Abstract
Production of the Zn-metalloprotease hemagglutinin (HA)/protease by Vibrio cholerae has been reported to enhance enterotoxicity in rabbit ileal loops and the reactogenicity of live cholera vaccine candidates. Expression of HA/protease requires the alternate sigma factor σS (RpoS), encoded by rpoS. The histone-like nucleoid structuring protein (H-NS) has been shown to repress rpoS expression in Escherichia coli. In V. cholerae strains of the classical biotype, H-NS has been reported to silence virulence gene expression. In this study we examined the role of H-NS in the expression of HA/protease and motility in an El Tor biotype strain by constructing a Δhns mutant. The Δhns mutant exhibited multiple phenotypes, such as production of cholera toxin in nonpermissive LB medium, reduced resistance to high osmolarity, enhanced resistance to low pH and hydrogen peroxide, and reduced motility. Depletion of H-NS by overexpression of a dominant-negative allele or by deletion of hns resulted in diminished expression of HA/protease. Epistasis analysis of HA/protease expression in Δhns, ΔrpoS, and Δhns ΔrpoS mutants, analysis of RpoS reporter fusions, quantitative reverse transcription-PCR measurements, and ectopic expression of RpoS in ΔrpoS and ΔrpoS Δhns mutants showed that H-NS posttranscriptionally enhances RpoS expression. The Δhns mutant exhibited a lower degree of motility and lower levels of expression of flaA, flaC, cheR-2, and motX mRNAs than the wild type. Comparison of the mRNA abundances of these genes in wild-type, Δhns, ΔrpoS, and Δhns ΔrpoS strains revealed that deletion of rpoS had a more severe negative effect on their expression. Interestingly, deletion of hns in the rpoS background resulted in higher expression levels of flaA, flaC, and motX, suggesting that H-NS represses the expression of these genes in the absence of σS. Finally, we show that the cyclic AMP receptor protein and H-NS act along the same pathway to positively affect RpoS expression.
Cholera is an acute diarrheal disease characterized by the passing of voluminous rice water stool. Vibrio cholerae of serogroups O1 and O139 continues to cause seasonal cholera outbreaks that affect highly populated regions in Asia, Africa, and Latin America. The bacterium can persist outside the human host and alternates between planktonic and biofilm community lifestyles. V. cholerae is a highly motile organism with a single sheathed polar flagellum. It colonizes the small intestine and expresses a variety of virulence determinants, such as the toxin-coregulated pilus (TCP) and cholera toxin (CT), encoded by tcpA (major subunit) and ctxA, respectively (13, 27). V. cholerae also secretes a Zn-dependent metalloprotease that displays a broad range of potentially pathogenic activities: hemagglutinin/protease (HA/protease), encoded by hapA (14, 18, 34, 59-61). HA/protease contributes to the reactogenicity of live cholera vaccine candidates and enhances enterotoxicity in the rabbit ileal loop model (4, 15, 51). Transcription of hapA requires the quorum-sensing regulator HapR (25), the cyclic AMP receptor protein (CRP) (3, 49, 31), and the alternative sigma factor S (RpoS), encoded by rpoS (49, 62).
The histone-like nucleoid structuring protein (H-NS), a global regulator of environmentally controlled gene expression, belongs to a family of small nucleoid-associated proteins that also include its paralog StpA, Fis, the heat-unstable protein (HU), and integration host factor (IHF) (13). Mutations that inactivate hns are highly pleiotropic, suggesting that H-NS influences a broad spectrum of physiological processes (1, 21). Escherichia coli H-NS (137 amino acids) consists of an N-terminal domain extending to residue 65 with oligomerization activity, connected by a flexible linker to a nucleic acid binding domain beginning at residue 90, whereas V. cholerae H-NS contains an additional oligomerization domain located within the first 24 amino acids of the protein's N terminus (1, 12, 44). Both oligomerization and DNA binding are required for the biological activity of H-NS (54). It has been suggested that H-NS functions primarily as a transcriptional repressor by binding to highly curved AT-rich DNA regions (46, 57).
Depletion of H-NS in V. cholerae of the classical biotype (by overexpression of a truncated hns allele lacking the DNA binding domain) induced pleiotropic effects such as the formation of mucoid colonies and reduced swarming in semisolid agar (56). Furthermore, V. cholerae hns mutants have been reported to exhibit diminished bile- and anaerobiosis-mediated repression of ctxA expression, motility, and intestinal colonization capacity (16, 30). H-NS has been shown to silence virulence gene expression by repressing transcription at different levels of the ToxR regulatory cascade, which include the toxT, tcpA, and ctxA promoters (45). At the level of the ctxA and tcpA promoters, the positive regulator ToxT binds to these promoters, displacing H-NS and allowing RNA polymerase (RNAP) to initiate transcription (63).
In E. coli, RpoS regulates the expression of more than 100 genes in response to starvation and other stresses such as osmotic shock, oxidative stress, and temperature (19). The intracellular concentration of σS is controlled at the levels of transcription, translation, and protein stability (19, 43, 58). At the level of transcription, the cyclic AMP-CRP complex has been shown to modulate the activity of an rpoS-lacZ transcriptional fusion by binding to CRP boxes in the rpoS promoter (19). Also, polyphosphate and guanosine tetraphosphate (ppGpp) have been shown to enhance rpoS transcription (19). At the level of translation, several trans-acting factors can bind to rpoS mRNA in response to high osmolarity, low temperature, and an acidic pH to enhance its translation (19). These factors include the RNA binding protein Hfq, the nucleoid-associated protein HU, and the small RNAs DsrA and RprA (19, 32). In addition, two riboregulators, OxyS and DsrA, enhance and diminish rpoS translation, respectively (19, 43, 58). At the protein level, the response regulator RssB is required for the degradation of σS by the housekeeping protease ClpXP (2, 19, 43, 58). H-NS has been shown to participate as a negative factor in the expression of RpoS by promoting rpoS mRNA degradation (6) and increasing the activity of RssB to enhance the proteolysis of σS (64).
V. cholerae rpoS mutants are more sensitive to starvation, high osmolarity, and oxidative stress than the wild type (62), suggesting an analogy to the role of RpoS in E. coli. However, very little is known about the regulation of RpoS expression in V. cholerae. Recently, it has been shown that multiple motility and chemotaxis genes are downregulated in an rpoS mutant, suggesting that RpoS could facilitate mucosal escape during infection by enhancing motility (42). Furthermore, V. cholerae rpoS mutants are affected in intestinal colonization (39), suggesting that RpoS could contribute to the ability of vibrios to overcome host-specific stresses in the gut. Since both H-NS and RpoS have been shown to affect cholera pathogenesis, we decided to examine the interaction between these global regulators. In this study we show that H-NS exerts positive regulation over RpoS and multiple RpoS-dependent genes, including hapA. Furthermore, we provide evidence that H-NS can affect the expression of some motility and chemotaxis genes through distinct and overlapping effects: (i) as a positive factor by posttranscriptionally enhancing RpoS expression and (ii) as a repressor or negative factor in the absence of σS. Finally, we show that CRP and H-NS act on the same pathway to enhance the expression of RpoS.
MATERIALS AND METHODS
Strains and media.
The V. cholerae and E. coli strains used in this study are shown in Table 1. For CT production, V. cholerae strains were grown in AKI (23) and LB media at 30°C. For HA/protease production, vibrios were grown in Bacto tryptic soy broth (TSB) at 37°C with shaking (250 rpm). For motility determination, strains were stabbed into LB medium containing 0.3% agar (swarm agar) and incubated at 30°C for 16 h. Plasmid DNA was introduced into V. cholerae by electroporation (33). Culture media were supplemented with ampicillin (Amp; 100 μg/ml), kanamycin (Km; 50 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 20 μg/ml), isopropyl-β-d-thiogalactopyranoside (IPTG; 20 μg/ml), or polymyxin B (PolB; 100 U/ml) as required.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| S17-1λPir | F−recA hsdR RP4-2 (Tc::Mu) (Km::Tn7) lysogenized with λpir phage | 9 |
| Vibrio cholerae | ||
| C7258 | Wild type, El Tor biotype | Peru, 1991 |
| C7258ΔlacZ | C7258 lacZ deletion mutant | This study |
| AJB80 | C7258ΔlacZ Δhns::Km | This study |
| AJB50ΔlacZ | C7258ΔlacZ ΔrpoS | This study |
| AJB51ΔlacZ | C7258ΔlacZ ΔhapR | This study |
| WL7258ΔlacZ | C7258ΔlacZ Δcrp | This study |
| AJB81 | AJB80 ΔrpoS | This study |
| AJB82 | AJB80 Δcrp | This study |
| Plasmids | ||
| pHNS12 | 0.9-kb SacI-BamHI DNA fragment 5′ of the hns ORF amplified with primers Hns120 and Hns1018 in pUC19 | This study |
| pHNS34 | 0.8-kb BamHI-SphI DNA fragment 3′ of the hns ORF amplified with primers Hns1467 and Hns2289 in pUC19 | This study |
| pΔHNS | BamHI-SphI fragments from pHNS34 in pHNS12 | This study |
| pΔHNSK | 1.2-kb BamHI Kmr cassette from pUC4K in pΔHNS | This study |
| pCVDΔHNSK | SacI-SphI Δhns::Km fragment from pΔHNSK in pCVD442 | This study |
| pHNS | 0.4-kb BamHI-HindIII DNA fragment carrying the hns ORF amplified with primers Hns162 and HnsD2 in pTTQ18 | This study |
| pTTHNS90 | 0.3-kb BamHI-HindIII fragment encoding 90 amino acids of the H-NS N terminus amplified with primers Hns162 and Hns429 in pTTQ18 | This study |
| pTTRpoS | 1.0-kb EcoRI-HindIII fragment carrying rpoS amplified with primers RpoS489 and RpoS1511 in pTTQ18 | This study |
| pTT3 | XbaI-PstI DNA fragment containing the rrnB T1T2 transcription terminator in pUC19 | 7, 52 |
| pTT3Lux | 0.3-kb SphI-HindIII DNA fragment carrying the V. harveyi luxC promoter amplified with primers LuxC84 and LuxC434 in pTT3 | This study |
| pLux-LacZ | 0.5-kb KpnI-HindIII fragment containing rrnB T1T2 and the luxC promoter fused to a promoterless lacZ gene in pKRZ1 | This study |
| pTT3PerA | 0.9-kb SphI-HindIII fragment carrying perA amplified with primers PerA498 and PerA987 in pTT3 | This study |
| pPerA-LacZ | 1.1-kb KpnI-HindIII fragment containing rrnB T1T2 and the perA promoter fused to a promoterless lacZ gene in pKRZ1 | This study |
| pRpoSLac5 | A SphI-HindIII DNA fragment containing the rpoS promoter region was cloned between the rrnB T1T2 transcription terminator and a promoterless lacZ gene in pKRZ1 | 49 |
| pLacZ12 | 0.9-kb SacI-BamHI DNA fragment 5′ of the lacZ ORF amplified with primers LacZ6 and LacZ859 in pUC19 | This study |
| pLacZ34 | 0.8-kb BamHI-SphI DNA fragment 3′ of the lacZ ORF amplified with primers LacZ3236 and LacZ4029 in pUC19 | This study |
| pΔLacZ | 0.8-kb BamHI-SphI DNA fragment from pLacZ34 in pLacZ12 | This study |
| pCVDΔLacZ | 1.6-kb SacI-SphI fragment from pΔLacZ in pCVD442 | This study |
Construction of mutants.
Deletion mutants were constructed by allelic exchange using strain C7258 as a wild-type precursor. V. cholerae target sequences were amplified from genomic DNA of strain C7258 by using the Advantage PCR system (BD Biosciences Clontech). All primers (Table 2) were designed based on the DNA sequence of the V. cholerae N16961 genome, downloaded from the TIGR database (http://cmr.tigr.org). Amplification products were directionally cloned into pUC19 using E. coli TOP10 as the host and were confirmed by sequencing of both DNA strands with M13 forward and reverse primers. In all cases, mutants were obtained by cloning a chromosomal DNA fragment containing a deletion of the target gene in pCVD442 (11). The resulting suicide vectors were constructed in E. coli S17-1λpir (9) and mobilized into the corresponding V. cholerae receptor strain by conjugation. Exconjugants were selected on LB agar containing PolB, Amp, or Km as required. Next, sucrose selection was used to isolate segregants retaining the mutant allele. Using this approach, strain C7258 and the isogenic mutants AJB50 (C7258 ΔrpoS), AJB51 (C7258 ΔhapR), and WL7258 (C7258 Δcrp), described previously (31), were modified by deletion of the chromosomal lacZ gene. The corresponding lacZ deletion mutants (Table 1) were confirmed by DNA sequencing. A similar strategy was followed to construct the hns deletion/insertion (Δhns::Km) mutant AJB80 (Table 1) starting from C7258ΔlacZ. Strain AJB80, containing the Δhns::Km allele, was confirmed by Southern blotting and sequencing of the V. cholerae chromosome-Km junctions. Strains AJB81 (ΔlacZ Δhns::Km ΔrpoS) and AJB82 (ΔlacZ Δhns::Km Δcrp) (Table 1) were similarly obtained from AJB80 by allelic exchange using the suicide vectors pCVDΔRpoS2 and pCVDΔCRP (31), respectively. Again, mutants were confirmed by DNA sequencing. A detailed description of the strain construction procedures is provided as supplemental material.
TABLE 2.
Primers
| Primer name | Sequencea |
|---|---|
| Bfr146 | 5′-TTGATGAGATGAAACACGCTGACCA |
| Bfr431 | 5′-GCTTGCAGATAATTTTGGATACCCG |
| CheR377 | 5′-TAGAAGTGCAGCAAAAACGTCCGG |
| CheR587 | 5′-CCATTAAGTTTTGCGGACGGAAGT |
| Cyt835 | 5′-GCTGTGGATATCATGGGGCGTTTA |
| Cyt948 | 5′-TCGTTGGATGGCGGTAAAATGGGT |
| FlaA40 | 5′-GCACAACGTTATCTGACCAAAGGC |
| FlaA291 | 5′-GAGTTGGTACCGTTCGCCGATTG |
| FlaC405 | 5′-CAAGCTGCTCAACGGTACATTCG |
| FlaC592 | 5′-GCCTTCGCGTTTGTCTTTCAGAGT |
| Hns120 | 5′-GAAGAGCTCGAAGATGGTGAACGTA |
| Hns162 | 5′-GAAGGATCCGGAAATGGTAATGTCG |
| Hns429 | 5′-GGCCAAGCTTTTTCGTTTTAGTTTCG |
| Hns1018 | 5′-GCAGGATCCATGAAGGTAAATCTCT |
| Hns1467 | 5′-GATGGATCCTGACAAGTTTTCGCTG |
| Hns2289 | 5′-GATGCATGCCCCTCTTTGACAAACA |
| HnsD2 | 5′-GGGAAAGCTTGGCAAAATTACAGAG |
| KatB594 | 5′-TACCGAACAAGGCAACTGGGATTT |
| KatB792 | 5′-GTTCGATAGCTTGCCGGTGTACT |
| LacZ6 | 5′-GCGGATCCAGCCGAGGAGTAAAGA |
| LacZ859 | 5′-GCGAGCTCCGAAAATGACTGTTGT |
| LacZ3236 | 5′-GCGGATCCAAAGCAAGAGCCA |
| LacZ 4029 | 5′-GCGCGCATGCAACTCGGCTATCGTCC |
| LuxC84 | 5′-GCGCAAGCTTAATCGATTTTCTTCAGTAG |
| LuxC434 | 5′-GCGCGCATGCACACTGTCACACATC |
| MotX253 | 5′-GGAGTGTGTGTGGATCAGGATGTT |
| MotX374 | 5′-GGATCGCGCGTTCTTTGTCTTGTT |
| PerA498 | 5′-GTTGCATGCCTTTACCACCTTGATC |
| PerA607 | 5′-GAAAATAGCCGTTACTCTGGTCAGC |
| PerA863 | 5′-CTAAGCCTTGTTCGTGCAGTTCAG |
| PerA987 | 5′-GCGAAGCTTATTGATTACTCCTTGC |
| RecA578 | 5′-GTGCTGTGGATGTCATCGTTGTTG |
| RecA863 | 5′-CCACCACTTCTTCGCCTTCTTTGA |
| RpoS576 | 5′-CGATTTTGAAGATGAAGCACTGGAAG |
| RpoS489 | 5′-GTTGAATTCGAGGCCGCTATGAGT |
| RpoS1003 | 5′-TGTTTGGTTCATCAGCGCACGTTC |
| RpoS1511 | 5′-GCGAAGCTTTTTTGGATGAGTCTGG |
| ToxR369 | 5′-GGAAACGGTTGAAGAAGAGATGGC |
| ToxR498 | 5′-TTATTCGTCACAACATTGGCTGGC |
Restriction sites used for directional cloning are underlined.
Plasmid constructions.
The complete hns open reading frame (ORF) was amplified from strain C7258 and cloned into pUC19 to yield plasmid pHNS (Table 1). A truncated hns allele (hns90) was amplified from C7258 to generate a DNA fragment encoding the first 90 amino acids of H-NS. The truncated allele was cloned into plasmid pTTQ18 (55) to create the expression vector pTTHNS90 (Table 1), expressing hns90 from the Tac promoter. Similarly, the complete rpoS ORF was amplified and cloned into pTTQ18 to yield the expression vector pTTRpoS (Table 1). The RpoS reporter plasmid pPerA-LacZ (Table 1) was constructed by cloning the perA promoter region downstream of the rrnB T1T2 transcription terminator into pTT3 (Table 1) and subsequently subcloning the terminator-promoter fragment upstream of a promoterless lacZ gene into plasmid pKRZ1 (48). An identical strategy was used to construct a HapR reporter plasmid. In this case, we used the lux promoter amplified from cosmid pBB1 containing the Vibrio harveyi lux operon (41) to generate plasmid pLux-LacZ (Table 1). The construction of the rpoS-lacZ promoter fusion pRpoSLac5 (Table 1) has been described previously (49).
RT-PCR.
V. cholerae strains were grown as described in each figure legend, and total RNA was isolated using the RNeasy kit (Qiagen Laboratories). The RNA samples were analyzed by quantitative real-time reverse transcription-PCR (qRT-PCR) using the iScript two-step RT-PCR kit with Sybr green (Bio-Rad Laboratories) as described previously (31). Relative expression values (R) were calculated as 2−(ΔCt target − ΔCt reference) where Ct is the fractional threshold cycle. recA mRNA was used as a reference. The following primer combinations were used: Bfr146 and Bfr431 for bfr mRNA, CheR377 and CheR587 for cheR-2 mRNA, Cyt835 and Cyt948 for c551 mRNA, FlaA40 and FlaA291 for flaA mRNA, FlaC405 and FlaC592 for flaC mRNA, KatB594 and KatB792 for katB mRNA, MotX253 and MotX374 for motX mRNA, PerA607 and PerA863 for perA mRNA, RecA578 and RecA863 for recA mRNA, RpoS576 and RpoS1003 for rpoS mRNA, and ToxR369 and ToxR498 for toxR mRNA. A control mixture lacking reverse transcriptase was run for each reaction to exclude chromosomal DNA contamination.
To assess the longevity of rpoS mRNA in the wild-type and Δhns::Km mutant backgrounds, the strains were grown to an optical density at 600 nm (OD600) of 1.5, and rifampin was added (150 μg/ml) to block transcription. Samples were taken at different time points after the addition of rifampin, and rpoS mRNA was detected using the Titanium One-Step RT-PCR kit by following the manufacturer's instructions, with 20 ηg of total RNA and 25 cycles of amplification. Since less rpoS mRNA is produced in the Δhns::Km mutant than in the wild type (see below), two additional cycles were provided for the mutant to generate a stronger starting signal.
Stress response assays.
V. cholerae strains were grown for 18 h (to stationary phase) in LB medium at 37°C. The cells were centrifuged, washed, and resuspended in 1 volume of LB medium. The bacterial suspension was then inoculated into fresh LB medium that either contained 2.4 M NaCl or 3 mM hydrogen peroxide or was brought to pH 4.5 so as to obtain a cell density of 106 to 107 cells per ml. Cultures were incubated at 37°C with shaking (250 rpm), and samples were taken at different time points for dilution plating.
Determination of CT levels.
CT levels were determined by a GM1 enzyme-linked immunosorbent assay using a standard curve of pure CT (Sigma Chemical Co.) as described previously (31).
Enzyme assays.
Production of HA/protease was measured using an azocasein assay as described previously (3). One azocasein unit is the amount of enzyme that produces an increase of 0.01 OD unit in this assay. β-Galactosidase activity was measured as described previously (40) using the substrate o-nitrophenyl-β-d-galactopyranoside (ONPG). Specific activities are given in Miller units and calculated as [1,000 (OD420/t × v × OD600)] where t is the reaction time and v is the volume of enzyme extract per reaction.
RESULTS
Construction of a V. cholerae El Tor biotype hns deletion/insertion mutant.
We have constructed an hns deletion/insertion mutant (Δhns::Km) of the El Tor biotype strain C7258ΔlacZ. The mutant was confirmed by Southern blot analysis and DNA sequencing. Since it has been reported that H-NS negatively influences virulence gene expression in classical biotype V. cholerae, we first determined whether or not H-NS had a similar effect in the El Tor biotype mutant. The El Tor biotype Δhns::Km mutant AJB80 produced 15.7 mg/liter/OD600 unit of CT in AKI cultures, compared to 2.6 mg/liter/OD600 unit produced by its wild-type precursor (C7258ΔlacZ) under identical conditions. Furthermore, while the wild-type strain did not produce detectable CT in nonpermissive LB medium, the Δhns::Km mutant produced 3.8 mg/liter/OD600 unit of CT in LB medium. Moreover, in contrast to its wild-type precursor, the Δhns::Km mutant expressed significant autoagglutination, an indicator of TCP expression (8), in LB medium (data not shown). These phenotypes were complemented by providing the hns gene in trans using plasmid pHNS. The results discussed above suggest that, as documented for hns mutants of the classical biotype (45), H-NS acts to silence virulence gene expression in the El Tor biotype. In addition, the Δhns::Km mutant exhibited additional phenotypes typical of hns mutants, such as reduced growth rate and motility (see below).
H-NS acts upstream of RpoS to positively regulate the production of HA/protease.
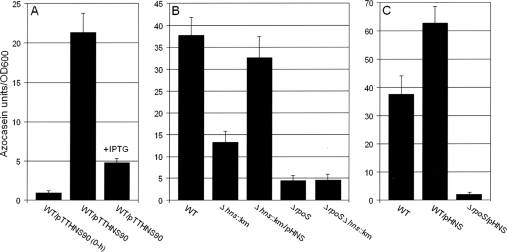
We have shown that transcription of hapA occurs in the stationary phase and requires RpoS (49). Since in E. coli H-NS has been reported to repress the expression of multiple RpoS-dependent genes (19, 20), we examined the role of H-NS in HA/protease expression. It has been reported that overexpression of a truncated hns allele lacking the DNA-binding domain leads to phenotypes reminiscent of those observed for hns mutants (56). Thus, we first examined if depletion of H-NS by overexpression of the truncated allele hns90 affected the production of HA/protease. As shown in Fig. 1A, induction of hns90 with IPTG had a strong negative effect on HA/protease expression relative to that by an uninduced control. In agreement with this result, elimination of hns by mutation in strain AJB80 (Δhns::Km) strongly diminished the production of HA/protease (Fig. 1B). This phenotype was complemented by providing the wild-type hns allele in trans (Fig. 1B). As shown in Fig. 1B, deletion of rpoS had a more severe effect on HA/protease production (P < 0.05 by the t test). In order to examine the relationship between H-NS and RpoS in the regulation of HA/protease production, we constructed the Δhns::Km ΔrpoS double mutant AJB81 and performed an epistasis analysis. Production of HA/protease by the Δhns::Km ΔrpoS double mutant was similar to that by the ΔrpoS mutant, indicating that RpoS acts downstream of H-NS to activate HA/protease production (Fig. 1B). In Fig. 1C, we show that ectopic expression of H-NS in wild-type V. cholerae results in increased HA/protease production (P < 0.05 by the t test). Consistent with our epistasis analysis, overexpression of H-NS did not restore HA/protease expression in a ΔrpoS background. Taken together, we conclude that H-NS positively affects HA/protease expression by acting upstream of RpoS.
FIG. 1.
Role of H-NS in HA/protease production. (A) Strain C7258ΔlacZ (wild type [WT]) containing pTTHNS90 was grown in TSB to an OD600 of 1, and the culture was divided in half. One half was induced by addition of IPTG, and the second half was used as a control. Cultures were further incubated for 3 h. (B) Strains C7258ΔlacZ (WT), AJB80 (C7258ΔlacZ Δhns::Km), AJB50ΔlacZ (ΔrpoS), and AJB81 (ΔlacZ Δhns::Km ΔrpoS) were grown to stationary phase (16 h) in TSB at 37°C. (C) Strain C7258ΔlacZ (WT) and strains C7258ΔlacZ and AJB50ΔlacZ (ΔrpoS) containing plasmid pHNS were grown as described above. Production of HA/protease (expressed in azocasein units) was measured as described in Materials and Methods. Each value is the average for three independent cultures. Error bars, standard deviations.
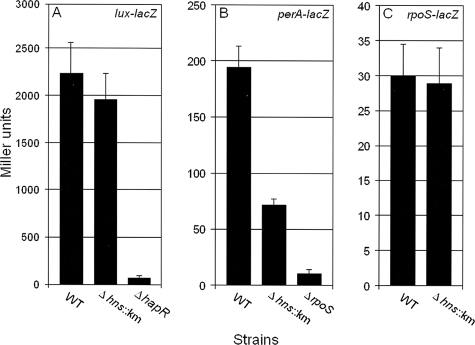
Since production of HA/protease requires both RpoS and the quorum-sensing regulator HapR, we examined if expression of these regulators was diminished in the Δhns::Km mutant AJB80. For HapR expression, we constructed the reporter plasmid pLux-LacZ (Table 1). Expression of β-galactosidase from the lux promoter is dependent on the production of active HapR protein. As shown in Fig. 2A, the Δhns::Km mutant produced a level of β-galactosidase activity similar to that of the wild type, while an isogenic ΔhapR mutant (AJB51ΔlacZ), used as a negative control, produced very little activity.
FIG. 2.
Expression of HapR and RpoS reporter lacZ fusions in V. cholerae hns mutants. (A) Strains C7258ΔlacZ (wild type [WT]), AJB80 (C7258ΔlacZ Δhns::Km), and AJB51ΔlacZ (ΔhapR) containing plasmid pLux-LacZ were grown to stationary phase (16 h) in TSB at 37°C. (B) Strains C7258ΔlacZ (WT), AJB80 (C7258ΔlacZ Δhns::Km), and AJB50ΔlacZ (ΔrpoS) containing plasmid pPerA-LacZ were grown as described above. (C) Strains C7258ΔlacZ (WT) and AJB80 (C7258ΔlacZ Δhns::Km) containing an rpoS-lacZ transcriptional fusion were grown as described above. β-Galactosidase activity was determined as described in Materials and Methods. Each value is the average for three independent cultures. Error bars, standard deviations.
Preliminary gene expression profiling of a ΔrpoS mutant of strain C7258 revealed that the VC1560 gene, annotated as catalase-peroxidase (perA), and VC0365 (bacterioferritin; bfr) were strongly downregulated in the ΔrpoS mutant (data not shown). Using the same strategy as that for the HapR reporter plasmid, we constructed a reporter plasmid (pPerA-LacZ) containing a perA-lacZ transcriptional fusion to monitor the production of active RpoS protein. As shown in Fig. 2B, significantly less RpoS activity was detected in the Δhns::Km mutant than in the wild type, while negligible activity was observed in an isogenic ΔrpoS mutant used as a negative control. These results indicate that the lower production of HA/protease by the Δhns::Km mutant is due to diminished expression of active RpoS protein.
Positive regulation of RpoS by H-NS is posttranscriptional.
To determine if the lower expression level of RpoS in the Δhns::Km mutant (Fig. 2B) was due to reduced transcription, we introduced the rpoS-lacZ transcriptional fusion pRpoSLac5 (49) into the wild-type and Δhns::Km mutant strains. As shown in Fig. 2C, the two strains produced similar β-galactosidase activities, suggesting that H-NS affects RpoS expression by a posttranscriptional mechanism. In E. coli, H-NS has been reported to affect rpoS expression at the levels of translation and protein stability (19, 64). Therefore, we used qRT-PCR to measure the abundances of rpoS mRNA and the RpoS-dependent genes perA and bfr in C7258ΔlacZ and the Δhns::Km mutant. As shown in Fig. 3, the Δhns::Km mutant produced a smaller amount of rpoS mRNA than the wild type, and this was reflected in diminished expression of the RpoS-dependent genes perA and bfr. These results suggest that H-NS has a strong positive effect on rpoS mRNA translation/stability. To further test the hypothesis that H-NS regulates rpoS expression posttranscriptionally, we constructed the pTTRpoS vector (Table 1), which expresses RpoS from the Tac promoter. In Fig. 4 we show that IPTG-induction of pTTRpoS in strain AJB50lacZ (ΔrpoS) complemented the RpoS defect, leading to HA/protease production. However, significantly less HA/protease could be detected upon induction of pTTRpoS in strain AJB81 (Δhns::Km ΔrpoS). Taken together, our data suggest that in contrast to E. coli, in V. cholerae H-NS posttranscriptionally enhances the expression of RpoS and multiple RpoS-dependent genes, such as hapA, perA, and bfr. We further examined the stability of rpoS mRNA in the wild-type and Δhns::Km mutant strains. As shown in Fig. 5, and consistent with a posttranscriptional regulatory mechanism, the rpoS mRNA signal was found to disappear more rapidly in the Δhns::Km mutant than in the wild-type after transcription had been blocked with rifampin.
FIG. 3.
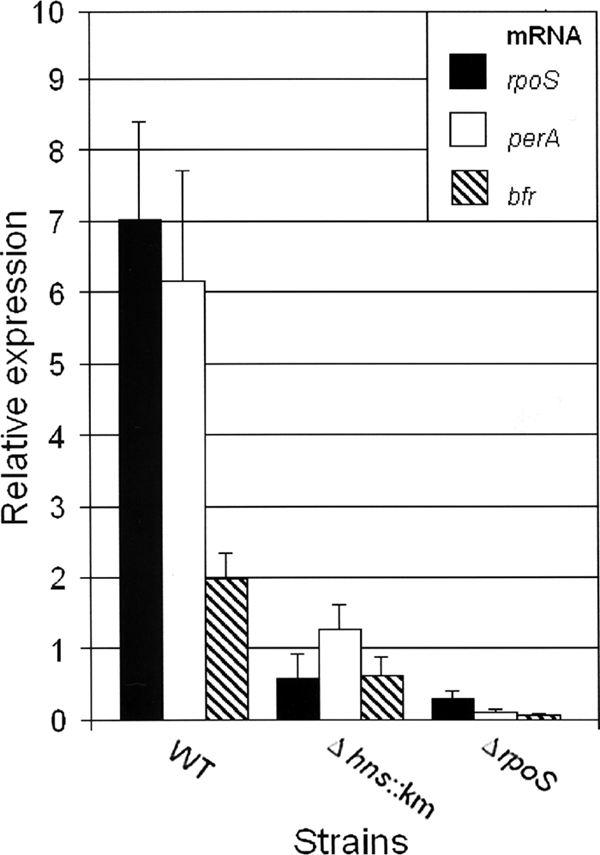
qRT-PCR analysis of rpoS expression in an hns mutant. Strains C7258ΔlacZ (wild type [WT]), AJB80 (C7258ΔlacZ Δhns::Km), and AJB50ΔlacZ (ΔrpoS) were grown to stationary phase (16 h) in TSB at 37°C. RNA was extracted, and rpoS, perA, and bfr mRNA abundances were determined by qRT-PCR as described in Materials and Methods. Results are averages for three independent cultures. Error bars, standard deviations.
FIG. 4.
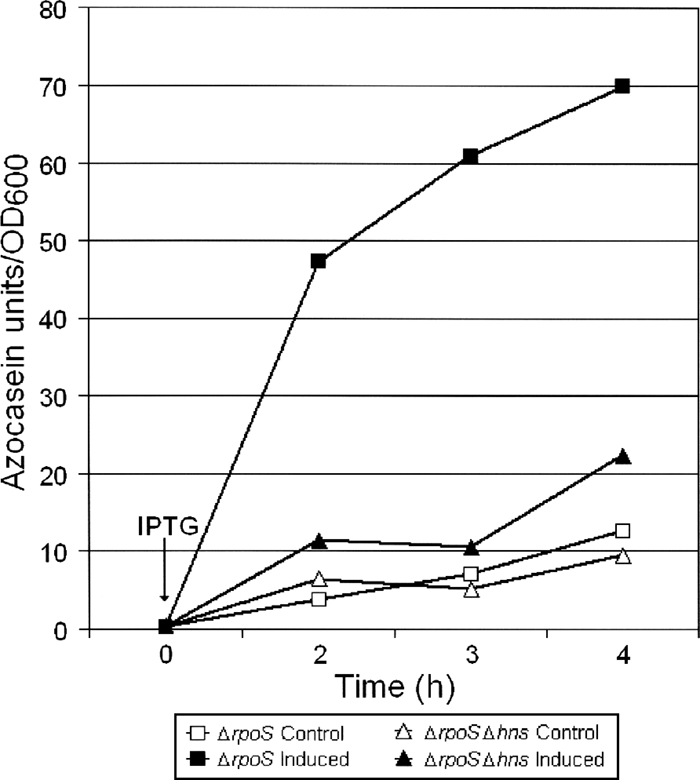
Production of HA/protease as an indicator of ectopic expression of RpoS in ΔrpoS and ΔrpoS Δhns mutants. Strains AJB50ΔlacZ (ΔrpoS) and AJB81 (Δhns::Km ΔrpoS) containing plasmid pTTRpoS were grown in TSB medium to an OD600 of 1.0 and divided in half. One half was induced by the addition of IPTG, and the other half (uninduced) was used as a control. Samples were taken at different times to determine the production of HA/protease by using the azocasein assay described in Materials and Methods. Each data point is the mean for three independent cultures. Error bars, standard deviations.
FIG. 5.
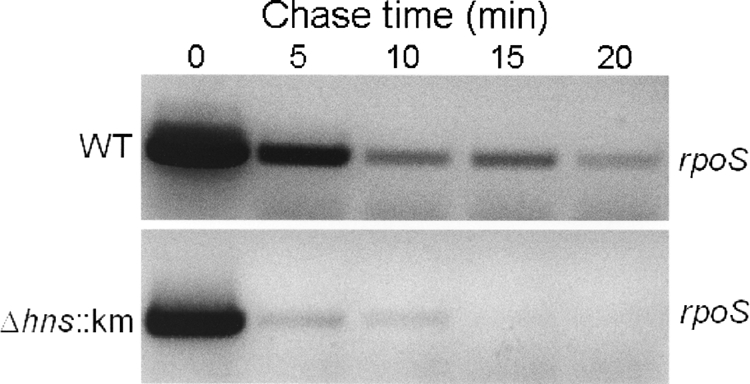
Analysis of rpoS mRNA longevity in a V. cholerae Δhns mutant. Strains C7258ΔlacZ (wild type [WT]) and AJB80 (C7258ΔlacZ Δhns::Km) were grown to an OD600 of 1.5 at 37°C; rifampin was added to block transcription; and samples were collected at different time points for RNA extraction and rpoS mRNA analysis as described in Materials and Methods.
H-NS affects the V. cholerae stress response.
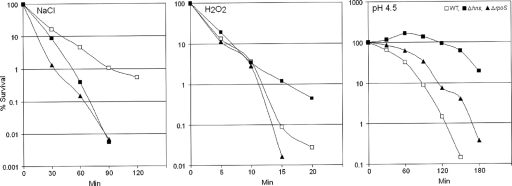
In V. cholerae, RpoS has been shown to mediate a general stress response (62). The results described above prompted us to investigate the role of H-NS in the V. cholerae response to environmental stressors. The Δhns::Km and ΔrpoS mutants were both more sensitive to 2.4 M NaCl than the wild type (Fig. 6). This result is in agreement with the lower expression of RpoS in the Δhns::Km mutant. However, the Δhns::Km mutant was more resistant than the wild-type strain and the ΔrpoS mutant to 3 mM hydrogen peroxide (Fig. 6). In order to explain this result, we used qRT-PCR to determine the relative expression of other catalase/peroxidase genes present in the V. cholerae genome. We observed that the Δhns::Km mutant expressed higher levels of VC0089, encoding c551 cytochrome/peroxidase, than the wild type (0.64 for the wild type and 4.7 for the Δhns::Km mutant). In addition, elevated expression of VC1585, encoding KatB catalase, was observed in the Δhns::Km mutant (levels were 0.030 for the wild type and 0.3 for the Δhns::Km mutant). These results suggest that H-NS is a repressor of c551 and katB. Elevated expression of c551 and katB in the Δhns::Km mutant explains its resistance to hydrogen peroxide in spite of expressing lower levels of the RpoS-dependent perA gene. The Δhns::Km mutant also exhibited greater resistance than the wild type to a pH of 4.5 (Fig. 6). We have not observed elevated expression of lysine decarboxylase, known to mediate the inorganic acid tolerance response (36-38), in the Δhns::Km mutant. However, we did observe that the Δhns::Km mutant expressed higher toxR mRNA levels than the wild type and the ΔrpoS mutant (toxR mRNA levels were 0.31 for the wild type, 0.25 for the ΔrpoS mutant, and 0.64 for the Δhns::Km mutant). Expression of ToxR has been reported to enhance resistance to organic acid shock (35).
FIG. 6.
Responses of a V. cholerae El Tor biotype hns mutant to environmental stresses. V. cholerae strains were grown and prepared as described in Materials and Methods. They were then subjected to either 2.4 M NaCl, 3 mM hydrogen peroxide, or a pH of 4.5 in LB medium. At different time points, samples were withdrawn and the viable count determined by dilution plating. Each point is the average for three experiments. WT, wild type.
Role of H-NS in the expression of motility and chemotaxis.
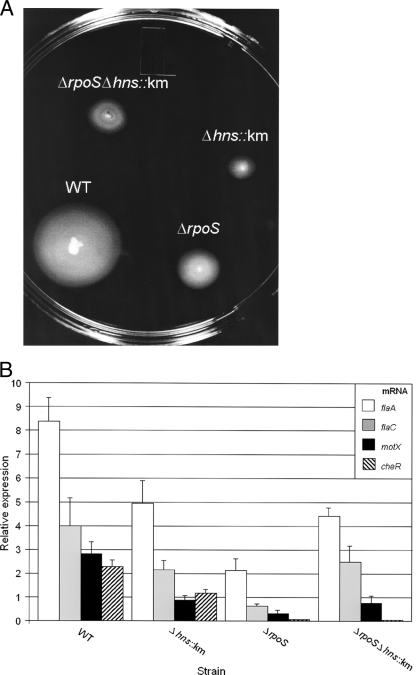
Gene expression profiling of V. cholerae C7258ΔrpoS revealed that several motility and chemotaxis genes were downregulated in the mutant background (data not shown). We found that both ΔrpoS and Δhns::Km mutants were less motile than the wild type by using a swarm assay in semisolid agar medium (Fig. 7A). These results prompted us to examine the relationship between H-NS and RpoS in the regulation of motility and chemotaxis gene expression. To investigate the reduced motility of the Δhns::Km mutant, we used qRT-PCR to measure the relative expression levels of the regulator genes flrA, flrB, flrC, fliA, and rpoN; the flagellin structural genes flaA and flaC; the motor genes motX and motY; and the chemotaxis genes cheA-2 (VC2063), cheR-2 (VC2201), and cheY-3 (VC2065). Inactivation of these genes, except flaC, impairs the ability of V. cholerae to swarm away from the inoculation site in semisolid agar (5, 17, 22, 47, 51). The Δhns::Km mutant expressed lower flaA, flaC, motX, and cheR-2 mRNA levels than the wild type (Fig. 7B). Compared to the deletion of rpoS, the deletion of hns had a smaller but still very significant negative effect on the expression of these genes. cheR-2 was found to be the most strongly RpoS dependent gene under the experimental conditions used. An interesting pattern was observed for flaA, flaC, and motX expression in the ΔrpoS Δhns double mutant. Although expression of these genes was lower in the ΔrpoS and Δhns::Km single mutants than in the wild type, deletion of hns in the ΔrpoS background significantly enhanced their expression (t tests for flaA, flaC, and motX expression yielded P values of <0.05) (Fig. 7B).
FIG. 7.
Role of H-NS in the expression of motility and chemotaxis. (A) Strains C7258ΔlacZ (wild type [WT]) and isogenic mutants AJB80 (C7258ΔlacZ Δhns::Km), AJB50ΔlacZ (ΔrpoS), and AJB81 (Δhns::Km ΔrpoS) were stabbed into LB medium containing 0.3% agar and incubated for 8 h at 30°C. (B) Strains C7258ΔlacZ (WT), AJB80 (C7258ΔlacZ Δhns::Km), AJB50ΔlacZ (ΔrpoS), and AJB81 (ΔlacZ Δhns::Km ΔrpoS) were grown in LB medium to an OD600 of 1.5 at 37°C. RNA was extracted, and the relative expression levels of flaA, flaC, motX, and cheR were determined by qRT-PCR as described in Materials and Methods, with recA mRNA as a reference. Results are averages for three independent cultures. Error bars, standard deviations.
CRP and H-NS act on the same pathway to regulate RpoS expression.
In a previous study, we showed that CRP positively modulates the expression of RpoS (49). Since we have shown above that H-NS also enhances the expression of RpoS, we asked if CRP and H-NS act along the same pathway to enhance RpoS. To investigate this question, we constructed the Δhns::Km Δcrp double mutant AJB82 and performed epistasis analysis of RpoS-dependent perA expression. We did not observe significant differences between the levels of perA expression in the Δhns::Km, Δcrp, and Δcrp Δhns::Km mutants (Fig. 8), suggesting that CRP and H-NS act along the same pathway to enhance RpoS expression. Next, we performed qRT-PCR to measure hns mRNA in wild-type and Δcrp strains. Strain WL7258ΔlacZ (Δcrp) was found to produce less hns mRNA than its wild-type precursor (0.49 for the wild type and 0.21 for the Δcrp mutant).
FIG. 8.
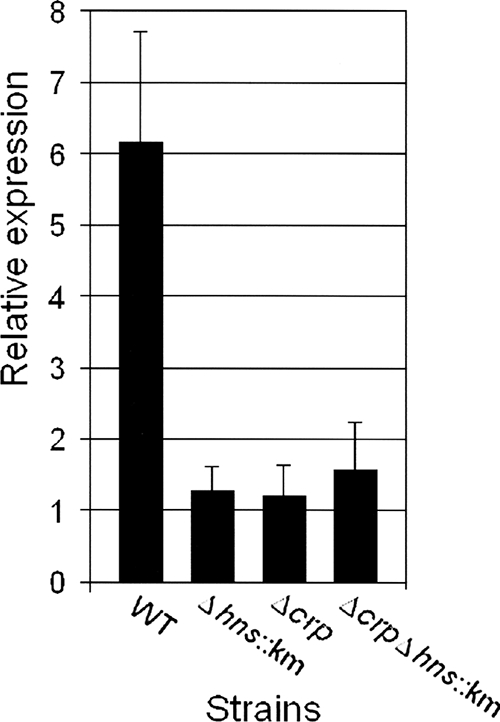
Analysis of perA transcription as a measure of RpoS expression in V. cholerae Δhns and Δcrp mutants. Strains C7258ΔlacZ, AJB80 (C7258ΔlacZ Δhns::Km), WL7258ΔlacZ (Δcrp), and AJB82 (ΔlacZ Δhns::Km Δcrp) were grown to stationary phase (16 h) in TSB at 37°C. Total RNA was extracted, and the abundance of perA mRNA was determined by qRT-PCR. Each value is the average for three independent cultures. Error bars, standard deviations.
DISCUSSION
The experiments described above increase our understanding of the complex regulatory interactions that control the expression of HA/protease, a factor proposed to enhance V. cholerae enterotoxicity and the reactogenicity of live vaccine candidates (4, 15, 51). By using HA/protease as an RpoS reporter activity, we show that in contrast to its action in E. coli, H-NS positively affects the expression of RpoS and multiple RpoS-dependent genes in V. cholerae. Recent studies have revealed important differences in the regulation of RpoS expression between E. coli and V. cholerae. For instance, we have shown that expression of RpoS in V. cholerae is positively affected by CRP (49), but the V. cholerae rpoS promoter does not contain the CRP boxes present in the E. coli promoter (see below). Also, V. cholerae mutants lacking relA and polyphosphate kinase (ppk), which are responsible for ppGpp and polyphosphate synthesis, respectively, produced wild-type levels of RpoS and HA/protease (24, 50). Moreover, deletion of Hfq, a factor that enhances rpoS translation in E. coli, did not affect RpoS expression in V. cholerae (10). These results clearly suggest that the regulation of RpoS expression in E. coli and V. cholerae has diverged to promote long-term colonization of different ecological niches. While E. coli is most commonly found in the gastrointestinal tracts of warm-blooded animals, V. cholerae can persist for longer periods in aquatic ecosystems.
Analysis of rpoS-lacZ transcriptional fusions and ectopic expression of rpoS in ΔrpoS and ΔrpoS Δhns mutants suggested that H-NS acts by a posttranscriptional mechanism. Our data suggest that positive regulation of rpoS expression by H-NS involves rpoS coding sequences cloned into the expression vector pTTRpoS (Fig. 4). In concurrence with the above data, rpoS mRNA was found to decay more rapidly in the Δhns::Km than in the wild-type background (Fig. 5). These findings are in agreement with multiple observations indicating that positive regulation by H-NS is by and large posttranscriptional (1, 12, 26). The reduced abundance of rpoS mRNA in the Δhns mutant suggests that H-NS could bind to rpoS mRNA, as reported for E. coli (6), or act indirectly to enhance rpoS transcript stability. However, our data do not rule out the possibility of H-NS additionally acting at the level of RpoS protein stability. Consistent with the positive effect of H-NS on RpoS expression, our hns mutant was more sensitive than the wild type to high osmolarity (Fig. 6). The surprising resistance of the Δhns mutant to hydrogen peroxide could be explained by elevated expression of other catalase/peroxidase enzymes encoded by V. cholerae. We do not, at the moment, have a clear explanation for the enhanced resistance of the Δhns mutant to a pH of 4.5. Possibly, acidification of LB medium could generate free organic acids to which the Δhns mutant could be more resistant due to elevated expression of ToxR (35). Alternatively, as reported for E. coli (21), H-NS could affect the expression of other amino acid decarboxylases, antiporter systems, or porins that could attenuate the deleterious effect of a low pH.
The Δhns:Km mutant was found to be less motile than the wild type and exhibited reduced expression of flaA, flaC, motX, and cheR-2. In contrast to a previous study using a classical biotype V. cholerae hns mutant (16), we did not observe reduced expression of the regulator flrA in our mutant. Deletion of rpoS negatively affected the expression of flaA, flaC, motX, and cheR-2. This result is in agreement with the finding that RpoS enhances motility in vitro and in vivo to facilitate mucosal escape (42). Analysis of the expression of flaA, flaC, and motX in Δhns, ΔrpoS, and Δhns ΔrpoS mutants revealed an interesting regulatory pattern. While elimination of rpoS had the strongest negative effect on flaA, flaC, and motX expression, elimination of hns was epistatic to rpoS. This result suggests that in the absence of σS, H-NS could act as a repressor of these genes. It has been proposed that H-NS can contribute to σS promoter specificity (20). For instance, initiation of transcription at some RpoS-dependent promoters in the stationary phase by RNAP containing σS can be more resistant to H-NS repression than transcription by the σ70-containing holoenzyme (20). Transcription of flaA and motX requires σ54 (RpoN), while that of flaC requires a σ28 analog (28, 47). Our studies suggest that transcription of flaA, motX, and flaC by RNAP containing σ54 or σ28 could be repressed by H-NS, while in the stationary phase RpoS could partially override H-NS repression. To further examine this possibility, we used the DNA Curvature Analysis program (http://www.lfd.uci.edu/∼gohlke/curve/) to predict the occurrence of curved AT-rich DNA sequences within the 5′ noncoding DNA preceding flaA, flaC, and motX. In all cases, we have found regions to which H-NS could potentially bind, with curvature indices similar to or higher than those calculated for the ctxA and toxT promoters, known to be repressed by H-NS (45, 63). Moreover, use of the Virtual Footprint program (http://www.prodoric.de/vfp/) identified high-scoring putative Fis sites within the 5′ noncoding sequences preceding flaA, flaC, and motX. Fis sites occur in many promoters repressed by H-NS, in which binding of Fis has been reported to hinder the interaction of H-NS with DNA and to antagonize repression (1, 12). Our results raise the intriguing question of how the different σ factors and H-NS interact to regulate the temporal expression of motility and chemotaxis. Knowledge of the temporal expression of RpoS and H-NS in V. cholerae is required to fully clarify their regulatory input on motility.
In a previous study, we demonstrated that CRP positively affects RpoS expression (49). Use of the Virtual Footprint program did not reveal high-scoring putative CRP binding sites, suggesting that CRP acts indirectly. Therefore, we analyzed whether or not CRP and H-NS act through the same pathway to positively regulate RpoS expression. To this end, we constructed an Δhns Δcrp double mutant and performed an epistasis analysis for the expression of RpoS-dependent perA mRNA. Simultaneous deletion of hns and crp did not significantly diminish perA expression more than deletion of hns or crp alone. Since CRP was found by qRT-PCR to positively affect the expression of H-NS, we propose that CRP positively affects RpoS expression in V. cholerae by enhancing H-NS expression (Fig. 9). The positive effect of CRP on H-NS expression is also consistent with the finding that crp mutants are less motile, express reduced flaA and flaC levels (31), and make more CT (31, 53) than the wild type. In fact, CRP can diminish CT expression by parallel mechanisms, which include enhancing H-NS expression, activating quorum sensing (31), and antagonizing the positive regulators AphA and AphB at the tcpPH promoter (29). As for the expression of HA/proteases, our data suggest that CRP impacts hapA transcription by activating quorum sensing (31) and by increasing the expression of H-NS to enhance RpoS biosynthesis.
FIG. 9.
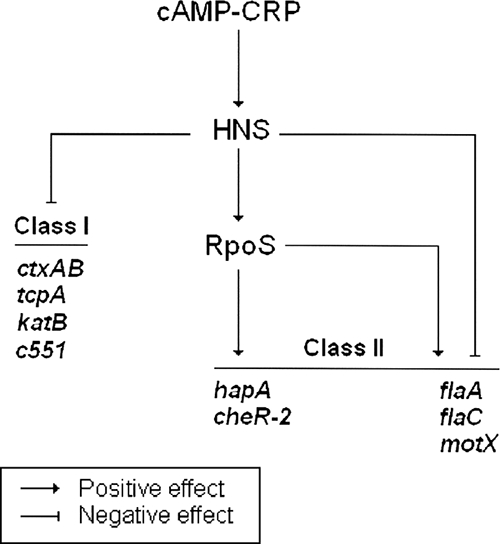
Model for the regulatory input of H-NS in virulence, the general stress response, and motility. We propose that CRP acts through H-NS to silence virulence gene expression and enhance RpoS expression. RpoS, in turn, activates genes involved in protease production, stress response, motility, and chemotaxis. For some RpoS-dependent genes (flaA, motX, and flaC), H-NS can also act as a repressor in the absence of an active rpoS gene.
In Fig. 9, we summarize the regulatory interactions between CRP, H-NS, and RpoS that could affect virulence, stress response, motility, and chemotaxis. The genes found to be affected by H-NS in this study can be divided into at least two categories. In class I genes, H-NS appears to affect gene expression by the well-documented mechanism of transcriptional silencing. These genes include ctxA, tcpA (45), katB, and c551. Expression of class II genes is positively affected by H-NS, which acts indirectly by enhancing the expression of RpoS. This class includes the RpoS-dependent genes hapA, cheR-2, flaA, flaC, and motX. Among these genes, flaA, flaC, and motX appear to constitute a subclass in which H-NS could also act as a transcriptional silencer in the absence of rpoS.
The ability of V. cholerae to respond to environmental changes is crucial to both intestinal colonization and survival outside the human host. This adaptation requires the concerted activity of multiple global regulators. A highly complex regulatory network controls the expression of the general stress response, motility, and chemotaxis to enhance V. cholerae environmental fitness. In this study, we provide evidence that H-NS regulates the stress response and motility by RpoS-dependent and -independent mechanisms.
Supplementary Material
Acknowledgments
The present study was supported by grant GM008248 from the National Institute of General Medical Sciences to A.J.S and by PHS grant AI63187 from the National Institute of Allergy and Infectious Disease to J.A.B.
Footnotes
Published ahead of print on 12 September 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 247-17. [DOI] [PubMed] [Google Scholar]
- 2.Becker, G., E. Klauck, and R. Hengge-Aronis. 1999. Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS. Proc. Natl. Acad. Sci. USA 966439-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benitez, J. A., A. J. Silva, and R. A. Finkelstein. 2001. Environmental signals controlling production of hemagglutinin/protease in Vibrio cholerae. Infect. Immun. 696549-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benítez, J. A., L. Garcia, A. J. Silva, H. Garcia, R. Fando, B. Cedre, A. Perez, J. Campos, B. L. Rodriguez, J. L. Perez, T. Valmaseda, O. Perez, A. Perez, M. Ramirez, T. Ledon, M. Diaz, M. Lastre, L. Bravo, and G. Sierra. 1999. Preliminary assessment of the safety and immunogenicity of a new CTXφ-negative hemagglutinin/protease-defective El Tor strain as a cholera vaccine candidate. Infect. Immun. 67539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boin, M. A., M. J. Austin, and C. C. Hase. 2004. Chemotaxis in Vibrio cholerae. FEMS Microbiol. Lett. 2391-8. [DOI] [PubMed] [Google Scholar]
- 6.Brescia, C. C., M. K. Kaw, and D. D. Sledjeski. 2004. The DNA binding protein H-NS binds to and alters the stability of RNA in vitro and in vivo. J. Mol. Biol. 339505-514. [DOI] [PubMed] [Google Scholar]
- 7.Brosius, J., A. Ullrich, M. A. Raker, A. Gray, T. J. Dull, R. R. Gutell, and H. F. Noller. 1981. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid 6112-118. [DOI] [PubMed] [Google Scholar]
- 8.Chiang, S. L., R. K. Taylor, M. Koomey, and J. J. Mekalanos. 1995. Single amino acid substitutions in the N-terminus of Vibrio cholerae TcpA affect colonization, autoagglutination, and serum resistance. Mol. Microbiol. 171133-1142. [DOI] [PubMed] [Google Scholar]
- 9.de Lorenzo, V., L. Eltis, B. Kessler, and K. N. Timmis. 1993. Analysis of the Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 12317-24. [DOI] [PubMed] [Google Scholar]
- 10.Ding, Y., B. M. Davis, and M. K. Waldor. 2004. Hfq is essential for Vibrio cholerae virulence and down regulates sigma expression. Mol. Microbiol. 53345-354. [DOI] [PubMed] [Google Scholar]
- 11.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive selection suicide vector. Infect. Immun. 594310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorman, C. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2391-400. [DOI] [PubMed] [Google Scholar]
- 13.Finkelstein, R. A. 1992. Cholera enterotoxin (choleragen): a historical perspective, p. 155-187. In D. Barua and W. B. Greenough (ed.), Cholera. Plenum Medical Book Company, New York, NY.
- 14.Finkelstein, R. A., M. Boesman-Finkelstein, and P. Holt. 1983. Vibrio cholerae hemagglutinin/lectin/protease hydrolyzes fibronectin and ovomucin: F.M. Burnet revisited. Proc. Natl. Acad. Sci. USA 801092-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García, L., M. D. Jidy, H. Garcia, B. L. Rodriguez, R. Fernández, G. Ano, B. Cedre, T. Valmaseda, E. Suzarte, M. Ramirez, Y. Pino, J. Campos, J. Menéndez, R. Valera, D. González, I. González, O. Perez, T. Serrano, M. Lastre, F. Miralles, J. Del Campo, J. L. Maestre, J. L. Perez, A. Talavera, A. Perez, K. Marrero, T. Ledon, and R. Fando. 2005. The vaccine candidate Vibrio cholerae 638 is protective against cholera in healthy volunteers. Infect. Immun. 733018-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh, A., K. Paul, and R. Chowdhury. 2006. Role of the histone-like nucleoid structuring protein in colonization, motility, and bile-dependent repression of virulence gene expression in Vibrio cholerae. Infect. Immun. 743060-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gosink, K. K., R. Kobayashi, I. Kawagishi, and C. C. Häse. 2002. Analysis of the role of the three cheA homologs in chemotaxis of Vibrio cholerae. J. Bacteriol. 1841767-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Häse, C. C., and R. A. Finkelstein. 1991. Cloning and nucleotide sequence of the Vibrio cholerae hemagglutinin/protease (HA/protease) gene and construction of an HA/protease-negative strain. J. Bacteriol. 1733311-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hengge-Aronis, R. 2002. Stationary phase gene regulation: what makes an Escherichia coli promoter σS-selective? Curr. Opin. Microbiol. 5591-595. [DOI] [PubMed] [Google Scholar]
- 21.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J.-P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 4020-36. [DOI] [PubMed] [Google Scholar]
- 22.Hyakutake, A., M. Homma, M. J. Austin, M. A. Boin, C. C. Häse, and I. Kawagishi. 2005. Only one of the five CheY homologs in Vibrio cholerae directly switches flagellar rotation. J. Bacteriol. 1878403-8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwanaga, M., K. Yamamoto, N. Higa, Y. Ichinose, N. Nakasone, and M. Tanabe. 1986. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol. Immunol. 301075-1083. [DOI] [PubMed] [Google Scholar]
- 24.Jahid, I. K., A. J. Silva, and J. A. Benitez. 2006. Polyphosphate stores enhance the ability of Vibrio cholerae to overcome environmental stresses in a low-phosphate environment. Appl. Environ. Microbiol. 727043-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jobling, M. G., and R. K. Holmes. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 261023-1034. [DOI] [PubMed] [Google Scholar]
- 26.Johansson, J., B. Dagberg, E. Richet, and B. E. Uhlin. 1998. H-NS and StpA proteins stimulate expression of the maltose regulon in Escherichia coli. J. Bacteriol. 1806117-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaper, J. B., G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 848-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klose, K. E., and J. J. Mekalanos. 1998. Differential expression of multiple flagellins in Vibrio cholerae. J. Bacteriol. 180303-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovacikova, G., and K. Skorupski. 2001. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol. Microbiol. 41393-407. [DOI] [PubMed] [Google Scholar]
- 30.Krishnan, H. H., A. Ghosh, K. Paul, and R. Chowdhury. 2004. Effect of anaerobiosis on expression of virulence factors in Vibrio cholerae. Infect. Immun. 723961-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang, W., A. Pascual-Montano, A. J. Silva, and J. A. Benitez. 2007. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology 1532964-2975. [DOI] [PubMed] [Google Scholar]
- 32.Majdalani, N., S. Chen, J. Murrow, K. St. John, and S. Gottesman. 2001. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 391382-1394. [DOI] [PubMed] [Google Scholar]
- 33.Marcus, H., J. M. Ketley, J. B. Kaper, and R. K. Holmes. 1990. Effect of DNase production, plasmid size, and restriction barriers on transformation of Vibrio cholerae by electroporation and osmotic shock. FEMS Microbiol. Lett. 56149-154. [DOI] [PubMed] [Google Scholar]
- 34.Mel, S. F., K. J. Fullner, S. Wimer-Mackin, W. L. Lencer, and J. J. Mekalanos. 2000. Association of protease activity in Vibrio cholerae vaccine strains with decrease in transcellular epithelial resistance of polarized T84 intestinal cells. Infect. Immun. 686487-6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merrell, D. S., C. Bailey, J. B. Kaper, and A. Camilli. 2001. The ToxR-mediated organic acid tolerance response of Vibrio cholerae requires OmpU. J. Bacteriol. 1832746-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merrell, D. S., and A. Camilli. 1999. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol. Microbiol. 34836-849. [DOI] [PubMed] [Google Scholar]
- 37.Merrell, D. S., and A. Camilli. 2002. Acid tolerance of gastrointestinal pathogens. Curr. Opin. Microbiol. 551-55. [DOI] [PubMed] [Google Scholar]
- 38.Merrell, D. S., D. L. Hava, and A. Camilli. 2002. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol. Microbiol. 431471-1491. [DOI] [PubMed] [Google Scholar]
- 39.Merrell, D. S., A. D. Tischler, S. H. Lee, and A. Camilli. 2000. Vibrio cholerae requires rpoS for efficient intestinal colonization. Infect. Immun. 686691-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, J. H. 1971. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110303-314. [DOI] [PubMed] [Google Scholar]
- 42.Nielsen, A. T., N. A. Dolganov, G. Otto, M. C. Miller, C. Y. Wu, and G. K. Schoolnik. 2006. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nogueira, T., and M. Springer. 2000. Post-transcriptional control by global regulators of gene expression in bacteria. Curr. Opin. Microbiol. 3154-158. [DOI] [PubMed] [Google Scholar]
- 44.Nye, M. B., and R. K. Taylor. 2003. Vibrio cholerae H-NS domain structure and function with respect to transcriptional repression of ToxR regulon genes reveals differences among H-NS family members. Mol. Microbiol. 50427-444. [DOI] [PubMed] [Google Scholar]
- 45.Nye, M. B., J. D. Pfau, K. Skorupski, and R. K. Taylor. 2000. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J. Bacteriol. 1824295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Owen-Hughes, T. A., G. D. Pavitt, D. S. Santos, J. M. Sidebotham, C. S. J. Hulton, J. C. D. Hinton, and C. F. Higgins. 1992. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell 71255-265. [DOI] [PubMed] [Google Scholar]
- 47.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 391595-1609. [DOI] [PubMed] [Google Scholar]
- 48.Rothmel, R. D., D. Shinabarger, M. Parsek, T. Aldrich, and A. M. Chakrabarty. 1991. Functional analysis of the Pseudomonas putida regulatory protein CatR: transcriptional studies and determination of the CatR DNA binding site by hydroxyl-radical footprinting. J. Bacteriol. 1734717-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silva, A. J., and J. A. Benitez. 2004. Transcriptional regulation of Vibrio cholerae hemagglutinin/protease by the cyclic AMP receptor protein and RpoS. J. Bacteriol. 1866374-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silva, A. J., and J. A. Benitez. 2006. A Vibrio cholerae relaxed (relA) mutant expresses major virulence factors, exhibits biofilm formation and motility, and colonizes the suckling mouse intestine. J. Bacteriol. 188794-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silva, A. J., G. J. Leitch, A. Camilli, and J. A. Benitez. 2006. Contribution of hemagglutinin/protease and motility to the pathogenesis of El Tor biotype cholera. Infect. Immun. 742072-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silva, A. J., K. Pham, and J. A. Benitez. 2003. Hemagglutinin/protease expression and mucin gel penetration in El Tor biotype Vibrio cholerae. Microbiology 1491883-1891. [DOI] [PubMed] [Google Scholar]
- 53.Skorupski, K., and R. K. Taylor. 1997. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin co-regulated pilus in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 94265-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spurio, R., M. Falconi, A. Brandi, C. L. Pon, and C. O. Gualerzi. 1997. The oligomeric structure of the nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA binding. EMBO J. 161795-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Starks, M. J. 1987. Multicopy expression vectors carrying the lac repressor for regulated high-level expression of genes in Escherichia coli. Gene 51255-267. [DOI] [PubMed] [Google Scholar]
- 56.Tendeng, C., C. Badaut, E. Krin, P. Gounon, S. Ngo, A. Danchin, S. Rimsky, and P. Bertin. 2000. Isolation and characterization of vicH, encoding a new pleiotropic regulator in Vibrio cholerae. J. Bacteriol. 1822026-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ueguchi, C., and T. Mizuno. 1993. The Escherichia coli nucleoid protein H-NS functions directly as a transcriptional repressor. EMBO J. 121039-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vicente, M., K. F. Chater, and V. De Lorenzo. 1999. Bacterial transcription factors involved in global regulation. Mol. Microbiol. 338-17. [DOI] [PubMed] [Google Scholar]
- 59.Wu, Z., D. Milton, P. Nybom, A. Sjö, and K. E. Magnusson. 1996. Vibrio cholerae hemagglutinin/protease (HA/protease) causes morphological changes in cultured epithelial cells and perturbs their paracellular barrier function. Microb. Pathog. 21111-123. [DOI] [PubMed] [Google Scholar]
- 60.Wu, Z., P. Nybom, and K. E. Magnusson. 2000. Distinct effects of Vibrio cholerae haemagglutinin/protease on the structure and localization of the tight junction-associated proteins occludin and ZO-1. Cell. Microbiol. 211-17. [DOI] [PubMed] [Google Scholar]
- 61.Wu, Z., P. Nybom, T. Sundqvist, and K. E. Magnusson. 1998. Endogenous nitric oxide in MDCK-I cells modulates the Vibrio cholerae haemagglutinin/protease (HA/P)-mediated cytotoxicity. Microb. Pathog. 24321-326. [DOI] [PubMed] [Google Scholar]
- 62.Yildiz, F. H., and G. K. Schoolnik. 1998. Role of rpoS in stress survival and virulence of Vibrio cholerae. J. Bacteriol. 180773-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu, R. R., and V. J. DiRita. 2002. Regulation of gene expression in Vibrio cholerae by toxT involves both antirepression and RNA polymerase stimulation. Mol. Microbiol. 43119-134. [DOI] [PubMed] [Google Scholar]
- 64.Zhou, Y., and S. Gottesman. 2006. Modes of regulation of RpoS by H-NS. J. Bacteriol. 1887022-7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.