Abstract
It has been shown that Escherichia coli harboring the derepressed IncFI and IncFII conjugative F plasmids form complex mature biofilms by using their F-pilus connections, whereas a plasmid-free strain forms only patchy biofilms. Therefore, in this study we investigated the contribution of a natural IncF conjugative F plasmid to the formation of E. coli biofilms. Unlike the presence of a derepressed F plasmid, the presence of a natural IncF F plasmid promoted biofilm formation by generating the cell-to-cell mating F pili between pairs of F+ cells (approximately two to four pili per cell) and by stimulating the formation of colanic acid and curli meshwork. Formation of colanic acid and curli was required after the initial deposition of F-pilus connections to generate a three-dimensional mushroom-type biofilm. In addition, we demonstrated that the conjugative factor of F plasmid, rather than a pilus synthesis function, was involved in curli production during biofilm formation, which promoted cell-surface interactions. Curli played an important role in the maturation process. Microarray experiments were performed to identify the genes involved in curli biosynthesis and regulation. The results suggested that a natural F plasmid was more likely an external activator that indirectly promoted curli production via bacterial regulatory systems (the EnvZ/OmpR two-component regulators and the RpoS and HN-S global regulators). These data provided new insights into the role of a natural F plasmid during the development of E. coli biofilms.
Surface-attached microbial communities (biofilms) have distinct morphological and biochemical properties that distinguish them from free-living planktonic cells (30). The formation of a three-dimensional (3D) mushroom-type mature biofilm by Pseudomonas sp. has been described as a stepwise process with at least four developmental stages: (i) initial reversible attachment of planktonic bacteria to the surface, (ii) a transition to irreversible attachment involving specific adhesive factors and the formation of microcolonies, (iii) maturation of the microcolonies into a thin film by production of extracellular polymeric substances in an adhesive matrix, and (iv) detachment of cells from the biofilm and return of cells to the planktonic state (41).
In addition to morphological descriptions of biofilm formation, there is also increasing interest in the horizontal gene transfer that controls biofilm formation (39). Mobile genetic elements mediate horizontal gene transfer between bacteria in natural habitats. These elements can be conjugative plasmids, transposons, or bacteriophages (24). Although a laboratory strain of Escherichia coli does not form extensive biofilms spontaneously, conjugative plasmid expression promotes the development of thick mature biofilms (18, 35). In previous studies of E. coli harboring a conjugative plasmid, the workers identified IncFI and IncFII conjugative plasmids that potentially promoted formation of a 3D mushroom-type mature biofilm similar to a Pseudomonas sp. biofilm (6, 34, 46). Notably, the plasmids previously tested (pOX38Km, an IncFI plasmid [34]; F′traD36, an IncFI plasmid [6]; and R1drd19, an IncFII plasmid [46]) are classified as derepressed conjugative plasmids which constitutively express F pili at all times (15). However, constitutively expressed F pili are required for development of highly organized mature E. coli biofilms (18) and support biofilm maturation even in the absence of flagella, type 1 fimbriae, curli, the Ag-43 autotransport protein, or Al-2-mediated cell-to-cell communication (34). These findings raise the question of how horizontal gene transfer can promote the formation of dense biofilms, especially via a natural IncF F plasmid, in which F-pilus formation and the conjugative mechanism are repressed. The mechanistic basis of the putative connection between natural F-pilus formation and biofilm maturation is poorly understood. Furthermore, it is also not known how natural IncF F-plasmid-containing biofilms are different in terms of ultrastructure and gene expression.
A natural IncF F plasmid that was originally obtained from a host E. coli K-12 strain is a pioneer conjugative plasmid belonging to the IncFI incompatibility group (15; H. Shimizu, Y. Saitoh, Y. Suda, K. Uehara, G. Sampei, and K. Mizobuchi, unpublished data). This plasmid is referred to as the fertility factor (also known as F factor or sex factor) that allows bacteria to produce F pili necessary for bacterial conjugation. All the sequences required for conjugative transmission (20 tra genes) of a natural IncF F plasmid are present in the 33.3-kb transfer region (the conjugative factor) of this 99.2-kb plasmid, while the other 65.9 kb (the nonconjugative factor) contains a number of other genetic sequences responsible for incompatibility, replication, and other functions (27).
In this study, we analyzed the temporal development of E. coli biofilms in continuous flow cell systems using confocal laser scanning microscopy (CLSM), scanning electron microscopy (SEM), and time-lapse video microscopy. Surprisingly, we demonstrated that two or three F pili could be expressed in a biofilm by a natural IncF F plasmid. The presence of F pili, induced by a natural IncF F plasmid, resulted in an increase in colanic acid and curli production during biofilm formation at 37°C, which is unusual for laboratory strains. We also examined the involvement of the conjugative factor and the nonconjugative factor in this plasmid in biofilm formation. To elucidate the mechanisms underlying the differences, we analyzed global transcriptional differences among a plasmid-free strain, a strain harboring a nonconjugative factor-containing plasmid (a derivative of a natural IncF F plasmid), and a strain harboring a natural IncF F plasmid. The results indicated that the natural IncF F plasmid had a global impact on the expression of genes involved in colanic acid and curli production. The importance of these genes in biofilm formation was confirmed by the results of loss-of-function experiments. In addition, we demonstrated that curli play an important role in generation of a 3D mushroom-type mature biofilm, so we characterized the contribution of a natural IncF F plasmid to curli biogenesis and regulation during biofilm maturation. Our results clearly demonstrated the effects of horizontal gene transfer on E. coli biofilm development and maturation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli laboratory strain MG1655, a strain harboring a natural IncF F factor (99.2-kb plasmid from a wild-type K-12 strain; GenBank accession number AP001918 [27; Shimizu et al., unpublished data]), and a strain harboring the nonconjugative factor of F plasmid (pNCF; a derivative of a natural IncF plasmid containing only 65.9 kb of the nonconjugative factor of the wild-type plasmid [see Text S1 in the supplemental material]) were used in this work. E. coli cpxA, cpxR, csgA, csgB, csgC, csgD, csgE, csgF, csgG, cspA, envZ, hns, lon, ompR, rpoS, cpsE, and cspE csgA deletion strains were also constructed by using the method of Datsenko and Wanner (13).
Luria-Bertani (LB) broth or agar was used for standard cultivation. A 0.1 M ZnSO4 stock solution was used as the source of Zn2+ to study the effect of Zn2+ on F-pilus formation in biofilms. Cells were grown in MOPS (morpholinepropanesulfonic acid) minimal media supplemented with 0.2% glucose (29). Samples were removed when the optical density at 600 nm was 0.5 (exponential growth phase) or 2.0 (stationary growth phase) and subjected to microarray and reverse transcription (RT)-PCR analyses. The experiments were repeated three times.
Flow cell experiments.
Biofilms were cultivated in single-channel flow cell reactors (model FC81; Biosurface Technologies Corp.) with individual channel dimensions of 2.54 by 12.7 by 50.8 mm. Flow cell chambers were incubated at 37°C with MOPS minimal medium supplemented with 0.2% glucose. Flow cells were inoculated with early-stationary-phase cultures of E. coli strains diluted with MOPS minimal medium to obtain an optical density at 600 nm of 0.1. After inoculation, the medium flow was stopped for 2 h to allow the inoculated bacteria to attach to the glass surface. Medium flow was then started at a constant rate of 0.3 ml min−1 using an Advantec PSM050DA microtube pump. The flow rate of this system was optimized to ensure that the dilution rate was greater than the doubling time of the bacteria used. Biofilms were harvested directly from the flow cell chamber with the Bacteria Protect reagent (Qiagen) and used for subsequent DNA and RNA experiments.
CLSM and image analysis.
Microscopic observation and image acquisition were performed with an LSM 510 CLSM (Zeiss). In order to study the spatial organization of the cells using CLSM, the bacterial strains were genetically marked by insertion of Ds-Red Express fluorescent proteins (Molecular Probes) into the chromosome using the protocol of Diederich et al. (14). Simulated fluorescence projection images and vertical cross sections were generated using the IMARIS software package (Bitplane AG). Time-lapse video images were captured by using an Axiovert 200 M inverted microscope (Zeiss) with a charge-coupled device video camera module (Sony). Representative video images were edited by using ImageJ (http://rsb.info.nih.gov/ij/index.html).
For biofilm quantification, the automatic threshold value and the connected volume filtration level for all image stacks were calculated by using he computer program PHLIP (28). Then the CSLM image stacks were analyzed by using the computer program COMSTAT (19).
Electron microscopy.
For detailed analysis of biofilm structure, cells were fixed with 1 M sodium cacodylate buffer (pH 7.2) containing 1% glutaraldehyde. The samples were then washed with the same buffer and postfixed in 1% osmium tetroxide in cacodylate buffer. The fixation and staining protocols were carried out by using the method of Prigent-Combaret et al. (32). The postfixed samples were dehydrated using a graduated ethanol series (30 to 100% ethanol), treated using the critical point drying method, and observed with a Hitachi S4000 scanning electron microscope (SEM). The bacterial structures are identified in Fig. S1 in the supplemental material.
Microarray experiment and data analysis.
An E. coli antisense GeneChip array (Affymetrix) was used to study the global gene expression pattern. Total RNA was isolated from biofilm cells using a Qiagen RNeasy Protect bacterial mini kit. cDNA was synthesized and labeled as described in the Affymetrix expression analysis technical manual. The labeled cDNA was hybridized to the arrays as recommended by the manufacturer. The arrays were scanned using a GeneChip scanner (Affymetrix). In each experiment, three biological replicates were analyzed. A statistical analysis was carried out by using the computer program DNA-Chip Analyzer (dChip) (25). The data were normalized using the invariant set method. A model-based expression value was calculated. The data were filtered by comparing samples in a pairwise fashion, and genes whose levels of expression changed more than twofold were picked. After statistical filtering, the transcripts were subjected to cluster analysis and functional classification analysis. An annotation analysis of the Affymetrix gene expression results was performed using the annotation information and Gene Ontology (GO) mining tools of the Affymetrix NetAffx Analysis Center (26). The data reported in this paper have been deposited in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession numbers E-MEXP-953 and E-MEXP-1145.
Real-time RT-PCR.
Real-time RT-PCR was used to validate the microarray gene expression data. Total DNA of the cultures tested were extracted by using a Qiagen DNeasy tissue kit. RNA was converted to cDNA using a QuantiTect RT kit (Qiagen) as described by the manufacturer. The remaining RNA was digested with RNase H, and first-strand cDNA was used directly as the template with Power SYBR green PCR master mixture for real-time PCR using an Applied Biosystems 7000 sequence detection system. The RT-PCR primers are shown in Table S5 in the supplemental material. A vector standard curve was obtained by using a background ftsZ housekeeping gene from the host genomic DNA, and the quantity used was equal to the quantity of the test RNA. The copy number was calculated by determining the number of cDNA copies per host genome copy detected for each of eight independent dilutions of the RNA samples. The high and low copy numbers were discarded to correct for pipetting error, and the average copy numbers were obtained by using the remaining six values.
Biofilm formation assay and curli production assay.
A biofilm was obtained by using LB medium and a 96-well polystyrene plate incubated at 37°C for 48 h. Biofilm formation was assayed by staining the cells attached to the polystyrene with crystal violet using a microplate washer (ImmunoWash 1575; Bio-Rad). The absorbance of crystal violet-binding biofilms was determined with a microplate reader (ARVO 1420 multilabel counter; Perkin Elmer). To determine curli-producing ability, biofilms were grown using the microplate method described above, and then cell suspensions were removed and washed using the microplate washer. The biofilm attached to the microplate wall was removed by using a small sterilized toothpick, and it was stabbed directly onto a Congo red assay plate. The Congo red phenotype assay was performed using LB medium as the basal medium as described in Ghigo's protocol (12), but the medium was incubated at 37°C for 48 h.
RESULTS
A natural IncF F-plasmid-containing E. coli strain produces F+ cell mating F pili, colanic acid, and curli during formation of a dense biofilm.
Several studies reported previously that a derepressed F-plasmid-containing E. coli strain formed a mushroom-type mature biofilm due to constitutive piliation within 24 h (18, 34). Unlike F pili of a derepressed F-plasmid-containing strain, F pili of a natural IncF F-plasmid-containing strain are not always produced and are controlled by a complex regulation system, including a repression system based on the products of the plasmid genes finO and finP (15). Therefore, it is important to investigate the contribution of a natural IncF F plasmid to biofilm formation.
In this study, we used an original natural IncF conjugative F plasmid from a wild-type E. coli K-12 strain (Shimizu et al., unpublished data). First, to elucidate the dynamics of biofilm formation by a natural IncF F-plasmid-containing E. coli strain over a 48-h period, we used CLSM and time-lapse video microscopy with continuous flow cell systems incubated at 37°C (Fig. 1A; see Video S1 in the supplemental material). We found that bacterial cells attached to a glass slide within 2 h. After attachment to the surface, E. coli formed microcolonies in about 6 to 8 h. It took another 12 to 16 h to generate young biofilms with an average thickness of 10 μm. Dense aggregates of bacterial cells covered the entire glass surface after 48 h. Our observations confirmed the strong biofilm formation ability a natural IncF F-plasmid-containing E. coli strain.
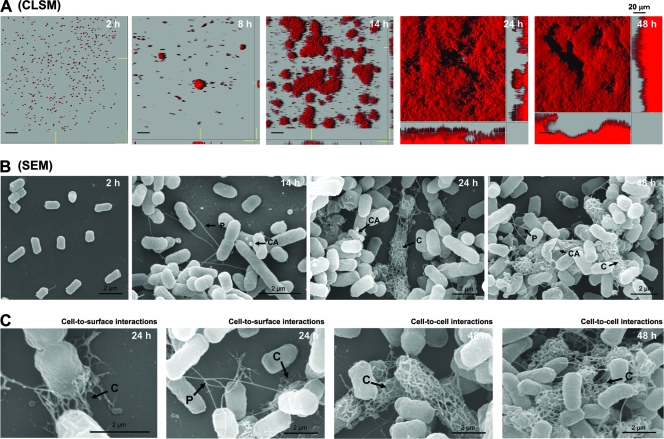
FIG. 1.
F-plasmid-containing E. coli forms complex biofilms that contain F+ cells with pili, colanic acid, and curli. (A) CSLM images showing time-dependent biofilm development for a natural F-plasmid-containing E. coli strain. At 2 h the bacteria formed microcolonies, which expanded into larger colonies at 8 h. By 24 h complex dense biofilms that covered most of the surface area had formed. At 48 h complex mature biofilms were observed. (B) SEM images of the biofilms at different time points. Note the conjugative pili (P) that formed connections among cells at 14 h. By 24 and 48 h, a more complex biofilm containing colanic acid (CA) and elaborated curli fibers (C) had formed. (C) Close-ups of different curli fiber structures that form cell-surface attachments at 24 h and cell-cell networks at 48 h. All experiments were repeated three times, and representative images are shown.
Extensive SEM analysis provided a more detailed view of multiple adherence factors (adhesins) involved in the maturation process (Fig. 1B). Adhesins were clearly identified as F pili, curli, or colanic acid based on comparison with previous findings (22, 32) and SEM analyses of strains overexpressing one of these adhesins (see Fig. S1 in the supplemental material). After 2 h of incubation, bacterial cells attached to the glass surface via the long side of the cells. Surprisingly, conjugative F pili were the first visible extracellular extrusions during the initial formation of a biofilm, and the capsular exopolysaccharide colanic acid was produced in the first 14 h. Colanic acid particles were attached either around the bacterial cells or to the glass surface at all time points. However, the F pili appeared to only link bacterial cells (approximately two to four pili per cell). Interestingly, curli formed networks around bacterial cells and appeared to mediate cell-surface interactions after 24 h (Fig. 1C). These filaments subsequently formed interconnecting meshes between cells (cell-cell interactions) after 48 h (Fig. 1C). Therefore, this experiment clearly showed that a natural IncF F plasmid could generate cell-cell F-pilus connections, indicating that there was same-sex F+ mating during biofilm formation. This phenomenon had characteristics similar to those of F− phenocopies (16), which express F pili that interact with other bacteria during stationary-phase-like conditions, while colanic acid and curli adhesins supported further biofilm development and maturation.
Absence of F plasmid and inhibition of F-pilus formation reduce the capacity to form biofilms.
To evaluate the contribution of a plasmid to biofilm formation, we compared the biofilm formation capacities of E. coli MG1655 carrying a natural IncF F plasmid [MG1655(F)], MG1655 carrying a nonconjugative variant of F plasmid, pNCF [MG1655(pNCF)], and a plasmid-free MG1655 strain in flow cell chambers at 14, 24, and 48 h (Fig. 2A). To quantify differences in biofilm structure (i.e., substratum coverage, average thickness, and roughness coefficient), the image stacks in Fig. 2A were analyzed by using COMSTAT (Fig. 2C). Biofilm roughness is an indicator of biofilm heterogeneity; if the value is close to 1, the biofilm has a mushroom-type structure (19, 23). MG1655(F) formed dense mature biofilms that covered the entire glass surface very rapidly (the substratum coverage was 80 to 100% at all times). Furthermore, MG1655(F) had the highest roughness coefficients during maturation at 24 and 48 h, because it formed a more mushroom-like biofilm than the other two strains (Fig. 2A). In contrast, MG1655 formed only a few discrete microcolonies that had the lowest average thickness and substratum coverage at 14 and 24 h (Fig. 2A). Thus, the plasmid-free strain had poor biofilm formation capacity. Although MG1655(pNCF) had a phenotype similar to that of MG1655 after 14 to 24 h of incubation, these strains could form thick flat mature biofilms at 48 h compared with the MG1655(F) biofilm (Fig. 2C).
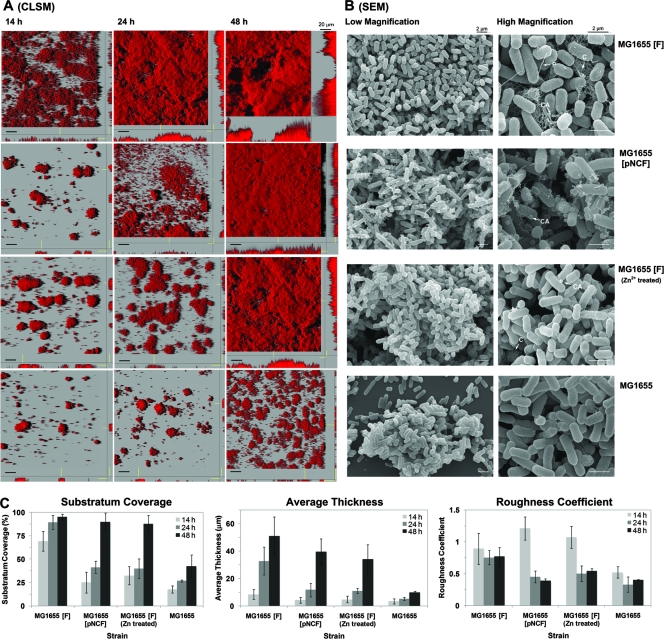
FIG. 2.
Absence of F plasmid or pilus formation delays the development of biofilms. The images show the biofilm growth patterns and morphologies of different bacterial strains, including F-plasmid-containing strain MG1655(F), nonconjugative F-plasmid-containing strain MG1655(pNCF), F-plasmid-containing strain MG1655(F) treated with Zn2+, and plasmid-free strain MG1655. (A) CSLM images of developing biofilms. MG1655 did not form dense mature biofilms at 48 h, in contrast to F-plasmid-containing strain MG1655(F), which formed mushroom-type biofilms. At 24 h, a clear delay in biofilm formation was observed when pilus formation was impaired by expression of a nonconjugative F plasmid [MG1655(pNCF)] or by Zn2+ treatment. However, both strains formed flat and uniform biofilms at 48 h. (B) SEM images of the different biofilms at 48 h. The MG1655 strain lacked adhesive appendages, while pili (P), colanic acid (CA), and curli meshwork (C) were abundant in MG1655(F) biofilms. Both the MG1655(pNCF) and MG1655(F) strains treated with Zn2+ lacked pili but contained colanic acid particles. Only MG1655(F) treated with Zn2+ could synthesize curli. All experiments were performed in duplicate, and representative images are shown. (C) Quantification of the substratum coverage, the average thickness, and the roughness coefficient of the biofilms. The values were calculated with the programs PHLIP and COMSTAT. The results are the means for 20 image stacks representing 10 image stacks from two independent experiments. The error bars indicate the standard deviations.
Based on SEM analyses, a meshwork consisting of F-pilus, colanic acid, and curli components was clearly visible in the MG1655(F) biofilm. In contrast, only colanic acid was produced in the MG1655(pNCF) biofilm (Fig. 2B). Thus, the presence of curli together with F-pilus connections promoted the formation of mushroom-type biofilms (Fig. 2A), while a strain with colanic acid formed only flat biofilms, like MG1655(pNCF) (Fig. 2A). Therefore, we hypothesized that E. coli lacking an F plasmid could not form dense mature biofilms because it could not produce F pili, colanic acid, and curli.
To confirm that the production of F pili contributes to the formation of dense biofilms by F-plasmid-containing E. coli, we inhibited pilus formation by adding a nonlethal concentration of Zn2+. Ou and Anderson (31) demonstrated that low doses of Zn2+ inhibited the formation of mating pairs or pilus construction and interrupted the conjugative system of F plasmid. We found that biofilm formation was delayed and that flat biofilms were formed after 48 h (Fig. 2C) when 10 mM ZnSO4 was added to the medium, which was similar to the results obtained for an MG1655(pNCF) biofilm (Fig. 2A). Even though the SEM analysis showed that there were not F pili around the cell surfaces, little colanic acid and curli were present (Fig. 2B). This suggested that the conjugative factor of F plasmid, rather than the F-pilus synthesis system, is involved in curli arrangement during biofilm formation. Therefore, we concluded that inhibiting pilus formation by genetic as well as chemical means delayed, but did not diminish, the development and maturation of dense biofilms. Our results also confirmed that F-pilus formation was indeed required for formation of a 3D mushroom-type biofilm and also stimulated the rapid formation of colanic acid and curli.
In a natural IncF F-plasmid-containing E. coli strain deposition of curli is required during development of a mature 3D biofilm.
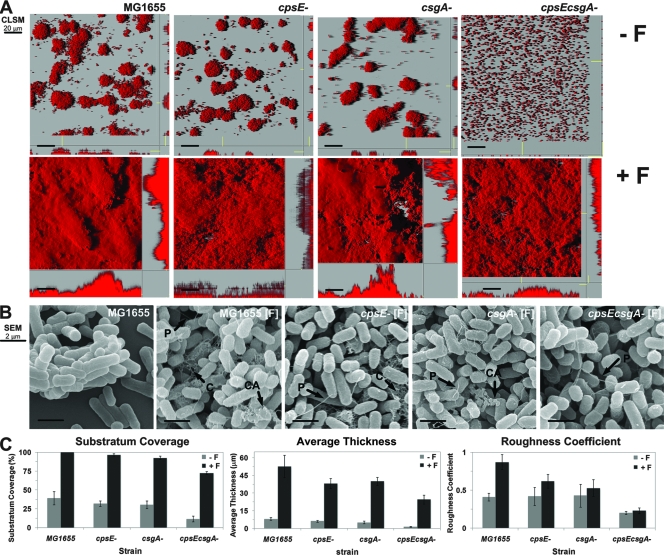
The wild-type strain and curli and colanic acid mutants (with the major curli subunit protein gene csgA deleted and with the colonic acid precursor gene cpsE deleted) were cultured in flow cell chambers, and after 48 h the biofilms were analyzed using CLSM and COMSTAT image analysis. As shown by representative CLSM micrographs (Fig. 3A) and by quantitative CLSM data analysis (Fig. 3C), the wild-type strain and the mutants (with csgA, cpsE, and both cpsE and csgA deleted) formed only discrete patchy biofilms when there was no natural IncF F plasmid (Fig. 3A, top panels). When F plasmid was introduced into the wild-type and single-mutant strains, the bacteria were able to form biofilms, which was shown by 90 to 100% substratum coverage (Fig. 3A, bottom panels). The wild-type strain formed mushroom-type biofilms, the cpsE F-plasmid mutant formed small hilly biofilms, the csgA F-plasmid mutant formed irregular biofilms with protruding structures, and the cpsE csgA F-plasmid mutant formed flat carpetlike biofilms (Fig. 3C). As expected, SEM analysis (Fig. 3B) revealed that the cpsE F-plasmid mutant produced F-pilus connections and a curli meshwork, but not colanic acid. The csgA F-plasmid mutant produced F-pilus appendages with colanic acid capsules, but no curli fibers. In contrast, the cpsE csgA F-plasmid mutant produced only a few F-pilus connections, resulting in a flat biofilm. These results suggested that the formation of colanic acid and curli is essential for generating the mushroom-type biofilm structure of E. coli harboring a natural F plasmid as deletion of both adhesins resulted in bacteria that could not form a mushroom-type biofilm (Fig. 3A). In addition, the presence of curli seemed to be more important in the later stage of biofilm formation (maturation), while deposition of colanic acid was found during the entire biofilm formation period and was likely required at the middle stage (colony formation). This might have been because curli could promote direct cell-cell and cell-surface interactions and frequently generated the meshwork with F pili and colanic acid (Fig. 1C). Therefore, a natural F plasmid stimulated the induction of curli and colanic acid in E. coli after the initial assembly of pili, leading to the formation of well-organized mature biofilms. To confirm this, we used gene expression approaches.
FIG. 3.
Colanic acid and curli are required for formation of complex biofilms by a natural F-plasmid-containing E. coli strain. Mature biofilms formed by the MG1655 parental strain and cpsE (colanic acid), csgA (curli), and cpsE csgD (curli and colanic acid) mutant derivatives were cultured with and without the F plasmid. (A and B) CSLM (A) and SEM (B) images of biofilms of the different bacterial strains with or without F plasmid at 48 h. Lack of curli resulted in a protruding biofilm. Lack of both colanic acid and curli in the double mutant resulted in a flat biofilm even though F pili were produced. (C) Substratum coverage, average thickness, and roughness of the individual biofilms, which were calculated with COMSTAT.
Expression of the genes involved in curli biosynthesis is increased in F-plasmid-containing E. coli biofilms.
To further characterize biofilm formation at the transcript level, we performed a global transcription analysis using an Affymetrix E. coli antisense microarray with cells harvested from biofilms grown in flow cells. Global gene expression data for E. coli MG1655, MG1655(F), and MG1655(pNCF) grown for 48 h were compared with data for corresponding planktonic cells in the exponential and stationary growth phases (see Table S1 in the supplemental material). Since our primary goal was to identify the genes and biological functions that are specifically activated in F-plasmid-containing biofilms during the maturation stage, we thoroughly compared the gene expression data for the biofilm samples using the F-plasmid-containing biofilm as the baseline in three subsequent steps. First, we compared the gene expression data for all biofilm samples [MG1655(F) versus MG1655 and MG1655(pNCF)] to determine the genes that were significantly induced by the presence of a natural F plasmid. Second, we compared the gene expression data for MG1655(F) and MG1655(pNCF) to identify the significant gene expression pattern due to the nonconjugative or conjugative genes of the F plasmid. Finally, we compared the gene expression data for MG1655(F) and MG1655 (which forms poor biofilms) to examine the genes significantly affected by only the conjugative factor of F plasmid. The data showed that there were significant differences (≥2-fold) in the expression of 165 transcripts (77 genes with classification functions and 88 intergenic regions [see Table S2 in the supplemental material]), 300 transcripts (248 genes with classification functions and 52 intergenic regions [see Table S3 in the supplemental material), and 206 transcripts (108 genes with classification functions and 98 intergenic regions [see Table S4 in the supplemental material]) in the comparisons described above, respectively. This indicates that about 3.9% of the E. coli genome (including 2.0% of the intergenic regions) was differentially expressed in the strain with the F plasmid at a statistically significant level (P ≤ 0.05). The most significant classes of natural F-plasmid-induced biofilm genes (Fig. 4A) are (i) genes involved in cell structures, such as curli biosynthesis genes (csgA and csgB), a colanic acid biosynthesis gene (wcaA), a glycogen biosynthesis gene (glgS), and outer membrane component genes (lpxC, mreC, and yfbS); (ii) genes involved in transport and binding of proteins (gatABC); and (iii) hypothetical and unclassified genes (yeaHG, yjbJ, and ycfR). The main classes of repressed genes include the genes involved in amino acid biosynthesis and metabolism, energy metabolism, cellular processes, and carbon compound metabolism (Fig. 4A).
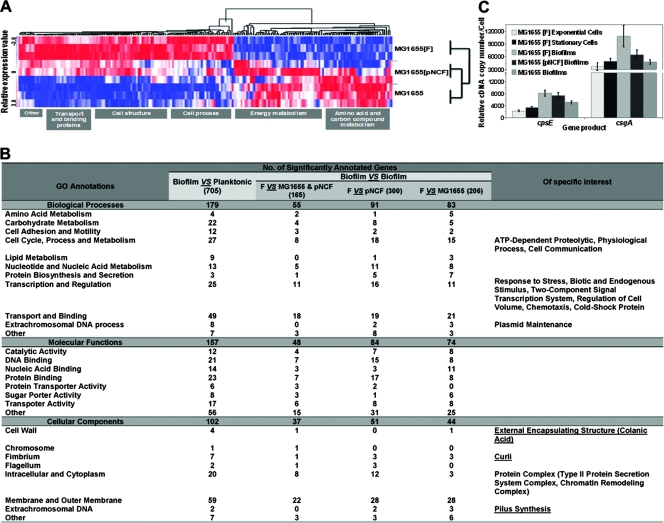
FIG. 4.
Genes regulating F-pilus, colanic acid, and curli formation are differentially expressed in biofilms containing a natural F plasmid. (A) Dynamic hierarchical clustering of E. coli gene expression during biofilm maturation at 48 h. Each row shows data for one culture form. Each column shows data for one gene. Red, upregulated; white, no change; blue, downregulated. The sample profiles were grouped into two clusters: MG1655(F) versus MG1655 (plasmid-free) and MG1655(pNCF) biofilms. (B) GO annotations overrepresented in an F-plasmid-containing biofilm compared with the other conditions tested. Note that an Affymetrix E. coli antisense GeneChip consists of 7,312 probe sets. These probe sets are placed into three ontology branches, the biological processes (2,979 probe sets), the molecular functions (3,228 probe sets), and the cellular components (1,999 probe sets), according to the NetAffx Analysis Center. The significant GO terms in this study are shown. The numbers of significantly differentially expressed genes are indicated. The last column indicates the specific GO characteristics of the significantly differentially expressed genes mediated by a natural conjugative F plasmid. (C) csgA and cpsE expression levels determined by quantitative RT-PCR. Note the increases in the csgA and cpsE expression levels in MG1655(F) and MG1655(pNCF) biofilms (P ≤ 0.05). All experiments were repeated three times, and the average levels of expression and standard derivations are indicated.
Furthermore, based on the comparisons described above, we identified the significantly expressed genes that were involved in biofilm maturation in a natural F-plasmid-carrying strain and compared the expression of these genes in planktonic growth phases using NetAffx GO annotation tools. The comparisons revealed the significant biological functions that make a difference in formation of biofilms by MG1655 carrying a conjugative factor or a nonconjugative factor of F plasmid (Fig. 4B). Based on this analysis, we showed that the F-pilus-, colanic acid-, and curli-related genes were induced in dense mature biofilms by the presence of both a conjugative factor and a nonconjugative factor on the plasmid.
To confirm our global transcriptional data, we validated the microarray gene expression data using real-time RT-PCR (Fig. 4C). The expression of csgA was significantly upregulated in mature biofilms formed by MG1655 carrying the F plasmid (P ≤ 0.05) compared with biofilms formed by a plasmid-free strain and with planktonic cells, whereas the expression of cpsE was slightly upregulated (P > 0.05) under the same conditions. This suggested that the expression of curli (csgA) requires assembly of F+ mating pili via both a conjugative factor and a nonconjugative factor on the F plasmid. In contrast, the expression of colanic acid (cpsE) is more independent since we found these particles at all times during biofilm formation (Fig. 1B). Furthermore, the production of colanic acid seemed to be related to the nonconjugative factor on the F plasmid (Fig. 2A).
A natural IncF F plasmid stimulates curli production via various host cell regulatory systems during biofilm maturation.
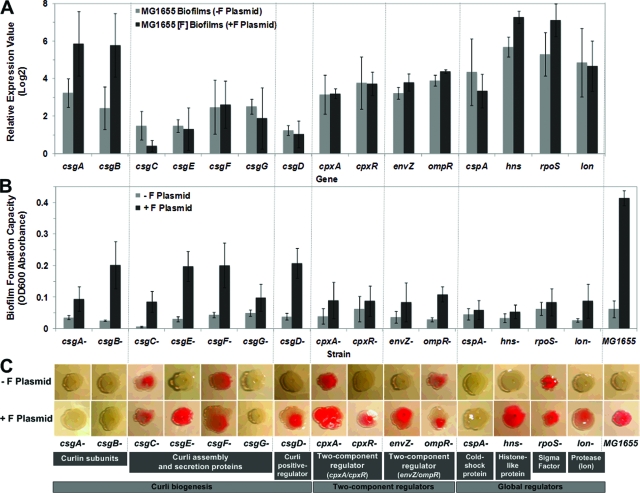
Recently, several studies showed that colanic acid was not essential for bacterial adhesion and further biofilm development (11, 32). However, we found that colanic acid seems to be necessary for formation of 3D mature biofilms by E. coli harboring the F plasmid. Furthermore, the formation of curli filaments plays an important role in cell-surface and cell-cell connections, promoting the formation of 3D mushroom-type mature biofilms (Fig. 1C). Therefore, it is important to examine how a natural F-plasmid-containing E. coli biofilm stimulates curli production. To further address the role of F plasmid in curli production, we examined the curli production and biofilm formation capacities of the wild-type E. coli and curli mutant strains. In this study, we focused on 15 curli biogenesis- or regulation-related genes that have been reported to be involved in E. coli curli production (8, 9, 33, 43), including cpxA and cpxR (encoding a two-component regulator via the response to envelope stress and/or misfolded periplasmic membrane), csgA and csgB (curlin subunits), csgD (positive transcriptional regulator), csgC, csgE, csgF, and csgG (curli assembly and secretion proteins), cspA (major cold shock protein), envZ and ompR (two-component regulator monitoring changes in osmolarity and membrane porin), hns (global regulator; histonelike protein), rpoS (global regulator; stationary-phase sigma factor), and lon (protein regulator; Lon protease). The microarray analysis data for these selected genes showed that they might have a global impact (≥2-fold changes) on either curli production or biofilm formation in a natural F-plasmid-containing E. coli strain (see Tables S1 to S4 in the supplemental material).
First, the relative gene expression values obtained from microarray data for genes mentioned above are compared in Fig. 5A. Curli subunit genes (csgA and csgB) were overexpressed in the F-plasmid-containing biofilm, a profile similar to the upregulated expression profiles of two global regulators (hns and rpoS). There was a slight change in the expression of the EnvZ/OmpR two-component system, but not in the expression of the CpxA/CpxR system. The microarray results also showed that the level of expression of csgC and cspA in the plasmid-free biofilm was higher than the level of expression in the F-plasmid-containing mature biofilm. Since we know that a microarray expression pattern suggests only a rough scheme for regulated pathways and not biochemical activities, we investigated biofilm formation ability using the microplate assay for individual strains with 1 of the 15 genes deleted either with or without a natural F plasmid (Fig. 5B). The results showed that the presence of the F plasmid dramatically increased the ability of most of the tested strains to form biofilms, but this was less true for the csgG, cpxA, cpxR, cspA, hns, rpoS, and lon mutants (P > 0.05). Furthermore, the Congo red phenotype assay was performed to identify which mutants produced curli (Fig. 5C). The biofilms examined in the previous experiments were directly used in this assay. The lack of curlin subunits (csgA and csgB), a curlin secretion protein (csgG), and a cold shock protein (cspA) did not encourage the Congo red-binding phenotype even though the F plasmid was introduced into the deletion strains. Our results indicated that some of the deletion strains (csgC, csgF, cpxA, ompR, and rpoS mutants) could produce curli fibers both in the presence and in the absence of F plasmid. Six deletion strains (csgE, csgD, cpxR, envZ, hns, and lon mutants), as well as the parental strain, were able to produce curli in the presence of F plasmid. Although an E. coli laboratory strain does not produce curli filaments at 37°C (4), we demonstrated that a natural F-plasmid-containing E. coli strain produced curli during biofilm formation even at 37°C. Thus, our results and the results of Bokranz et al. (7) and Kikuchi et al. (22) showed that curli could be expressed in an E. coli biofilm at 37°C, which may be important for infection and pathogenesis.
FIG. 5.
Natural F plasmid helped E. coli produce curli during biofilm maturation. (A) Levels of expression of 15 curli biogenesis- and regulation-related genes obtained from microarray analysis. The bars indicate the averages for three replicates, and the error bars indicate the standard deviations; the data are representative of the data from two independent experiments. Asterisk, P < 0.05 compared with a plasmid-free biofilm. (B) Crystal violet biofilm formation assay showing that most curli biogenesis- and regulation-related mutants were likely dependent on the presence of F plasmid during biofilm formation, but the csgG, cpxA, cpxR, cspA, hns, rpoS, and lon mutants were less dependent (P > 0.05). The bars indicate the averages for eight replicates, and the error bars indicate the standard deviations; the data are representative of the data from 10 independent experiments. Asterisk, P < 0.05 compared with a plasmid-free biofilm. OD600, optical density at 600 nm. (C) Congo red assay showing that curli production was induced by introducing F plasmid only into cpxR, csgD, csgE, envZ, hns, and lon mutants at 37°C. Assays were repeated 10 times, and representative images of Congo red binding are shown.
DISCUSSION
Steps in formation of a biofilm by a natural IncF F-plasmid-containing E. coli strain.
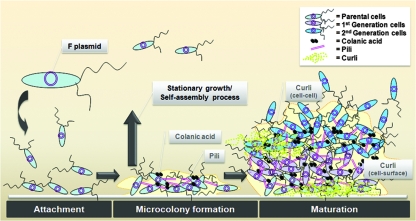
The stepwise formation of a biofilm by a natural IncF conjugative F-plasmid-containing E. coli strain (Fig. 1) had similarities to the formation of biofilms by Pseudomonas aeruginosa and derepressed F-plasmid-containing E. coli, although the biofilms were stabilized by different adhesion factors (34, 41). The proposed steps in biofilm formation are shown in Fig. 6. After the initial attachment of bacterial cells, microcolonies were formed in 6 to 8 h, during which bacteria made cell-cell contact with F pili that branched out in random directions. Microcolony formation lasted for only a few hours, which was consistent with the observation showing that it takes 1 to 2 h for pilus synthesis (15). No cell-surface contact by the F pili was observed at this stage. Although a donor crossing with a donor (F+ × F+) is an unusual phenomenon, it happened during biofilm development in the same way as the F− phenocopy phenomenon (16). Biofilms are thought to have a stationary-phase-like character (20, 38), and this may be the reason why we observed unusual F-pilus formation by F+ × F+ mating cell pairs in interactions with other bacteria in a biofilm. Along with F-pilus mating we also observed capsular colanic acid deposition at the microcolony formation stage. This colanic acid deposition might be important for maintaining the biofilm architecture, but it is unlikely to be the major adhesive factor at the initial stage because the colanic acids were attached mainly to the glass surface rather than to other bacterial cells. Therefore, the first step in biofilm formation requires F+ cell pairs and F-pilus-mediated cell-cell interactions.
FIG. 6.
Proposed biofilm development and maturation for a natural conjugative F-plasmid-containing E. coli strain. The diagram shows the following steps: (i) an initial microcolony biofilm is formed by stationary-growth-phase-like sessile bacteria expressing conjugative F pili, which promote only cell-cell contact; (ii) the cells simultaneously produce capsular colanic acid to support clonal growth of the newly generated populations; and (iii) after completion of the initial step, the stationary-growth-phase-like bacterial cells generate the meshwork of curli fibers to expand complex 3D biofilm structures, which promote adherence of cells to the surface and later reinforce cell-cell interactions.
F-pilus formation and colanic acid deposition in the microcolonies were completed after 8 to 14 h. In the maturation phase (20 to 48 h), complex meshworks of curli consisting of hundreds of long thin filaments were formed. These thin filaments packed bacterial cells in clusters and interacted with the substratum. The curli filaments thus appear to reinforce the cell-surface adhesion, which we did not observe in the earlier phase (2 to 20 h) of biofilm formation. The delay in biofilm formation (14 to 24 h) could be explained by progressive assembly outside the bacterial cells by more mature stationary-phase-like cells, since curli genes are expressed maximally during the late stationary phase (2). Thus, we assumed that the curli-producing cells or colonies at 24 h were the F-pilus-producing cells or colonies that were present at 14 h. We speculate that these sessile bacteria used their stationary-phase characteristics (4) to activate the cell-surface interaction that generates curli meshwork later. After 36 to 48 h, dense mature biofilms were finally formed with complex meshwork consisting of cell-cell curli filaments, F pili, and colanic acid. Since we observed curli formation only at a relatively late stage, we suggest that curli synthesis might be required for stronger attachment to the surface and better cohesion among the cells. A study of curli-overproducing strains also indicated that a similar curli arrangement mediated either cell-surface or cell-cell contact during biofilm formation at initial attachment and microcolony formation phases (42). However, the presence of a natural IncF F plasmid stimulated curli filaments only at the later stage. Therefore, we propose that most of the natural conjugative plasmids have the same effect on biofilm formation since the pili are repressed, but the bacteria still could form mature biofilms. According to Ghigo (18), 22 repressed conjugative plasmid-containing strains could form thick biofilms via F+ mating pili, which usually are expressed during the early step in biofilm formation. We show here that pili are involved in cell-cell interactions only at the beginning of biofilm development and then stimulate the production of curli and colanic acid at later stages of development. Thus, when pilus formation is complete, the pili recede and are replaced by adhesion factors that maintain the coherence of biofilms.
Role of conjugative F pili, colanic acid, and curli in formation of a biofilm by natural IncF F-plasmid-containing E. coli.
Based on a comparison of the effects of different types of plasmids (Fig. 2) and the results of a loss-of-function analysis (Fig. 3), we concluded that (i) only unmodified IncF F plasmid (containing both the conjugative and nonconjugative factors) promotes rapid formation of a mushroom-type biofilm, (ii) F pili are important, especially at the beginning of biofilm formation (microcolony formation) because they are the only initially effective cell-cell adhesion factor, (iii) the nonconjugative factor of F plasmid is able to induce mainly colanic acid and little curli, but not F pili, resulting in a flat biofilm, and (iv) the curli meshwork is required for further development of a dense mature biofilm, which is stimulated by the presence of F plasmid, and supports the development of a 3D mushroom-type biofilm involving both a conjugative factor and a nonconjugative factor of F plasmid.
Gene expression pattern in a natural IncF F-plasmid-containing E. coli biofilm.
It is important to understand which genes are expressed during biofilm formation (Fig. 4). Previous studies did not identify consistent expression profiles even when the same bacterial strain was used (5). When we compared our results with previous results, we did not find significant overlaps for the expressed genes, although certain expression cassettes were shared. However, when we compared upregulated genes in the three different mature biofilms that we studied [biofilms formed by MG1655, MG1655(F), and MG1655(pNCF)], we identified a general trend for biofilm-specific genes, especially overexpression of F-pilus-, colanic acid-, and curli-related genes in the presence of F plasmid (Fig. 4B). For this reason, we evaluated the regulated genes based on their biological functions and found that different plasmids and biofilm growth conditions were associated with different global gene expression profiles (Fig. 4A). The presence of F plasmid in biofilms had a global impact on E. coli gene expression. Notably, the first comparison between F-plasmid-containing and plasmid-free E. coli biofilms (using strain TG1) was performed recently by Beloin et al. (6). There were a number of our results that did not overlap with results reported previously by Beloin et al. (6), which were likely due to the different plasmids used. The F plasmid in TG1 was mutated to delete the DNA transfer ability (traD36 genotype), which resulted in derepressed F-pilus formation (1, 47). Beloin et al. found that 33% of the genes induced in a TG1 biofilm were also statistically significantly overexpressed in a biofilm formed by TG, an F-plasmid-free derivative of TG1. However, in this study with a natural F plasmid, we found that about 43.5% of the genes were induced in a similar comparison. We found that 41.5% of the genes overlapped (27 of 65 overexpressed genes reported by Beloin et al.), including members of the cpx, rbs, fim, ycf, yoa, yqc, ycc, yfc, ygh, and yce operons (most of whose functions have not been characterized). Therefore, it is very difficult to determine a universal gene expression pattern for bacterial biofilms based on a comparison of the array data obtained in different laboratories and with different array platforms. It is important to standardize the experimental protocols so that results obtained in different laboratories can be directly compared.
Involvement of a natural IncF F plasmid in curli production during E. coli biofilm formation.
Genes involved in curli production are organized in the csgBAC and csgDEFG operons, but the functions of only a few of these genes have been fully determined (10). Although these gene clusters are highly conserved among E. coli strains (37), many laboratory strains do not produce curli due to the silencing repression of the csgD promoter (9). The expression of the csgD promoter is affected by several transcriptional regulators, including RpoS, H-NS, OmpR, and CpxR (33). Even though we used a microarray and several molecular approaches in association with a biofilm and curli formation assay to evaluate the impact of a natural IncF F plasmid on curli production during biofilm formation, we obtained almost no clear evidence of how F plasmid stimulates curli production. This was probably due to the fact that curli gene regulation is extraordinarily complex (17) and curli gene expression is very responsive to many environmental cues (4). However, we identified 15 genes that are thought to be the genes that most influence curli production in the presence of F plasmid (Fig. 5). Seven of these genes are curli biogenesis genes and are located in the same operon. The curlin subunits (csgBA) were important for curli production and for formation of a mushroom-type biofilm, as indicated by overexpression of both factors in the presence of F plasmid. The csgDEFG operon includes csgEFG encoding curli assembly and secretion proteins (4). Our results showed that the presence of F plasmid induced curli production and biofilm formation, especially with csgE and csgD mutants, but had no effect on the levels of csgDEFG transcription. Since CsgD is apparently required for csgBAC transcription (12, 17), this suggested that the regulation of the csgD transcript in biofilms is likely affected by outside transcriptional regulators, including genes on the F plasmid. In addition, CsgE is a periplasmic protein and interacts with the bacterial outer membrane (10, 36), and accordingly the presence of F pili might induce curli protein assembly. However, how F plasmid suppresses the csgE and csgD phenotypes is not understood at this time, and thus the possibilities described above are currently being investigated.
Several bacterial regulatory systems seemed to be involved in stimulation of curli production in F-plasmid-containing biofilms. One of the two-component regulatory systems, EnvZ/OmpR, seems to have a significant effect on curli gene regulation. The EnvZ/OmpR system responds to changes in osmolarity and regulates several porins (21). Expression of the EnvZ/OmpR genes was significantly upregulated in the presence of F plasmid. In addition, cells were able to form biofilms and curli formation was stimulated when EnvZ/OmpR mutants contained an F plasmid. Thus, the EnvZ/OmpR system might positively regulate curli production in an F-plasmid-containing biofilm. Recently, analysis of the global impact of derepressed IncFI and IncFII conjugative plasmids on E. coli biofilm gene expression revealed that there was significant upregulation of related osmolarity and porin genes, especially genes encoding Omp proteins, indicating the importance of the involvement of these regulatory systems during biofilm formation (6, 46). Although results indicated that the Cpx regulatory system was activated to interrupt the membrane and induced constitutively to promote F-pilus formation, we did not find any significant function for this system in a natural IncF plasmid. In contrast, we found that the amount of biofilm was not different even when the curli was induced when a cpxR mutant contained an F plasmid. This was probably due to the derepressed effect on the plasmid, which contributed to the expression of curli and biofilm formation. Several global regulators were also tested in this study. One of these regulators, RpoS, the stationary-phase sigma factor, also seemed to play an important role, as expression of the RpoS gene was upregulated in the presence of F plasmid, while F plasmid did not significantly promote biofilm formation. In addition, the histone-like protein HN-S also may play a role in the biofilm. HN-S has a key role in the conjugative regulator of a conjugative plasmid and is a regional silencer during pilus mating pair formation (40, 44, 45). The HN-S factor was upregulated in the presence of F plasmid, interrupted biofilm formation, and induced the production of curli. In our view, HN-S might be the most important key for switching F-pilus and curli formation on and off during biofilm development since it has often been referred to as either F plasmid conjugation or curli production dependent (3, 44, 45).
In conclusion, our study provided experimental evidence that the mating F pili between pairs of F+ cells of E. coli harboring a natural IncF F plasmid are required for formation of the 3D mushroom-type biofilms because they stimulate colanic acid and curli production. The biosynthesis of curli played an important role in the maturation process and the structure of mature biofilms. These findings provide a basis for the connection between natural horizontal gene transfer and biofilm formation.
Supplementary Material
Acknowledgments
We thank Maarten Leyssen of the Flanders Institute for Biotechnology in Belgium, Andreas Reisner of the University of Graz in Austria, Tim Tolker-Nielsen of the Biocentrum-DTU in Denmark, and Akinobu Ito of our laboratory for valuable recommendations and critical comments on the manuscript, Yoshinobu Nodasaka for excellent technical assistance with electron microscope analyses, Keiko Ito and Seiichi Yasuda of the National Institute of Genetics in Japan for kindly providing E. coli K-12 and derivative strains, Hirotada Mori of the Nara Institute of Science and Technology in Japan for supplying the mutants, Philippe Lejeune of INSA de Lyon in France for providing PHL628, Claus Sternberg and Janus Haagensen of the Biocentrum-DTU for suggestions about flow cell setup and COMSTAT analysis, and Cheng Li of the Harvard School of Public Health in the United States for providing the dChip software and comments on the statistical analysis of the data.
Footnotes
Published ahead of print on 12 September 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Achtman, M., N. Willetts, and A. J. Clark. 1971. Beginning a genetic analysis of conjugational transfer determined by the F factor in Escherichia coli by isolation and characterization of transfer-deficient mutants. J. Bacteriol. 106529-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnqvist, A., A. Olsen, and S. Normark. 1994. Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 131021-1032. [DOI] [PubMed] [Google Scholar]
- 3.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 247-17. [DOI] [PubMed] [Google Scholar]
- 4.Barnhart, M. M., and M. R. Chapman. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60131-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beloin, C., and J. M. Ghigo. 2005. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 1316-19. [DOI] [PubMed] [Google Scholar]
- 6.Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. J. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J. M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51659-674. [DOI] [PubMed] [Google Scholar]
- 7.Bokranz, W., X. Wang, H. Tschape, and U. Romling. 2005. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Med. Microbiol. 541171-1182. [DOI] [PubMed] [Google Scholar]
- 8.Brombacher, E., A. Baratto, C. Dorel, and P. Landini. 2006. Gene expression regulation by the curli activator CsgD protein: modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J. Bacteriol. 1882027-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brombacher, E., C. Dorel, A. J. B. Zehnder, and P. Landini. 2003. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 1492847-2857. [DOI] [PubMed] [Google Scholar]
- 10.Chapman, M. R., L. S. Robinson, J. S. Pinkner, R. Roth, J. Heuser, M. Hammar, S. Normark, and S. J. Hultgren. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295851-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 1823593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Da Re, S., and J. M. Ghigo. 2006. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J. Bacteriol. 1883073-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diederich, L., L. J. Rasmussen, and W. Messer. 1992. New cloning vectors for integration into the λ attachment site attB of the Escherichia coli chromosome. Plasmid 2814-24. [DOI] [PubMed] [Google Scholar]
- 15.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Mol. Biol. Rev. 58162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frost, L. S., and J. Manchak. 1998. F− phenocopies: characterization of expression of the F transfer region in stationary phase. Microbiology 1442579-2587. [DOI] [PubMed] [Google Scholar]
- 17.Gerstel, U., C. Park, and U. Romling. 2003. Complex regulation of csgD promoter activity by global regulatory proteins. Mol. Microbiol. 49639-654. [DOI] [PubMed] [Google Scholar]
- 18.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412442-445. [DOI] [PubMed] [Google Scholar]
- 19.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 1462395-2407. [DOI] [PubMed] [Google Scholar]
- 20.Ito, A., T. May, K. Kawata, and S. Okabe. 2008. Significance of rpoS during maturation of Escherichia coli biofilms. Biotechnol. Bioeng. 991462-1471. [DOI] [PubMed] [Google Scholar]
- 21.Jubelin, G., A. Vianney, C. Beloin, J. M. Ghigo, J. C. Lazzaroni, P. Lejeune, and C. Dorel. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 1872038-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kikuchi, T., Y. Mizunoe, A. Takade, S. Naito, and S. Yoshida. 2005. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol. Immunol. 49875-884. [DOI] [PubMed] [Google Scholar]
- 23.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 481511-1524. [DOI] [PubMed] [Google Scholar]
- 24.Koonin, E. V., K. S. Makarova, and L. Aravind. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 9831-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, G., A. E. Loraine, R. Shigeta, M. Cline, J. Cheng, V. Valmeekam, S. Sun, D. Kulp, and M. A. Siani-Rose. 2003. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res. 3182-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manwaring, N. P., R. A. Skurray, and N. Firth. 1999. Nucleotide sequence of the F plasmid leading region. Plasmid 41219-225. [DOI] [PubMed] [Google Scholar]
- 28.Mueller, L., J. de Brouwer, J. Almeida, L. Stal, and J. Xavier. 2006. Analysis of a marine phototrophic biofilm by confocal laser scanning microscopy using the new image quantification software PHLIP. BMC Ecol. 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 5449-79. [DOI] [PubMed] [Google Scholar]
- 31.Ou, J. T., and T. F. Anderson. 1972. Effect of Zn2+ on bacterial conjugation: inhibition of mating pair formation. J. Bacteriol. 111177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prigent-Combaret, C., G. Prensier, T. T. Le Thi, O. Vidal, P. Lejeune, and C. Dorel. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2450-464. [DOI] [PubMed] [Google Scholar]
- 33.Prüβ, B. M., C. Besemann, A. Denton, and A. J. Wolfe. 2006. A complex transcription network controls the early stages of biofilm development by Escherichia coli. J. Bacteriol. 1883731-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reisner, A., J. A. J. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48933-946. [DOI] [PubMed] [Google Scholar]
- 35.Reisner, A., B. M. Höller, S. Molin, and E. L. Zechner. 2006. Synergistic effects in mixed Escherichia coli biofilms: conjugative plasmid transfer drives biofilm expansion. J. Bacteriol. 1883582-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson, L. S., E. M. Ashman, S. J. Hultgren, and M. R. Chapman. 2006. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 59870-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romling, U., Z. Bian, M. Hammar, W. D. Sierralta, and S. Normark. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schembri, M. A., K. Kjaergaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48253-267 [DOI] [PubMed] [Google Scholar]
- 39.Sorensen, S. J., M. Bailey, L. H. Hansen, N. Kroer, and S. Wuertz. 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3700-710. [DOI] [PubMed] [Google Scholar]
- 40.Starčič-Erjavec, M., J. P. M. Putten, W. Gaastra, B. J. A. M. Jordi, M. Grabnar, and D. Žgur-Bertok. 2003. H-NS and Lrp serve as positive modulators of traJ expression from the Escherichia coli plasmid pRK100. Mol. Genet. Genomics 27094-102. [DOI] [PubMed] [Google Scholar]
- 41.Tolker-Nielsen, T., U. C. Brinch, P. C. Ragas, J. B. Andersen, C. S. Jacobsen, and S. Molin. 2000. Development and dynamics of Pseudomonas sp. biofilms. J. Bacteriol. 1826482-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidal, O., R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman, and P. Lejeune. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 1802442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, X., D. R. Smith, J. W. Jones, and M. R. Chapman. 2007. In vitro polymerization of a functional Escherichia coli amyloid protein. J. Biol. Chem. 2823713-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Will, W. R., and L. S. Frost. 2006. Characterization of the opposing roles of H-NS and TraJ in transcriptional regulation of the F-plasmid tra operon. J. Bacteriol. 188507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Will, W. R., J. Lu, and L. S. Frost. 2004. The role of H-NS in silencing F transfer gene expression during entry into stationary phase. Mol. Microbiol. 54769-782. [DOI] [PubMed] [Google Scholar]
- 46.Yang, X., Q. Ma, and T. K. Wood. 2008. The R1 conjugative plasmid increases Escherichia coli biofilm formation through an envelope stress response. Appl. Environ. Microbiol. 742690-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.