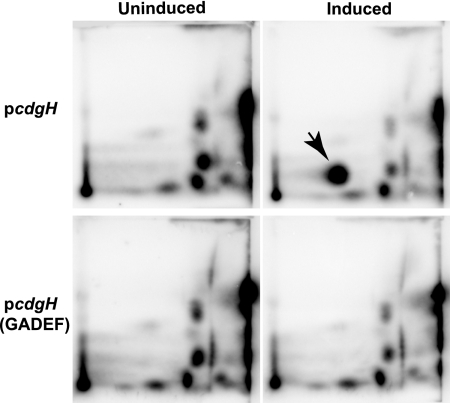
CdgH is predicted to have a GGDEF domain (PF00990). Sequence alignments showed that CdgH carries the conserved GGDEF residues in the active site (A-site) of the enzyme, which is reported to be required for the DGC activity (9). To test the enzymatic activity of CdgH, we overexpressed cdgH using an arabinose-inducible expression vector (pBAD/myc-His). To ensure pcdgH produces fully functional CdgH, we determined that pcdgH is able to complement the RΔcdgH mutant phenotype (data not shown). We also generated the overexpression plasmid [pcdgH(GADEF)] harboring the gene with point mutations converting GGDEF residues to GADEF in CdgH. We transformed pcdgH and pcdgH(GADEF) into smooth, where c-di-GMP levels are low, and measured intracellular c-di-GMP levels. The cells were incubated with [32P]orthophosphate under inducing and noninducing conditions, and total nucleotides were analyzed by using 2D-TLC. Overexpression of cdgH resulted in a high amount of c-di-GMP accumulation in the cell, while overexpression of cdgH(GADEF) did not result in increased c-di-GMP production (Fig. 3). Consistent with the phenotypic changes observed in high cellular c-di-GMP levels, overexpression of cdgH in the smooth genetic background led to the formation of rugose colonies and a decrease in motility (data not shown). Taken together, these results show that CdgH functions as a DGC.
FIG. 3.
Analysis of enzymatic activity of CdgH. 2D-TLC analysis of total nucleotides extracted from smooth strains harboring pcdgH or pcdgH(GADEF) grown in MOPS minimal medium with [32P]orthophosphate was performed. The arrow indicates the spot corresponding to c-di-GMP according to its Rf values of 0.16 in the NH4CO3 dimension and 0.37 in the KH2PO4 dimension. The result shown are representative of two independent experiments.
CdgG negatively regulates rugosity-associated phenotypes.
The second locus that we identified in our screen to be involved in rugosity was VC0900 (cdgG). The rugose cdgG mutant (RΔcdgG) formed “super-rugose” colonies with increased corrugation compared to rugose (Fig. 4A). However, no significant differences were observed between RΔcdgG biofilms and rugose biofilms when analyzed using CLSM (Fig. 4B). COMSTAT analysis revealed that RΔcdgG biofilms did not significantly differ from rugose biofilms (Table 2). To elucidate mechanism by which cdgG affects colony corrugation, we analyzed the transcription of vpsL, vpsR, and vpsT using vpsL-lacZ, vpsR-lacZ, and vpsT-lacZ transcriptional fusions via β-galactosidase assays. The results indicated that CdgG does not significantly affect the transcription of these genes (Fig. 4C). We reasoned that CdgG could regulate VPS production by posttranslational mechanisms. Therefore, we compared exopolysaccharide production in RΔcdgG and rugose. No significant difference in VPS production between rugose and RΔcdgG was observed (data not shown). We also determined the role of CdgG on motility by measuring the migration zone formed on LB soft agar plates. There was a small but consistent decrease in the motility of RΔcdgG compared to rugose (Fig. 4D). Altogether, our results indicate that CdgG modulates colony corrugation and motility but does not affect vps gene expression.
FIG. 4.
Phenotypic characterization of CdgG in the rugose genetic background. (A) Colony morphologies of rugose and RΔcdgG strains that were grown for 24 and 48 h on LB agar plates at 30°C. (B) Three-dimensional biofilm structures of rugose and RΔcdgG strains that are formed 8 h postinoculation under static conditions at 30°C. Images were acquired with CLSM, with top-down (large panes) and orthogonal (side panels) views of biofilms shown. Scale bar, 30 μm. (C) Transcription of vpsL-lacZ, vpsR-lacZ, and vpsT-lacZ fusions determined in rugose and RΔcdgG strains by measuring β-galactosidase activity in cells that were grown to mid-exponential phase in LB medium at 30°C. The results shown are representative of three independent experiments. Error bars represent the standard deviations. (D) Diameter of migration zone of rugose, RΔcdgG, RΔvps-I, and RΔflaA strains measured in LB soft agar plates (0.3%) after 18 h of incubation at 30°C. The data shown are representative of three independent experiments. Error bars represent the standard deviations.
In the smooth genetic background, the deletion of cdgG (SΔcdgG) markedly altered colony morphology and biofilm formation. SΔcdgG formed compact colonies with a slight corrugation in the center (Fig. 5A). There was no difference in the growth rate of the cdgG deletion mutant and smooth (see Fig. S1 in the supplemental material). Hence, colony compactness is not due to a growth defect. SΔcdgG formed significantly thicker and more structured biofilms in a flow cell biofilm setup compared to its smooth parent (Fig. 5B). Quantitative analysis of these biofilms with the COMSTAT program verified increased roughness and maximum thickness (Table 2). We also measured vpsL, vpsR, and vpsT transcription via β-galactosidase assays. cdgG mutants had slightly increased vpsL transcription (1.5-fold) relative to smooth strains. There was no detectable difference in the transcription of vpsR or vpsT in these two strains (Fig. 5C). We also determined that SΔcdgG exhibits a slight decrease in motility (Fig. 5D). Taken together, our results indicate that CdgG acts mainly at posttranscriptional level to affect rugosity, biofilm formation, and motility-associated phenotypes.
FIG. 5.
Phenotypic characterization of CdgG in the smooth genetic background. (A) Colony morphologies of smooth and SΔcdgG strains that were grown for 24 h on LB agar plates at 30°C. (B) Three-dimensional biofilm structures of smooth and SΔcdgG strains that are formed 24 h postinoculation in a once-through flow cell system. Images were acquired with CLSM, with top-down (large panes) and orthogonal (side panels) views of biofilms shown. Scale bar, 30 μm. (C) Transcription of vpsL-lacZ, vpsR-lacZ, and vpsT-lacZ fusions determined in smooth and SΔcdgG strains by measuring β-galactosidase activity in cells that were grown to mid-exponential phase in LB medium at 30°C. The results shown are representative of three independent experiments. Error bars represent the standard deviations. (D) Diameter of migration zone of smooth, SΔcdgG, SΔvps-I, and SΔflaA strains measured in LB soft agar plates (0.3%) after 18 h of incubation at 30°C. The data shown are representative of three independent experiments. Error bars represent the standard deviations.
CdgG does not act as a DGC.
CdgG is predicted to have a GGDEF domain (PF00990). However, CdgG carries SGEEF residues in the active site of this domain, suggesting that it may not act as a DGC. To test the enzymatic activity of CdgG, we constructed two overexpression plasmids: pcdgG, harboring a wild-type copy of cdgG, and pcdgG(SGAAF), harboring cdgG with point mutations converting SGEEF residues to SGAAF in CdgG. We transformed each of the overexpression plasmids into smooth strains and measured intracellular c-di-GMP levels as described above. Under either inducing or noninducing conditions, overproduction of CdgG or CdgG-SGAAF did not lead to an accumulation of c-di-GMP (Fig. 6). Furthermore, plasmids overexpressing CdgG and its mutant derivative CdgG-SGAAF can complement the cdgG phenotype (see Fig. 11). Taken together, these findings suggest that CdgG does not function as a DGC.
FIG. 6.
Analysis of enzymatic activity of CdgG. 2D-TLC analysis of total nucleotides extracted from smooth strains harboring pcdgG or pcdgG(SGAAF) grown in MOPS minimal medium with [32P]orthophosphate was performed. The results shown are representative of two independent experiments.
FIG. 11.
Characterization of A-site and I-site of CdgG. (A) Multiple sequence alignment of C. crescentus protein PleD and CdgG at the predicted I-site and A-site of GGDEF domain is shown. Conserved residues are highlighted, and the marked residues (Arg434, Asp437, Glu445, and Glu446) were changed to alanine residues via site-directed mutagenesis. (B) Colony morphologies of RΔcdgG strains harboring pBAD/myc-His, pcdgG, pcdgG(SGAAF), or pcdgG(ADSA) that were grown for 24 h at 30°C on LB agar plates containing 0.02% (wt/vol) l-arabinose. (C) Three-dimensional biofilm structures of SΔcdgG strains harboring pBAD/myc-His, pcdgG, pcdgG(SGAAF), or pcdgG(ADSA) that are formed 24 h postinoculation in a once-through flow cell system. Images were acquired with CLSM, with top-down (large panes) and orthogonal (side panels) views of biofilms shown. Scale bar, 30 μm.
CdgG and CdgH act in parallel pathways to control rugosity-associated phenotypes.
To decipher possible epistatic interactions between cdgG and cdgH, we generated ΔcdgG ΔcdgH double knockout mutants in the smooth and rugose genetic backgrounds and analyzed their colony morphologies and motility phenotypes. RΔcdgGΔcdgH formed colonies with intermediate corrugation (increased compared to RΔcdgH colonies and decreased compared to RΔcdgG colonies) (Fig. 7A). RΔcdgGΔcdgH displayed increased motility compared to RΔcdgG and decreased motility compared to RΔcdgH (Fig. 7B). Epistasis analysis in the smooth genetic background revealed similar results where SΔcdgGΔcdgH exhibited intermediate motility phenotype (Fig. 7B). Taken together, results indicate that cdgG and cdgH act in parallel pathways to modulate colony corrugation and motility in both genetic backgrounds.
FIG. 7.
Epistasis analysis of cdgG and cdgH. (A) Colony morphologies of rugose, RΔcdgG, RΔcdgH, RΔcdgGΔcdgH, smooth, SΔcdgG, SΔcdgH, and SΔcdgGΔcdgH strains that were grown for 24 h on LB agar plates at 30°C. (B) Diameter of migration zone of rugose, RΔcdgG, RΔcdgH, RΔcdgGΔcdgH, smooth, SΔcdgG, SΔcdgH, and SΔcdgGΔcdgH strains measured in LB soft agar plates (0.3%) after 18 h of incubation at 30°C. The data shown are representative of three independent experiments. Error bars represent the standard deviations.
CdgH acts in a parallel pathway to other DGCs to control rugosity-associated phenotypes.
Previously, we identified two other DGCs that regulate rugosity: VpvC and CdgA (4, 6, 32). VpvC is responsible for the smooth-to-rugose phase variation in our prototype rugose V. cholerae variant (6). Loss of vpvC in the rugose genetic background leads to a decrease in c-di-GMP levels and formation of smooth colonies and creates strains that are similar to the smooth variant regarding biofilm formation and vps transcription. To investigate the epistatic interactions between vpvC and cdgH, we generated the RΔvpvCΔcdgH mutant. Colony morphology of RΔvpvCΔcdgH was similar to that of RΔvpvC (Fig. 8A). We also performed motility assays to decipher epistatic interactions between vpvC and cdgH and observed that, while single deletions of vpvC or cdgH led to an increase in motility, RΔvpvCΔcdgH exhibited motility that was higher than either single mutant (Fig. 8B). Taken together, while vpvC is epistatic to cdgH with respect to colony corrugation phenotype, cdgH and vpvC have an additive effect on motility.
FIG. 8.
Epistasis analysis of cdgH, vpvC, and cdgA. (A) Colony morphologies of rugose, RΔcdgH, RΔvpvC, and RΔvpvCΔcdgH strains that were grown for 24 h on LB-agar plates at 30°C. (B) Diameter of migration zone of rugose, RΔcdgH, RΔvpvC, and RΔvpvCΔcdgH strains measured in LB soft agar plates (0.3%) after 18 h of incubation at 30°C. The data shown are representative of three independent experiments. Error bars represent the standard deviations. (C) Colony morphologies of smooth, SΔcdgA, SΔcdgH, SΔcdgAΔcdgH, SΔhapR, SΔhapRΔcdgA, SΔhapRΔcdgH, and SΔhapRΔcdgAΔcdgH that were grown for 48 h on LB agar plates at 30°C.
CdgA was identified in our search for the gene responsible for rugosity in the hapR mutant (4). We showed that deletion of cdgA in SΔhapR leads to the formation of smooth-looking colonies and a decrease in vps expression. It is noteworthy that VpvC is not involved in HapR-mediated rugosity as SΔhapRΔvpvC forms rugose colonies (data not shown). To elucidate the role of cdgH in HapR-mediated rugosity and to investigate the epistatic interactions between cdgA and cdgH, we generated SΔhapRΔcdgH, SΔcdgAΔcdgH, and SΔhapRΔcdgAΔcdgH strains and analyzed their colony morphologies. Interestingly, deletion of cdgH in the SΔhapR genetic background also caused a decrease in the colony corrugation and yielded smooth-looking colonies (Fig. 8C), suggesting that CdgH, like CdgA, also regulates HapR-mediated rugosity. However, the colonies of SΔhapRΔcdgH were more compact than those of SΔhapRΔcdgA. To further investigate the contribution of CdgA and CdgH to HapR-mediated rugosity, we generated a triple-deletion mutant, SΔhapRΔcdgAΔcdgH. The colony morphology of SΔhapRΔcdgAΔcdgH was similar to that of SΔhapRΔcdgA (Fig. 8C), suggesting that although both CdgH and CdgA contribute to rugosity in SΔhapR, cdgA is epistatic to cdgH.
CdgG acts in parallel pathways with MbaA and CdgC, and rocS is epistatic to cdgG with respect to rugosity.
In addition to DGCs, several PDEs regulate vps expression and rugosity in V. cholerae (7, 32, 41, 52). We previously characterized three genes (cdgC, rocS, and mbaA) encoding PDEs for their contribution to rugosity. Consistent with an increase in c-di-GMP due to the loss of key PDEs, cdgC, rocS, and mbaA mutants formed super-rugose colonies that are more opaque and wrinkled than the rugose variant (32). Using epistasis analysis, we determined that both in the rugose genetic background (32) and in the smooth genetic background (see Fig. S2 in the supplemental material) CdgC, RocS, and MbaA act through parallel pathways to control rugosity.
SΔcdgC, SΔrocS, and SΔmbaA strains form more compact colonies than the parent smooth strain. Similarly, SΔcdgG strain also forms colonies that are more compact than its smooth parent. We thus tested whether CdgG interacts with CdgC, RocS, or MbaA to exert its effect on colony morphology. To this end, we created double deletion mutants of cdgG with rocS, mbaA, and cdgC (SΔrocSΔcdgG, SΔmbaAΔcdgG, and SΔcdgCΔcdgG). We observed that deletion of cdgG in the SΔmbaA or SΔcdgC genetic backgrounds increased the colony compactness (Fig. 9), suggesting that CdgG acts in parallel pathways with CdgC and MbaA to regulate colony corrugation. However, deletion of cdgG in the SΔrocS genetic background resulted in colonies that matched the rocS single mutant (Fig. 9), indicating that rocS is epistatic to cdgG in controlling colony compactness and corrugation. Similarly, deletion of cdgG in SΔrocSΔmbaAΔcdgC did not alter further alter colony morphology, possibly due to an epistatic interaction between rocS and cdgG. Whether CdgG and RocS physically interact and form complexes remains to be elucidated.
FIG. 9.
Epistasis analysis of rocS, mbaA, cdgC and cdgG in the smooth genetic background. Colony morphologies of smooth, SΔrocS, SΔmbaA, SΔcdgC, SΔrocΔmbaAΔcdgC, SΔcdgG, SΔrocSΔcdgG, SΔmbaAΔcdgG, SΔcdgCΔcdgG, and SΔrocSΔmbaAΔcdgCΔcdgG strains that were grown for 48 h on LB agar plates at 30°C.
Plz proteins have minimal effect on the rugosity of V. cholerae.
Earlier studies identified proteins with PilZ domains as c-di-GMP binding proteins (11, 36, 40, 45). In V. cholerae there are four genes encoding proteins with conserved PilZ domains: VC0697 (plzA), VC2344 (plzC), VCA0042 (plzD), and VCA0735 (plzE) (2). A study done by Pratt et al. characterized two of these genes, plzC and plzD, and an additional gene VC1885 (plzB) encoding a remote PilZ domain protein that does not have the residues critical for c-di-GMP binding. These authors showed that PlzB, PlzC, and PlzD affected biofilm formation, motility, and intestinal colonization in the V. cholerae O1 classical biotype. Furthermore, it was shown that PlzC and PlzD are capable of binding to c-di-GMP (40).
The effect of plzC and plzD on biofilm formation in the classical biotype prompted us to look at the effect of plz genes on colony corrugation in the other V. cholerae biotype, El Tor. We were able to delete each gene encoding Plz proteins singly and in combination, indicating that they are not essential in our V. cholerae O1 El Tor strain. We observed that RΔplzA, RΔplzD, and RΔplzE formed colonies similar to those of rugose (Fig. 10A). However, deletion of plzC in the rugose genetic background slightly increased the colony corrugation. The colony morphology of the quadruple deletion mutant RΔplzACDE was also similar to that of RΔplzC, indicating that PlzC is the only Plz protein that affects colony corrugation. In addition, deletion of plzB in rugose or in the RΔplzACDE did not affect colony morphology (Fig. 10A). We also analyzed the role of Plz proteins in biofilm formation. RΔplzACDE biofilms were indistinguishable from rugose biofilms (Fig. 10B) under the condition we tested. This finding is somewhat expected, since there was a slight difference in colony corrugation between the rugose and RΔplzACDE. Altogether, these results indicate that Plz proteins are not essential for rugosity and the formation of three-dimensional biofilms in the El Tor biotype background.
FIG. 10.
Characterization plz genes for their contribution to rugosity. (A) Colony morphologies of rugose, RΔplzA, RΔplzB, RΔplzC, RΔplzD, RΔplzE, RΔplzACDE, and RΔplzABCDE strains that were grown for 48 h on LB agar plates at 30°C. (B) Three-dimensional biofilm structures of rugose and RΔplzACDE strains that are formed at 24 h postinoculation in a once-through flow cell system. Images were acquired with CLSM, with top-down (large panes) and orthogonal (side panels) views of biofilms shown. The white bar equals 30 μm. (C) Colony morphologies of rugose, RΔcdgH, RΔvpvC, RΔplzC, RΔcdgHΔplzC, and RΔvpvCΔplzC strains that were grown for 24 h on LB agar plates at 30°C. (D) Diameter of migration zone of rugose, RΔplzC, RΔcdgH, RΔcdgHΔplzC, RΔvpvC, and RΔvpvCΔplzC strains measured in LB soft agar plates (0.3%) after 18 h of incubation at 30°C. The data shown are representative of three independent experiments. Error bars represent the standard deviations.
As described above, proteins that create rugosity alter cellular levels of c-di-GMP. To affect these processes, c-di-GMP must be sensed. Since only RΔplzC had increased colony corrugation and PlzC binds c-di-GMP (40), we questioned whether PlzC acts downstream of DGCs CdgH and VpvC, which control rugosity. To address this possibility, we generated RΔcdgHΔplzC and RΔvpvCΔplzC double mutants and analyzed colony morphology and motility phenotypes. The double mutants exhibited an intermediate phenotype compared to corresponding single mutants (Fig. 10C and D), indicating that PlzC is not a cognate c-di-GMP binding partner of either VpvC or CdgH. Taken together, these results indicate that PlzC and other Plz proteins contribute minimally to rugosity-associated phenotypes and that non-PilZ domain c-di-GMP receptor proteins are likely to be involved in controlling rugosity and biofilm formation in V. cholerae.
RXXD motif, which can bind c-di-GMP, is necessary for the function of CdgG.
As described above, the V. cholerae Plz proteins are minimally involved in rugosity, and so we searched for other potential c-di-GMP binding proteins. Recently, a non-PilZ domain c-di-GMP receptor protein, PelD, was identified (30). This protein possesses an RXXD motif similar to the ones found in the I-site (inhibition site) of DGCs, such as Caulobacter crescentus protein PleD (10, 30). RXXD motifs of PelD and PleD are able to bind c-di-GMP (10, 30). CdgG does not have a DGC activity but has the conserved RXXD motif (Fig. 11A). We hypothesized that CdgG may be one of the non-PilZ domain c-di-GMP receptor proteins and the RXXD motif is critical for its function. To evaluate the importance of the RXXD motif to CdgG function, we generated an overexpression plasmid carrying cdgG with point mutations converting RXXD (RDSD) residues to AXXA (ADSA). We introduced this plasmid, pcdgG(ADSA), pBAD/myc-His, pcdgG, and pcdgG(SGAAF) (described above, converts the GGDEF domain SGEEF residues to SGAAF) into the RΔcdgG strain and tested each clone for its capacity to complement the RΔcdgG colony rugosity mutant phenotype under inducing conditions. While pcdgG or pcdgG(SGAAF) were able to complement the mutant phenotype, no complementation was seen in the strains carrying pBAD/myc-His or pcdgG(ADSA) (Fig. 11B). We also introduced these plasmids into the SΔcdgG and tested for complementation of the biofilm phenotype. Overexpression of pcdgG and pcdgG(SGAAF) in SΔcdgG yielded biofilms with decreased thickness and structural complexity, a finding indicative of complementation, compared to the strains carrying pBAD/myc-His or pcdgG(ADSA) (Fig. 11C). Taken together, these findings indicate that while SGEEF residues in the A-site of CdgG are not required, RDSD residues at the I-site, which is predicted to bind c-di-GMP, are essential for the function of CdgG. We speculate that CdgG may be a c-di-GMP binding protein controlling rugosity.
We then questioned whether CdgG acts downstream of DGC, VpvC. To address this possibility, we generated RΔvpvCΔcdgG double mutant and analyzed the colony morphology phenotype. Double mutants exhibited an intermediate phenotype compared to the single mutants (see Fig. S3 in the supplemental material), indicating that CdgG and VpvC act in parallel pathways to control rugosity. As discussed earlier, CdgG and CdgH also act in parallel pathways, indicating that CdgG is not a cognate partner of either VpvC or CdgH.
DISCUSSION
c-di-GMP has emerged as a ubiquitous second messenger in bacteria, controlling the transition from a free-living, motile lifestyle to a biofilm lifestyle (42). In the V. cholerae genome, there are 62 genes encoding proteins with GGDEF, EAL, or HD-GYP domains (18). We have previously characterized 10 of these genes for their importance in rugosity-associated phenotypes (4-6, 32). These phenotypes include enhanced capacity to produce biofilm matrix materials (VPS and biofilm matrix proteins), resulting in colony corrugation, increased biofilm formation, and decreased motility. In the present study, our main objective was to identify additional genes encoding DGCs or PDEs that are involved in rugosity and biofilm formation. To this end, we generated in-frame deletion mutants of each gene encoding a protein with GGDEF and/or EAL domains and analyzed the mutants for colony corrugation and biofilm formation phenotypes under standard growth conditions (growth in LB medium at 30°C). We identified two additional genes, cdgH and cdgG, involved in colony corrugation and biofilm formation phenotypes. Deletion of cdgH resulted in decreased colony corrugation, biofilm formation, and increased motility. Consistent with these phenotypic characteristics, CdgH exhibits DGC activity. In contrast, deletion of cdgG resulted in increased colony corrugation and decreased motility. Interestingly, we had identified cdgG as a negative regulator of biofilm formation in an independent screen designed to identify mutants with enhanced biofilm forming capacities (S. Beyhan and F. H. Yildiz, unpublished result).
Although both CdgH and CdgG have GGDEF domains, they had opposite effects on rugosity-associated phenotypes. CdgG does not have the conserved residues in the active site of the domain (A-site); instead, it has SGEEF. Mutational analyses of GGDEF domains in DGCs showed that GG[DE]EF residues are essential for the function of the DGC enzymes (9, 35). However, it has also been shown that proteins with “imperfect residues” could have a DGC activity or that the imperfect residues are critical for the function of the protein. For example, NVDEF residues in the GGDEF domain of MxdA were shown to be required for the DGC activity and biofilm formation and detachment in Shewanella oneidensis (51). In addition, GDSIF residues of Pseudomonas aeruginosa protein FimX (26), GEDEF residues of C. crescentus protein CC3396 (12), GGDQF residues of P. aeruginosa BifA (28), and GVGEW residues of V. cholerae CdpA (50) are critical for modulating the activity of these proteins. We observed that changing the A-site residues of CdgG to SGAAF does not alter the function of this protein, as determined by complementation assay using the colony corrugation and biofilm phenotypes (Fig. 11B).
V. cholerae has 31 genes that encode proteins with GGDEF domains (without an EAL domain) and, of 31 GGDEF domain-containing proteins, 27 are predicted to have conserved GG[DE]EF residues at the A-site of the enzyme, indicating a DGC activity. VpvC, CdgA, and CdgH are all active DGCs, and all have conserved residues of GGDEF in the A-site. Under the experimental conditions we utilized, only mutants of vpvC, cdgA, or cdgH exhibited altered colony corrugation and biofilm phenotypes. This finding indicates that these proteins are not functionally redundant. It is possible that the remaining genes involved in c-di-GMP signaling systems may not be transcribed under the experimental conditions we utilized. In addition, c-di-GMP signaling systems can act as a link between environmental signals, and some of the c-di-GMP signaling networks are only activated by a specific environmental stimulus. For example, the activity of MbaA, an inner membrane protein with cytoplasmic GGDEF-EAL domains, is regulated by norspermidine (25). In V. cholerae, the polyamine norspermidine enhances biofilm formation in an NpsS-MbaA-dependent manner. It was proposed that MbaA activity is regulated by the interaction of NpsS or norspermidine-NpsS with the periplasmic domain of MbaA (25). It is likely that under the experimental condition we utilized, some of the DGCs or PDEs may not be activated due to a lack of environmental stimuli. Therefore, we did not observe any changes in the phenotypic properties of some of the DGC or PDE mutants.
In addition to DGCs and PDEs, other essential components of the c-di-GMP signaling system include the c-di-GMP receptor proteins. In earlier studies, PilZ domain-containing proteins were shown to bind c-di-GMP (11, 36, 40, 45). These proteins control flagellar motility in C. crescentus (11) and Salmonella enterica serovar Typhimurium (45) and alginate biosynthesis in P. aeruginosa (36). In V. cholerae, they control motility, biofilm formation, and virulence (40). Therefore, we wanted to understand the contribution of PilZ domain-containing proteins to rugosity-associated phenotypes. We observed that only PlzC affected colony corrugation (albeit slightly) in V. cholerae. We tested the possibly that PlzC acts downstream of DGC's VpvC or CdgH and observed that PlzC is not a c-di-GMP binding partner of either VpvC or CdgH and acts in parallel pathways with them. Therefore, additional non-PilZ c-di-GMP binding proteins are involved in the rugosity-associated c-di-GMP signaling system of V. cholerae.
Suzuki et al. recently showed that CsrD, which is predicted to carry a GGDEF and an EAL domain with imperfect residues in both domains (HRSDF instead of GGDEF and ELM instead of EAL), controls the degradation of CsrB/C RNAs (49), indicating that proteins with imperfect GGDEF/EAL motifs can have alternative functions. CdgG also has an imperfect GGDEF domain and does not have a DGC activity. We determined that CdgG posses an inhibitory site (I-site) with a conserved RXXD motif that is necessary for its function. In other proteins, this set of amino acids is capable of binding c-di-GMP (10). It has yet to be determined, however, whether CdgG binds c-di-GMP and relays the signal to effector proteins or downstream processes.
Significant information gaps remain in our understanding of the mechanisms by which c-di-GMP signaling operates. Although c-di-GMP was first identified as an allosteric regulator affecting mainly protein activity (43), recent studies indicate that c-di-GMP can regulate gene expression by interacting with transcriptional factors (23) and through cyclic di-GMP riboswitches (48). We have studied c-di-GMP signaling proteins that affect colony rugosity. Several of these alter gene expression, including VpvC, CdgA, CdgH, CdgC, RocS, and MbaA (4, 6, 33), while CdgG apparently does not. These findings indicate that both transcriptional and posttranscriptional modes of regulation operate in V. cholerae c-di-GMP signaling systems. V. cholerae has 62 genes predicted to encode proteins with GGDEF, EAL, or HD-GYP domains and faces another challenge, namely, how an organism with a large number of c-di-GMP signaling proteins can prevent cross talk or noise in signaling. Spatial sequestration of GGDEF/EAL proteins to microcompartmentalize c-di-GMP levels in the cell is one of the proposed mechanisms. VpvC, CdgA, and CdgH are predicted to be localized to the cytoplasmic membrane. We propose that they could form independent c-di-GMP signaling clusters in different regions of the cell, together with their effector proteins (transcriptional regulators or proteins that can change activity or function of transcriptional regulators). It is likely that each of these proteins generate a different c-di-GMP pool that can be degraded by cognate PDEs (Fig. 12). c-di-GMP signaling complexes could be dynamic and effector proteins could be released from these complexes and participate directly or indirectly in gene expression. Alternatively, some portion of c-di-GMP may freely diffuse in the cytoplasm, and c-di-GMP might interact with cytoplasmically localized transcriptional regulators, thereby regulating gene expression. We undertook systematic mutational and phenotypic analyses of c-di-GMP signaling proteins in V. cholerae and identified critical c-di-GMP signaling proteins required for rugosity-associated phenotypes. There is much to be discovered about the mechanisms by which the c-di-GMP signaling system regulates rugosity and interacts with other regulatory networks controlling rugosity, including two component regulatory systems and quorum sensing. A better understanding of the mechanism of c-di-GMP signaling and biofilm formation and the importance of these processes in V. cholerae biology will prove useful for the development of future strategies for predicting and controlling cholera epidemics.
FIG. 12.
A model of c-di-GMP signaling systems modulating rugosity-associated phenotypes in V. cholerae. VpvC, CdgA, and CdgH are localized in the cytoplasmic membrane, generating different pools of c-di-GMP upon receiving environmental cues. Intracellular c-di-GMP is degraded by membrane localized MbaA and cytoplasmically localized RocS and CdgC. The function of CdgG in the system is unclear, although it is likely to interact with RocS. CdgG can act as a c-di-GMP binding protein. Through Plz proteins and other c-di-GMP binding proteins and then, via effector proteins, c-di-GMP signaling systems affect colony corrugation, biofilm formation, VPS production, and motility in V. cholerae.
Supplementary Material
Acknowledgments
We thank Bentley Lim for constructing pFY-572; Vanessa Soliven for generating Fy_Vc_337 and Fy_Vc_339; and Karen Ottemann, Manel Camps, and Yildiz laboratory members for their valuable comments on the manuscript.
This study was supported by NIH grant AI055987.
Footnotes
Published ahead of print on 12 September 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alam, M., M. Sultana, G. B. Nair, R. B. Sack, D. A. Sack, A. K. Siddique, A. Ali, A. Huq, and R. R. Colwell. 2006. Toxigenic Vibrio cholerae in the aquatic environment of Mathbaria, Bangladesh. Appl. Environ. Microbiol. 722849-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amikam, D., and M. Y. Galperin. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 223-6. [DOI] [PubMed] [Google Scholar]
- 3.Bao, Y., D. P. Lies, H. Fu, and G. P. Roberts. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109167-168. [DOI] [PubMed] [Google Scholar]
- 4.Beyhan, S., K. Bilecen, S. R. Salama, C. Casper-Lindley, and F. H. Yildiz. 2007. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J. Bacteriol. 189388-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beyhan, S., A. D. Tischler, A. Camilli, and F. H. Yildiz. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 1883600-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beyhan, S., and F. H. Yildiz. 2007. Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol. Microbiol. 63995-1007. [DOI] [PubMed] [Google Scholar]
- 7.Bomchil, N., P. Watnick, and R. Kolter. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 1851384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casper-Lindley, C., and F. H. Yildiz. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 1861574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan, C., R. Paul, D. Samoray, N. C. Amiot, B. Giese, U. Jenal, and T. Schirmer. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. USA 10117084-17089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christen, B., M. Christen, R. Paul, F. Schmid, M. Folcher, P. Jenoe, M. Meuwly, and U. Jenal. 2006. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 1044112-4117. [DOI] [PubMed] [Google Scholar]
- 11.Christen, M., B. Christen, M. G. Allan, M. Folcher, P. Jeno, S. Grzesiek, and U. Jenal. 2007. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 1044112-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christen, M., B. Christen, M. Folcher, A. Schauerte, and U. Jenal. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 28030829-30837. [DOI] [PubMed] [Google Scholar]
- 13.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235386-405. [DOI] [PubMed] [Google Scholar]
- 14.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 621301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fong, J. C., K. Karplus, G. K. Schoolnik, and F. H. Yildiz. 2006. Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J. Bacteriol. 1881049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fong, J. C., and F. H. Yildiz. 2007. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J. Bacteriol. 1892319-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fullner, K. J., and J. J. Mekalanos. 1999. Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect. Immun. 671393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 20311-21. [DOI] [PubMed] [Google Scholar]
- 19.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50101-104. [DOI] [PubMed] [Google Scholar]
- 20.Haugo, A. J., and P. I. Watnick. 2002. Vibrio cholerae CytR is a repressor of biofilm development. Mol. Microbiol. 45471-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1726557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146(Pt. 10)2395-2407. [DOI] [PubMed] [Google Scholar]
- 23.Hickman, J. W., and C. S. Harwood. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69376-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Islam, M. S., M. I. Jahid, M. M. Rahman, M. Z. Rahman, M. S. Islam, M. S. Kabir, D. A. Sack, and G. K. Schoolnik. 2007. Biofilm acts as a microenvironment for plankton-associated Vibrio cholerae in the aquatic environment of Bangladesh. Microbiol. Immunol. 51369-379. [DOI] [PubMed] [Google Scholar]
- 25.Karatan, E., T. R. Duncan, and P. I. Watnick. 2005. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 1877434-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kazmierczak, B. I., M. B. Lebron, and T. S. Murray. 2006. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 601026-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovacikova, G., W. Lin, and K. Skorupski. 2005. Dual regulation of genes involved in acetoin biosynthesis and motility/biofilm formation by the virulence activator AphA and the acetate-responsive LysR-type regulator AlsR in Vibrio cholerae. Mol. Microbiol. 57420-433. [DOI] [PubMed] [Google Scholar]
- 28.Kuchma, S. L., K. M. Brothers, J. H. Merritt, N. T. Liberati, F. M. Ausubel, and G. A. O'Toole. 2007. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 1898165-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lauriano, C. M., C. Ghosh, N. E. Correa, and K. E. Klose. 2004. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J. Bacteriol. 1864864-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, V. T., J. M. Matewish, J. L. Kessler, M. Hyodo, Y. Hayakawa, and S. Lory. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 651474-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang, W., A. J. Silva, and J. A. Benitez. 2007. The cyclic AMP receptor protein modulates colonial morphology in Vibrio cholerae. Appl. Environ. Microbiol. 737482-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim, B., S. Beyhan, J. Meir, and F. H. Yildiz. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60331-348. [DOI] [PubMed] [Google Scholar]
- 33.Lim, B., S. Beyhan, and F. H. Yildiz. 2007. Regulation of Vibrio polysaccharide synthesis and virulence factor production by CdgC, a GGDEF-EAL domain protein, in Vibrio cholerae. J. Bacteriol. 189717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipp, E. K., A. Huq, and R. R. Colwell. 2002. Effects of global climate on infectious disease: the cholera model. Clin. Microbiol. Rev. 15757-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malone, J. G., R. Williams, M. Christen, U. Jenal, A. J. Spiers, and P. B. Rainey. 2007. The structure-function relationship of WspR, a Pseudomonas fluorescens response regulator with a GGDEF output domain. Microbiology 153980-994. [DOI] [PubMed] [Google Scholar]
- 36.Merighi, M., V. T. Lee, M. Hyodo, Y. Hayakawa, and S. Lory. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65876-895. [DOI] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1972. Assay of β-galactosidase. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 38.Moorthy, S., and P. I. Watnick. 2004. Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol. Microbiol. 52573-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris, J. G., Jr., M. B. Sztein, E. W. Rice, J. P. Nataro, G. A. Losonsky, P. Panigrahi, C. O. Tacket, and J. A. Johnson. 1996. Vibrio cholerae O1 can assume a chlorine-resistant rugose survival form that is virulent for humans. J. Infect. Dis. 1741364-1368. [DOI] [PubMed] [Google Scholar]
- 40.Pratt, J. T., R. Tamayo, A. D. Tischler, and A. Camilli. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 28212860-12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rashid, M. H., C. Rajanna, A. Ali, and D. K. Karaolis. 2003. Identification of genes involved in the switch between the smooth and rugose phenotypes of Vibrio cholerae. FEMS Microbiol. Lett. 227113-119. [DOI] [PubMed] [Google Scholar]
- 42.Romling, U., and D. Amikam. 2006. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9218-228. [DOI] [PubMed] [Google Scholar]
- 43.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinbergerohana, R. Mayer, S. Braun, E. Devroom, G. Vandermarel, J. Vanboom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter-xylinum by cyclic digualylic acid. Nature 325279-281. [DOI] [PubMed] [Google Scholar]
- 44.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 1036712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Ryjenkov, D. A., R. Simm, U. Romling, and M. Gomelsky. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 28130310-30314. [DOI] [PubMed] [Google Scholar]
- 46.Ryjenkov, D. A., M. Tarutina, O. V. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 1871792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt, A. J., D. A. Ryjenkov, and M. Gomelsky. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 1874774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sudarsan, N., E. R. Lee, Z. Weinberg, R. H. Moy, J. N. Kim, K. H. Link, and R. R. Breaker. 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321411-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki, K., P. Babitzke, S. R. Kushner, and T. Romeo. 2006. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 202605-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamayo, R., S. Schild, J. T. Pratt, and A. Camilli. 2008. Role of cyclic di-GMP during El Tor biotype Vibrio cholerae infection: characterization of the in vivo-induced cyclic di-GMP phosphodiesterase CdpA. Infect. Immun. 761617-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thormann, K. M., S. Duttler, R. M. Saville, M. Hyodo, S. Shukla, Y. Hayakawa, and A. M. Spormann. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 1882681-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tischler, A. D., and A. Camilli. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 735873-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wai, S. N., Y. Mizunoe, A. Takade, S. I. Kawabata, and S. I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 643648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 1831716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yildiz, F. H., X. S. Liu, A. Heydorn, and G. K. Schoolnik. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53497-515. [DOI] [PubMed] [Google Scholar]
- 58.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 964028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu, J., and J. J. Mekalanos. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5647-656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.