Abstract
Time-lapse imaging of Streptomyces hyphae revealed foci of the essential protein DivIVA at sites where lateral branches will emerge. Overexpression experiments showed that DivIVA foci can trigger establishment of new zones of cell wall assembly, suggesting a key role of DivIVA in directing peptidoglycan synthesis and cell shape in Streptomyces.
Gram-positive bacteria of the genus Streptomyces grow by tip extension and form branched hyphae and mycelia (8, 11, 12). This polarized cell wall growth is strikingly different from the mode of growth of, e.g., Escherichia coli and Bacillus subtilis, which like most rod-shaped bacteria extend the cell and acquire their rod shape by intercalatory insertion of new peptidoglycan units along the lateral wall (5, 6). This is dependent on the actin-like MreB proteins, which form helical filaments extending along the cell and acting via interaction with membrane proteins to organize the cell wall assembly (1, 2, 16, 19). In contrast, Streptomyces tip extension appears to occur by an mreB-independent mechanism (22) and is also independent of FtsZ and cell division (23). The Streptomyces coelicolor genome contains two mreB genes, but they are involved primarily in sporulation and have no overt impact on tip extension in the vegetative mycelium (22; G. Muth, University of Tübingen, Germany, personal communication). In fact, most rod-shaped relatives of Streptomyces within the phylum Actinobacteria, like mycobacteria and corynebacteria, lack mreB genes and assemble their cell walls at the cell poles (3, 5, 15, 24).
This mreB-independent and polarized growth in Actinobacteria involves the coiled-coil protein DivIVA. In S. coelicolor, DivIVA is essential for growth and accumulates at growing hyphal tips, and the effects of partial depletion and ectopic overexpression revealed a strong impact on tip extension and cell shape determination (10). Among other Actinobacteria, the DivIVA orthologues, also named antigen 84 and Wag31, in Mycobacterium tuberculosis, Mycobacterium smegmatis, and Corynebacterium glutamicum are polarly localized and appear to be essential and, when overproduced, have a very similar effect on cell shape to that seen in S. coelicolor (17, 24, 25). Recently, DivIVA was found to be required for polar cell elongation and acquisition of rod shape in C. glutamicum and M. smegmatis (17, 20). Furthermore, Streptomyces and Mycobacterium DivIVA could restore polar growth to a C. glutamicum strain depleted for DivIVA, while orthologues from the phylum Firmicutes (e.g., Bacillus subtilis) could not (20) and are known to be associated with different cellular functions (9, 21, 26, 29). While these findings suggest a role for Streptomyces DivIVA in tip extension, its exact function is not known. In this study, we have investigated the subcellular targeting of S. coelicolor DivIVA and its involvement in the establishment of tip extension during hyphal branching.
DivIVA is a molecular marker of new branch sites in S. coelicolor.
Apart from the striking apical localization of S. coelicolor DivIVA, occasional smaller foci can be seen along the lateral walls of hyphae (10). In order to examine the significance and dynamics of such DivIVA foci, we have developed a system for live cell imaging, in which Streptomyces hyphae can be monitored during several hours of growth. Hyphae of S. coelicolor A3(2) strain K112 [divIVA+/Φ(divIVA-egfp)Hyb], which produces a DivIVA-EGFP (enhanced green fluorescent protein) fusion protein (10), were growing on 1% agarose pads with Oxoid antibiotic medium no. 3 that were sealed on the bottom by an oxygen-permeable Lumox Biofoil 25 membrane (Greiner Bio-One) and on the top by a coverslip. Samples were incubated at 24 to 27°C, and images were captured every 6 min using a Zeiss Axio Imager Z1 microscope, a 9100-02 EM-CCD camera (Hamamatsu Photonics), and Volocity 3DM software (Improvision). The average rate of tip extension was 6 ± 2 μm/h in this system. The fluorescence images were processed by Volocity, including adjustment of the grayscale limits to exclude most of the autofluorescence signal of the medium and to saturate the brightest pixels from the EGFP signal. Noise was reduced using the fine median filter in Volocity.
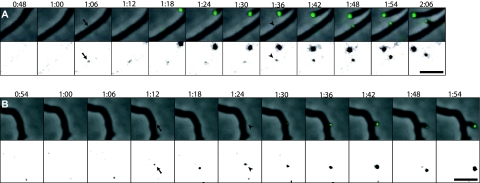
In addition to the DivIVA-EGFP fluorescence associated with each elongating hyphal tip, branching was frequently observed, and for 36 branches the process was captured in sufficient detail to evaluate the localization of DivIVA-EGFP foci throughout the branch emergence. Interestingly, in all such cases, a signal from DivIVA-EGFP appeared at the future branching site before any sign of lateral outgrowth was visible (Fig. 1; see Video S1 in the supplemental material). The average time between detection of the DivIVA focus and a visible protuberance indicating branch outgrowth was 27 ± 11 min (n = 36). Thus, DivIVA appeared to mark the sites where branches subsequently emerged. The fluorescent foci that were seen along hyphae were initially relatively faint, and not all of them developed into branches. A number of small foci appeared for a period of time and then disappeared, suggesting that assembly at this stage is dynamic. Those foci that developed into branches expanded and increased in intensity around the time when the new branch started to grow out from the lateral wall (Fig. 1; see Video S1 in the supplemental material), suggesting that establishment of cell wall synthesis or formation of the new cell pole led to recruitment of further DivIVA molecules and stabilization of a larger apical cluster.
FIG. 1.
DivIVA marks sites of hyphal branching. Hyphae of strain K112 expressing divIVA-egfp were grown on agarose pads, and images were captured every 6 min. Time-lapse series of representative branching events are shown as overlays of fluorescence signal (green) on the phase contrast images (top) and as the fluorescence channel with the gray scale inverted and adjusted to clearly visualize the initially weak DivIVA-EGFP signals (bottom). The first time points when DivIVA-EGFP foci were detected at future branching sites are marked with arrows. The subsequent time points at which outgrowth of the branches can be seen are indicated by arrowheads. Time is shown in hours and minutes after starting time-lapse acquisition. Bars, 5 μm. The full time-lapse sequence from which the frames in panel A are derived is shown in Video S1 in the supplemental material.
S. coelicolor DivIVA is, as reported here, the earliest recognized marker of the site where a new cell pole will be created during hyphal branching. Thereby, it also marks a new axis of cell polarity. In other organisms that have been investigated, DivIVA accumulates at cell poles and is recruited there already during a late stage of the division process (9, 20, 21, 24). The only documented exception is during spore germination of B. subtilis, when there are no nascent division sites and DivIVA is recruited directly to the preformed cell pole (13, 14). This observation, and the accumulation of the heterologously expressed B. subtilis protein at cell poles of E. coli (7), have fueled the speculation that DivIVA may have a preference for some general property of the cell pole. However, in the subapical cells in S. coelicolor mycelium, where branching typically occurs, DivIVA does not accumulate at the two available poles—the hyphal crosswalls that limit the cell—even though a faint signal can be seen at some septa (10). Instead, novel DivIVA foci are initiated on the inside of the lateral hyphal wall, and new branches subsequently emerge from such foci (Fig. 1).
DivIVA foci and branches form preferentially at curved hyphal walls.
In a very simple model, DivIVA can start de novo assembly at a new site along the hyphal side wall through oligomerization and cooperative interactions without preference for certain sites or preexisting nucleation points. However, we noted in the time-lapse analyses (Fig. 1; see Video S1 in the supplemental material) an interesting preference of branches to appear on the outer side of bent or slightly curved hyphae. These were not curvatures that arose as part of the branching, but rather hyphal shapes established already during growth (note that the hyphal walls are laid down during growth at the tip and then remain more or less inert). Of a total of 120 branching events that were observed, 68% emerged from the outer convex side of curved hyphae, and only two branches appeared to emerge from the inner concave side (data not shown). A total of 15% of the branches emerged from hyphal segments that looked straight, and 15% of the cases were not possible to clearly classify. It should be noted that we were able to monitor hyphal curvature in only two planes (x and y), and that the latter two classes are likely to include cases were curvature escaped detection since it was in the z plane. This bias in selection of site may reflect a preference of Streptomyces DivIVA for curved membrane surfaces. However, this does not target DivIVA directly to an existing pole in the subapical cell. Instead, DivIVA appears involved in establishing tip extension at a new site in the lateral wall, thereby creating a new cell pole.
DivIVA can trigger establishment of new sites of cell wall assembly and lateral branching.
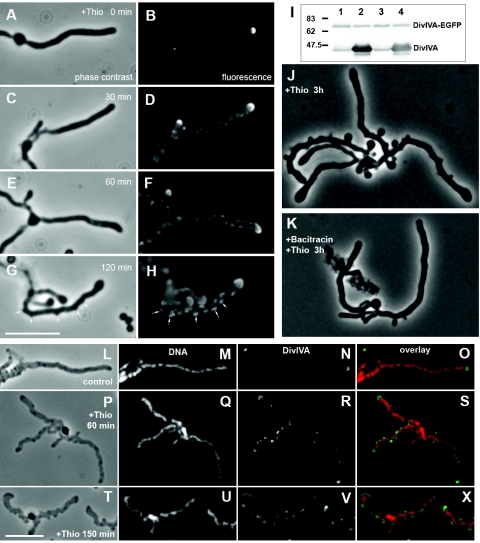
Induction of divIVA overexpression in S. coelicolor hyphae results in the emergence of multiple branch-like lateral outgrowths (10). This could be caused if the overproduced DivIVA titrated a branching inhibitor away from potential branching sites. Alternatively, DivIVA may play a direct and positive role by recruiting components required for establishing a branch to new sites. In order to distinguish between these models, we determined how DivIVA was localized during such overexpression experiments. Plasmid pKF59 carrying the Φ(divIVA-egfp)Hyb fusion was integrated into the divIVA locus of S. coelicolor A3(2) strain K114, which carries a thiostrepton-inducible copy of divIVA controlled by the tipAp promoter integrated at the attpSAM2 site (10), resulting in the new strain K121 (divIVA::pKF59[Φ(divIVA-egfp)Hyb] attBpSAM2::pKF58[tipAp-divIVA]). In the absence of the inducer in complex yeast extract-malt extract (YEME) medium (18), this strain grew normally, accumulated DivIVA-EGFP at each hyphal tip (Fig. 2A), and contained approximately equal amounts of EGFP-tagged and normal DivIVA (Fig. 2I). After the addition of thiostrepton (10 μg/ml), the normal DivIVA was strongly overproduced while the DivIVA-EGFP level was not affected (Fig. 2I). The shape of the apical cluster of DivIVA gradually became broader and lined the apical part of the swelling hyphal tip (Fig. 2). Furthermore, the signal intensity of the apical clusters decreased while fluorescence along the hyphal cells gradually increased, suggesting a redistribution of DivIVA-EGFP to new sites, presumably mixed with the overexpressed nontagged DivIVA. The signal soon developed into discrete foci of fluorescence along the hyphae (typical examples are shown in Fig. 2). By 2 h, multiple lateral bulges or outgrowths were readily visible, and they were always associated with DivIVA-EGFP clusters. In order to exclude that these patterns were artifacts caused by the EGFP tag, we repeated the experiments with strain K114 (divIVA+ attBpSAM2::pKF58[tipAp-divIVA]) (10), and visualized DivIVA with immunofluorescence microscopy. This revealed a very similar behavior with multiple lateral foci detected after 1 h and lateral bulges associated with such foci 2.5 h after addition of the inducer (Fig. 2L to X). We have previously used fluorescently labeled vancomycin to visualize active cell assembly at such lateral outgrowths resulting from divIVA overexpression (10). The new finding that the multiple outgrowths emerge at sites where overexpressed DivIVA has formed foci shows that DivIVA can act to recruit or activate components necessary to initiate branching. It remains unclear why this does not lead to normally shaped branches under overexpression conditions. The large amount of DivIVA that accumulated at the new tips may disturb cell shape determination (10), but it is also possible that other factors needed for proper elongation at these tips become limiting.
FIG. 2.
Ectopic overexpression of divIVA leads to redistribution of apical DivIVA, assembly of multiple lateral foci, and eventually initiation of cell wall assembly at sites coinciding with such foci. (A to H) Representative images of strain K121 (divIVA::pKF59[Φ(divIVA-egfp)Hyb] attBpSAM2::pKF58[tipAp-divIVA]) before and at intervals after induction of overexpression of tipAp-divIVA by addition of the inducer thiostrepton (Thio) to 10 μg/ml in YEME medium. Phase contrast images (A, C, E, and G) and fluorescence images of live cells show distribution of the DivIVA-EGFP protein (B, D, F, and H). Arrows indicate examples of the branch-like lateral outgrowths emerging at DivIVA foci. Immunoblotting with an anti-DivIVA antiserum (I) showed that the amount of DivIVA-EGFP did not change in response to induction of the tipAp promoter, while there was strong overproduction of normal DivIVA. Two independent cultures were each split in two halves, and thiostrepton (10 μg/ml) was added to one of the halves (lanes 2 and 4) while the other was uninduced for 2 h (lanes 1 and 3). A molecular weight standard is indicated. (J and K) Phase contrast images of cells from a similar experiment as described for panels A to H, except that 5 min prior to thiostrepton addition, bacitracin was added to block cell wall synthesis (K), or a mock addition of a corresponding amount of 0.1 M HCl was made (J). (L to X) Immunofluorescence microscopy images showing the relocalization of DivIVA in strain K114 (tipAp-divIVA) in response to overexpression induced by addition of thiostrepton (10 μg/ml) in YEME. Hyphae were attached to poly-l-lysine-coated slides and prepared for immunofluorescence essentially as described previously (27). DNA was stained with 7-aminoactinomycin D, and DivIVA was visualized using an anti-DivIVA antiserum and a secondary anti-rabbit immunoglobulin G antibody conjugated to Alexa Fluor 488 (Molecular Probes). Bars, 6 μm.
In order to confirm that formation of these branch-like outgrowths required new cell wall synthesis and were not caused merely by local weakening of the cell wall, divIVA overexpression was induced in strain K121 shortly after blocking cell wall growth by bacitracin (50 μg/ml), which arrests export of the peptidoglycan precursor lipid II (28) and rapidly stops hyphal growth in S. coelicolor. In this case, no outgrowths or bulges emerged from the lateral walls (Fig. 2K). Thus, the morphological effects of divIVA overexpression required de novo peptidoglycan synthesis and were not caused only by autolytic processes.
The finding that S. coelicolor DivIVA formed multiple new foci upon overexpression, and that at least some of these foci appeared to trigger peptidoglycan assembly, is consistent with a model in which DivIVA foci can recruit the cell wall synthesis machinery to new sites. Alternatively, DivIVA could activate or stabilize already positioned enzyme complexes and therefore give rise to multiple new branches after overproduction. In both scenarios, DivIVA would act to promote branching and play a positive role in establishment of new zones of cell wall growth. A recent report showed interaction with an apically localized cellulose-synthase-like protein of unclear function (30), but no direct interactions have yet been demonstrated between S. coelicolor DivIVA and any proteins involved in export and polymerization of peptidoglycan units. While the well-characterized DivIVA protein in B. subtilis has no documented role in cell wall growth, it is intriguing that B. subtilis has another DivIVA-like protein, GpsB, that is involved in controlling cell wall synthesis and interacts directly with penicillin-binding protein 1 (4). The results reported here strongly suggest that Streptomyces DivIVA directly or indirectly acts to recruit or activate cell wall synthesis enzymes. This is likely to be an essential function for hyphal growth and could explain why divIVA deletions are lethal in S. coelicolor (10).
Supplementary Material
Acknowledgments
We are grateful to Nora Ausmees, Mark Buttner, and Justin Nodwell for critical reading of the manuscript.
This work was supported by a postdoctoral stipend from Wenner-Gren Foundations to S.-B.W. and grants from the Swedish Research Council (no. 621-2004-4454), O.E. och Edla Johanssons Vetenskapliga Stiftelse, and Åke Wibergs Stiftelse to K.F. J.A.G. acknowledges a grant (B102008-00519) from the Ministerio de Ciencia y Technología. The foundations Crafoordska Stiftelsen, Carl Tesdorpfs Stiftelse, and Maja och Erik Lindqvists Forskningsstiftelse are gratefully acknowledged for support that allowed purchase of the fluorescence microscopy system.
Footnotes
Published ahead of print on 19 September 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Cabeen, M. T., and C. Jacobs-Wagner. 2005. Bacterial cell shape. Nat. Rev. Microbiol. 3601-610. [DOI] [PubMed] [Google Scholar]
- 2.Carballido-López, R. 2006. The bacterial actin-like cytoskeleton. Microbiol. Mol. Biol. Rev. 70888-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chauhan, A., H. Lofton, E. Maloney, J. Moore, M. Fol, M. V. Madiraju, and M. Rajagopalan. 2006. Interference of Mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol. Microbiol. 62132-147. [DOI] [PubMed] [Google Scholar]
- 4.Claessen, D., R. Emmins, L. W. Hamoen, R. A. Daniel, J. Errington, and D. H. Edwards. 2008. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol. Microbiol. 681029-1046. [DOI] [PubMed] [Google Scholar]
- 5.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113767-776. [DOI] [PubMed] [Google Scholar]
- 6.de Pedro, M. A., J. C. Quintela, J.-V. Höltje, and H. Schwarz. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 1792823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards, D. H., H. B. Thomaides, and J. Errington. 2000. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J. 192719-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliot, M. A., M. J. Buttner, and J. R. Nodwell. 2008. Multicellular development of Streptomyces, p. 419-438. In D. E. Whitworth (ed.), Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC.
- 9.Fadda, D., A. Santona, V. D'Ulisse, P. Ghelardini, M. G. Ennas, M. B. Whalen, and O. Massidda. 2007. Streptococcus pneumoniae DivIVA: localization and interactions in a MinCD-free context. J. Bacteriol. 1891288-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flärdh, K. 2003. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol. Microbiol. 491523-1536. [DOI] [PubMed] [Google Scholar]
- 11.Flärdh, K. 2003. Growth polarity and cell division in Streptomyces. Curr. Opin. Microbiol. 6564-571. [DOI] [PubMed] [Google Scholar]
- 12.Flärdh, K., and M. J. Buttner. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol., in press. [DOI] [PubMed]
- 13.Hamoen, L. W., and J. Errington. 2003. Polar targeting of DivIVA in Bacillus subtilis is not directly dependent on FtsZ or PBP 2B. J. Bacteriol. 185693-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harry, E. J., and P. J. Lewis. 2003. Early targeting of Min proteins to the cell poles in germinated spores of Bacillus subtilis: evidence for division apparatus-independent recruitment of Min proteins to the division site. Mol. Microbiol. 4737-48. [DOI] [PubMed] [Google Scholar]
- 15.Hett, E. C., and E. J. Rubin. 2008. Bacterial growth and cell division: a mycobacterial perspective. Microbiol. Mol. Biol. Rev. 72126-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104913-922. [DOI] [PubMed] [Google Scholar]
- 17.Kang, C. M., S. Nyayapathy, J. Y. Lee, J. W. Suh, and R. N. Husson. 2008. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology 154725-735. [DOI] [PubMed] [Google Scholar]
- 18.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 19.Kruse, T., J. Bork-Jensen, and K. Gerdes. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 5578-89. [DOI] [PubMed] [Google Scholar]
- 20.Letek, M., E. Ordónez, J. Vaquera, W. Margolin, K. Flärdh, L. M. Mateos, and J. A. Gil. 2008. DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum. J. Bacteriol. 1903283-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marston, A. L., H. B. Thomaides, D. H. Edwards, M. E. Sharpe, and J. Errington. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 123419-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazza, P., E. E. Noens, K. Schirner, N. Grantcharova, A. M. Mommaas, H. K. Koerten, G. Muth, K. Flärdh, G. P. van Wezel, and W. Wohlleben. 2006. MreB of Streptomyces coelicolor is not essential for vegetative growth but is required for the integrity of aerial hyphae and spores. Mol. Microbiol. 60838-852. [DOI] [PubMed] [Google Scholar]
- 23.McCormick, J. R., E. P. Su, A. Driks, and R. Losick. 1994. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol. Microbiol. 14243-254. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen, L., N. Scherr, J. Gatfield, A. Walburger, J. Pieters, and C. J. Thompson. 2007. Antigen 84, an effector of pleiomorphism in Mycobacterium smegmatis. J. Bacteriol. 1897896-7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramos, A., M. P. Honrubia, N. Valbuena, J. Vaquera, L. M. Mateos, and J. A. Gil. 2003. Involvement of DivIVA in the morphology of the rod-shaped actinomycete Brevibacterium lactofermentum. Microbiology 1493531-3542. [DOI] [PubMed] [Google Scholar]
- 26.Rigden, M. D., C. Baier, S. Ramirez-Arcos, M. Liao, M. Wang, and J. A. R. Dillon. 2008. Identification of the coiled-coil domains of Enterococcus faecalis DivIVA that mediate oligomerization and their importance for biological function. J. Biochem. 14463-76. [DOI] [PubMed] [Google Scholar]
- 27.Schwedock, J., J. R. McCormick, E. A. Angert, J. R. Nodwell, and R. Losick. 1997. Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomyces coelicolor. Mol. Microbiol. 25847-858. [DOI] [PubMed] [Google Scholar]
- 28.Stone, K. J., and J. L. Strominger. 1971. Mechanism of action of bacitracin: complexation with metal ion and C55-isoprenyl pyrophosphate. Proc. Natl. Acad. Sci. USA 683223-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomaides, H. B., M. Freeman, M. El Karoui, and J. Errington. 2001. Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev. 151662-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu, H., K. F. Chater, Z. Deng, and M. Tao. 2008. A cellulose synthase-like protein involved in hyphal tip growth and morphological differentiation in Streptomyces. J. Bacteriol. 1904971-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.