Abstract
Enteroaggregative Escherichia coli (EAEC) is a pathogen implicated in several infant diarrhea or diarrheal outbreaks in areas of endemicity. Although multiple genes involved in EAEC pathogenesis have been identified, the overall mechanism of virulence is not well understood. Recently, a novel secretion system, called type VI secretion (T6S) system (T6SS), has been identified in EAEC and most animal or plant gram-negative pathogens. T6SSs are multicomponent cell envelope machines responsible for the secretion of at least two putative substrates, Hcp and VgrG. In EAEC, two copies of T6S gene clusters, called sci-1 and sci-2, are present on the pheU pathogenicity island. In this study, we focused our work on the sci-1 gene cluster. The Sci-1 apparatus is probably composed of all, or a subset of, the 21 gene products encoded on the cluster. Among these subunits, some are shared by all T6SSs identified to date, including a ClpV-type AAA+ ATPase (SciG) and an IcmF (SciS) and an IcmH (SciP) homologue, as well as a putative lipoprotein (SciN). In this study, we demonstrate that sciN is a critical gene necessary for T6S-dependent secretion of the Hcp-like SciD protein and for biofilm formation. We further show that SciN is a lipoprotein, as shown by the inhibition of its processing by globomycin and in vivo labeling with [3H]palmitic acid. SciN is tethered to the outer membrane and exposed in the periplasm. Sequestration of SciN at the inner membrane by targeting the +2 residue responsible for lipoprotein localization (Gly2Asp) fails to complement an sciN mutant for SciD secretion and biofilm formation. Together, these results support a model in which SciN is an outer membrane lipoprotein exposed in the periplasm and essential for the Sci-1 apparatus function.
The virulence of most bacterial pathogens relies on a subset of processes including biofilm formation, interaction with the host cell, and release of toxin proteins (35). This latter mechanism requires dedicated machineries called secretion systems. Seven secretion systems have been described so far, which assemble from 3 to >20 subunits. These secretion systems derive from or coevolved with bacterial organelles, such as ABC transporters (type I), type IV pili (type 2), flagella (type 3), or conjugative machines (type IV) (12, 23, 43, 44). The newly identified type VI secretion (T6S) system (T6SS) is present in most pathogens that have contact with animals, plants, or humans (8, 10, 21, 33). T6SSs have been identified in several bacteria but are not yet well characterized. These secretion systems are involved in numerous pathogenic processes, including resisting predation by amoebae for Vibrio cholerae and Burkholderia cenocepacia (4, 56), inhibiting the macrophage phagocytizing activity of macrophages for Aeromonas hydrophila (67), favoring persistence within the host for Salmonella enterica (54), biofilm formation for Vibrio parahaemolyticus (29), rotting of potato stems for Pectobacterium atrosepticum (46), or the global virulence of the fish pathogen Edwardsiella tarda (57). T6SSs assemble at the bacterial cell envelope and form an apparatus dedicated to the secretion of essentially two proteins into the culture supernatant (8, 10, 33, 75). These proteins, called Hcp (hemolysin coregulated protein) and VgrG (valine glycine repeat), were found in culture supernatants of bacteria carrying T6S gene clusters (28, 52, 55, 56, 67), although whether these proteins are real substrates of the machine or are released in the supernatant following mechanical shearing in liquid culture is still matter of debate (10, 55). However, the presence of Hcp and VgrG in culture supernatants constitutes a good indicator of T6SS function. In a recent systematic deletion approach, 11 of the 13 evp genes of the E. tarda T6SS have been found to be necessary for secretion of Hcp and VgrG into culture supernatant (75). The products of these genes may thus assemble as a secretion apparatus to deliver substrates to the medium or the host cell. Among these genes, several are shared by all T6SSs identified so far and are called “core components.” These genes encode an AAA+ ATPase of the ClpV family, a putative lipoprotein, several putative cytoplasmic proteins of unknown function, and inner membrane (IM) proteins sharing similarities with the type IVb secretion IcmF and IcmH proteins (8, 10, 33). The ClpV-like ATPases are thought to assemble as a ring structure capable of hydrolyzing nucleoside triphosphate to energize machine assembly or substrate secretion (59). In the Legionella pneumophila Dot/Icm T4bSS, the IcmF and IcmH (or DotU) proteins interact and stabilize the transport apparatus (62). Interestingly, most T6S-associated IcmH proteins carry C-terminal extensions resembling the peptidoglycan binding domain of the OmpA/Pal/MotB family and may thus anchor the apparatus to the cell wall.
Two gene clusters encoding T6S machines, namely sci-1 and sci-2, are present on the pheU pathogenicity island of enteroaggregative Escherichia coli (EAEC) strains (28). EAEC strains define an emerging E. coli pathotype responsible for severe and persistent diarrhea of infants, young children, or immunocompromised individuals (39, 53). Although not well understood, the EAEC virulence mechanism is based on colonization of the intestinal mucosa involving both the flagella and adhesive structures called fimbriae, formation of a thick biofilm at the surface of the colon epithelia, and secretion of entero- and cytotoxins (45). These processes involve plasmid-borne and chromosomal virulence factors that had not been identified yet. However, several pathogenic determinants are encoded within the pheU pathogenicity island, and the two sci T6S gene clusters are interesting candidates. Besides the presence of genes encoding homologues of Hcp and VgrG, these clusters possess characteristic features of functional T6S machines, including core components that are conserved in all clusters identified so far. The sci-2 gene cluster expression has been shown to be controlled by the AraC family transcriptional activator AggR, which is a master regulator of EAEC virulence (28). It assembles a T6SS machine responsible for the secretion of the Hcp homologue AaiC. However, mutagenesis studies showed that no virulence defects are linked to sci-2 mutations. The sci-1 gene cluster has not been characterized, but the Sci-1 T6S proteins assemble and function under the laboratory conditions (28). The sci-1 cluster encodes typical T6S-associated proteins, including the SciD Hcp-like protein, a VgrG homologue, the SciG AAA+ ATPase, the SciP IcmH-like protein, the SciS IcmF-like protein, and SciN, a probable lipoprotein.
In the present work, we initiated studies on the EAEC sci-1 gene cluster by targeting the sciN gene. We showed that an sciN mutant is unable to secrete the Hcp-like SciD substrate in the supernatant, which is accompanied by a decrease in biofilm formation, and is thus an essential component of the T6SS machine. Although SciN is predicted to be a lipoprotein based on its consensus N-terminal sequence for recognition by the signal peptidase II and lipid modification, its localization and acylation have not been determined experimentally. Herein, we performed experiments that demonstrated that SciN is a lipoprotein localized in the outer membrane (OM). We further showed that SciN is exposed in the periplasmic space. Mutational analyses showed that proper localization at the OM is required for SciN function.
MATERIALS AND METHODS
Bacteria, plasmids, chemicals, and growth conditions.
E. coli K-12 DH5α was used for cloning procedures. The lpp deletion strain JE5505 (68) was used for copurification experiments. The EAEC strain 17-2 (kindly provided by Arlette Darfeuille-Michaud, University of Clermont-Ferrand, France) was used for this study. Unless otherwise indicated, strains were routinely grown in LB broth at 37°C, with aeration. Plasmids were maintained by the addition of ampicillin (100 μg/ml for K-12 and 200 μg/ml for EAEC), kanamycin (50 μg/ml for K-12, 50 μg/ml for chromosomal insertion on EAEC, and 100 μg/ml for plasmid-bearing EAEC), and chloramphenicol (40 μg/ml). Plasmid pAMR43L, allowing production of LolA with the mutation R43L and a six-His tag [His6-LolA(R43L); kindly provided by Hajime Tokuda, University of Tokyo, Japan) has been described previously (51). [9,10(n)-3H]palmitate was purchased from NEN (Perkin Elmer, MA). Globomycin was kindly provided by Danièle Cavard (Institute of Structural Biology and Microbiology, Marseille, France).
Plasmid construction.
PCRs were performed with a Biometra thermocycler, using Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA). Oligonucleotides were synthesized by Eurogentec. Plasmid pSciD-FLAG was constructed by a double PCR technique (70), allowing amplification of the sciD gene, carrying a FLAG epitope coding sequence at the 3′ end, flanked by extensions annealing to the target vector (pUC12) (74). The product of the first PCR was then used as oligonucleotides for a second PCR using the target vector as a template. The oligonucleotides used were 5′-ggaaacagctatgaccatgattacgaatATGGCAATTCCAGTTTATCTGTGGC and 5′-tagaggatccccgggcgagctcgattaTTTATCATCCTCATCTTTATAATCCGCGGTGGTACGCTCACTCCAT (italicized uppercase letters indicate bases complementary to gene of interest [first PCR], lowercase letters indicate bases complementary to target vector [second PCR], and underlined uppercase letters indicate the FLAG epitope coding sequence). pSciN-HA (where HA is hemagglutinin) was constructed by amplification of the sciN gene using purified EAEC chromosomal DNA as a template and the oligonucleotides 5′-GTCGGAATTCGCGATTATCGCTGGTAAGGCTGG (EcoRI restriction site is underlined) and 5′-CTGCCTCGAGTTTATCCTTCACCGGTAACAGGGTCAG (XhoI restriction site is underlined). The PCR product was digested by EcoRI and XhoI and cloned into the same sites of the pMS600 vector (a derivative of the pOK12 cloning vector [72] allowing in-frame fusion with a C-terminal HA coding sequence) (M. S. Aschtgen, unpublished data). Plasmid pSciN(G2D) with a G2D mutation in SciN was constructed by site-directed mutagenesis using plasmid pSciN-HA as the template and oligonucleotides 5′-TCCTGATAACGACAGGGAAAACAACGCAATAATCAGACCGTAACC and 5′-GTTTTCCCTGTCGTTATCAGGATGCGATCTGACGCAAAGAGTGGCAGACG. All constructs have been verified by restriction analyzes and DNA sequencing (Genome Express).
Construction of EAEC isogenic mutants.
Isogenic mutants were generated using a modified protocol of a one-step inactivation procedure (22). The kanamycin resistance cassette was generated by PCR using the pKD4 plasmid as a template and a pair of oligonucleotides carrying 50-nucleotide extensions homologous to regions adjacent to the target gene (underlined in the following oligonucleotide sequences). Oligonucleotides used in this study were 5′-GATGCGCCTGGGTTACGGCGAAGGGCGGTAGCGTGCCTGACGGTCTCCTGACAGTGATGTGTAGGCTGGAGCTGCTTCG and 5′-GCCTTCTTCCCTTTGATTTTTACCTGCGCCAGCGTCCTGATTTCTGCCATAGCCGGTCATATGAATATCCTCCTTAGTTC for deletion of the sciG (clpV) gene and 5′-GGAACATTATCAACGTAAGGAGACTCAGGAAAATGGCGATTATCGCTGGTAAGGCTGTGTGTAGGCTGGAGCTGCTTCG and 5′-TCCAGCCCGCCAGTGAAATTGCCATAGAGAATTTCATAAAGGGAAGGACGCGGCATATGAATATCCTCCTTAGTTC for deletion of the sciN gene. The PCR product was then electroporated into EAEC cells carrying the pKOBEG plasmid (16), as described previously (18). Replacement of the gene by the kanamycin cassette flanked by two Flp recombinase target sequences was confirmed by PCR. The resulting strain was then transformed with the pCP20 plasmid (22) and incubated for 24 h at 30°C, allowing excision of the cassette by the Flp recombinase. Plasmid pCP20 was then eliminated at 37°C, and the cassette excision was verified by PCR.
Test for biofilm formation.
An adherence assay was performed in 24-well polystyrene microtiter dishes or glass tubes after incubation in Dulbecco's modified Eagle medium (high glucose; Sigma) at 30°C without agitation for 20 h. Attached bacteria were stained with 1% crystal violet for 15 min and washed twice with water. For quantification, the ring of stained bacteria was collected with 500 μl of 95% ethanol and diluted in the same volume of water. The absorbance of the suspension was then measured at 600 nm using a Jenway spectrophotometer.
Separation of supernatant and cell fractions.
A total of 2 × 109 cells grown in high-glucose Dulbecco's modified Eagle medium were harvested and collected by centrifugation at 2,000 × g for 5 min. The supernatant fraction was then subjected to a second low-speed centrifugation and then to centrifugation at 16,000 × g for 15 min. The supernatant was then filtered on sterile polyester membranes with a pore size of 0.2 μm (membrex 25 PET; membraPure GmbH) before precipitation with 15% trichloroacetic acid (TCA). Cells and precipitated supernatant were then resuspended in loading buffer and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
Globomycin treatment.
EAEC cells producing SciN-HA from the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter were grown to an optical density at 600 nm of ∼0.6 before addition of 50 μg/ml of globomycin. After 10 min of incubation, IPTG was then added at a final concentration of 100 μM, and cells were further incubated for 30 min at 37°C. Cells were harvested, and samples were analyzed by SDS-PAGE and immunoblotting.
In vivo acylation assay.
Lipoproteins from 3 × 109 exponentially growing EAEC cells producing SciN-HA were labeled with 100 μCi of [9,10(n)-3H]palmitate for 1 h at 37°C. Proteins of interest were solubilized by a procedure using SDS-Triton X-100 and immunoprecipitated with specific antibodies coupled to protein A-Sepharose CL-4B beads (GE Healthcare) as previously described (15). Immunoprecipitated samples were analyzed by SDS-PAGE and autoradiography for 4 to 6 days at −80°C.
Spheroplast formation and fractionation.
Cells were converted to spheroplasts as previously described (14). Briefly, a pellet of 2 × 109 exponentially growing cells was resuspended in 1 ml of 10 mM Tris-HCl (pH 8.0)-20% sucrose and incubated for 10 min on ice. After the addition of 100 μg/ml of lysozyme and 0.5 mM EDTA and further incubation for 25 min on ice, the periplasm and spheroplast fractions were separated by centrifugation. Spheroplasts were washed with 10 mM Tris-HCl (pH 8.0)-20%, sucrose, resuspended in 1 ml of 10 mM Tris-HCl-20% sucrose, and then subjected to five cycles of freezing and thawing before centrifugation to remove unbroken cells and to ultracentrifugation (40 min at 100,000 × g) to separate cytoplasm and membrane fractions. Membranes were washed with 10 mM Tris-HCl (pH 8.0)-2 mM MgCl2. Periplasmic and cytoplasmic fractions were precipitated with 15% TCA and resuspended in loading buffer prior to analysis by SDS-PAGE and immunoblotting.
Proteinase K accessibility.
A total of 109 spheroplasts or whole cells resuspended in 1 ml of 20 mM Tris-HCl (pH 8.0)-10 mM MgCl2 were treated with proteinase K at a final concentration of 100 μg/ml for 20 min on ice, collected by centrifugation, and then analyzed by SDS-PAGE and immunoblotting.
Sarkozyl differential extraction.
Membranes prepared from 1010 cells using the fractionation protocol were resuspended in 1 ml of 10 mM Tris-HCl (pH 8.0) supplemented with 2% sodium N-lauroyl sarcosinate (SLS; Sigma Aldrich), and incubated on a rotating wheel for 1 h at room temperature. Insoluble and soluble fractions were recovered by ultracentrifugation at 100,000 × g for 40 min before analysis by SDS-PAGE and immunoblotting.
Membrane sedimentation analyses.
Inner and outer membranes were separated using discontinuous sedimentation sucrose gradients. A total of 2 × 1011 EAEC cells producing SciN-HA or SciN(G2D)-HA were harvested, suspended in 1.5 ml of 10 mM Tris-HCl (pH 7.4), 20% sucrose and 10 μg/ml RNase and lysed by French press treatment. Total membranes were recovered by centrifugation at 100,000 × g for 40 min and resuspended in 0.5 ml of 20% sucrose containing a protease inhibitor cocktail (Complete EDTA-free; Roche). The membrane fraction was then loaded on the top of a discontinuous sucrose gradient composed of the superposition of 1.5 ml of 30, 35, 40, 45, 50, 55, and 60% sucrose solutions (from top to bottom). Gradients were centrifuged at 90,000 × g for 100 h, and 500-μl fractions were collected from the top. The composition of these fractions was analyzed by an NADH oxidase enzymatic test and by SDS-PAGE and immunoblotting with the anti-AcrA and anti-OmpA polyclonal antibodies. The NADH oxidase activity was measured in 96-well polystyrene microtiter dishes, using 20 μl of each fraction diluted in 180 μl of 50 mM Tris-HCl (pH 7.5), 0.2 mM dithiothreitol, and 0.5 mM NADH. The decrease in absorbance of the NADH at 340 nm, which reflects the activity of the NADH oxidase, was measured after 15 min of incubation at 25°C. Each fraction was tested in triplicate.
Six-histidine pull-down experiments.
A total of 5 × 109 EAEC cells producing SciN-HA (from pSciN-HA plasmid) or both SciN-HA and His6-LolA(R43L) (from plasmid pAMR43L) were harvested and treated for spheroplast formation. The supernatant (periplasm fraction) was collected, and NaCl (100 mM final), imidazole (10 mM final), and protease cocktail inhibitors were added; the suspension was mixed with 100 μl of cobalt-coupled agarose beads (Talon; Clontech) and further incubated on a rotating wheel for 4 h at 25°C. The unbound fraction was collected, and beads were washed twice with 1 ml of buffer A containing 10 mM imidazole before elution with 1 ml of buffer A supplemented with 500 mM imidazole. Eluted material was then precipitated with TCA at a final concentration of 15% and resuspended in loading buffer before analysis by SDS-PAGE and immunoblotting.
Miscellaneous.
Caenorhabditis elegans killing assays were performed as follows. A total of 108 bacteria to be tested were spotted on the center of nematode growth medium plates and were incubated overnight at 37°C. Twenty-five worms were then transferred to each plate and kept at 25°C. The number of living versus dead or paralyzed worms was scored every 24 h for 12 days. Worms were considered dead when they were nonmotile and did not respond to a stimulus such as probing with a wire pick. Each strain was tested in triplicate. Proteins suspended in loading buffer were subjected to SDS-PAGE. For detection by immunostaining, proteins were transferred onto nitrocellulose membranes, and immunoblots were probed with antibodies and goat secondary antibodies coupled to alkaline phosphatase and developed in alkaline buffer in the presence of 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium. Anti-OmpA, -Pal, -TolB, -TolR, -TolA, -EfTu, -AcrA, and -MalE antibodies were from our laboratory collection. Anti-FLAG (Sigma-Aldrich), anti-HA (Roche), anti-His5 (Qiagen), and alkaline phosphatase-conjugated goat anti-rabbit, mouse, or rat antibodies (Millipore) were purchased as indicated.
RESULTS
SciN is required for T6S.
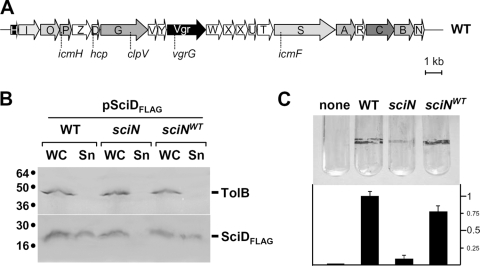
In all T6SSs described so far, two proteins were found in culture supernatants: (i) the Hcp or Hcp-like proteins, which form hexameric rings; and (ii) the VgrG proteins, which resemble the tail spike of bacteriophage T4. An Hcp-like protein and a VgrG homologue are encoded within the EAEC sci-1 gene cluster (Fig. 1A). We first tested whether the Hcp-like protein SciD is released from EAEC 17-2 wild-type (WT) cells under our conditions, as previously reported (28). We thus constructed a plasmid allowing IPTG-inducible expression of sciD. This SciD protein carries a FLAG epitope at the C terminus. As expected, SciD-FLAG was found in culture supernatants of EAEC cells but was not released from E. coli K-12 DH5α cells (Fig. 1B and data not shown). Immunodetection of the periplasmic TolB protein (47) further demonstrated that no lysis or periplasmic leakage occurred during the experiment. To verify that this was not a nonspecific release, we constructed an EAEC mutant with a deletion of the sciG (clpV) gene. ClpV proteins are ATPases of the AAA+ family linked to T6SS and have been shown to be essential for T6S in V. cholerae, P. aeruginosa, and E. tarda (52, 56, 75). SciD-FLAG was not recovered in culture supernatants of EAEC sciG cells, demonstrating the specificity of secretion (data not shown).
FIG. 1.
SciN is required for T6S. (A) Schematic organization of the EAEC sci-1 gene cluster. Conserved genes are indicated by gradations of gray to black. White genes are not conserved among T6S gene clusters. The generic names of conserved genes characterized are indicated below. (B) Effect of the sciN mutation on Hcp-like SciD protein secretion. SciD-FLAG secretion was assessed by separating whole cells (WC) and supernatant (Sn) fractions from WT, sciN, and complemented sciN (sciNWT) strain cultures. A total of 2 × 108 cells and the TCA-precipitated material of the supernatant from 5 × 108 cells were loaded on a 15% acrylamide SDS-PAGE gel and subjected to immunodetection using anti-FLAG monoclonal antibody (lower panel) and the anti-TolB polyclonal antibodies (upper panel). (C) Effect of the sciN mutation on biofilm formation. Biofilms formed in static cultures of WT, sciN, and complemented sciN cells (none, LB medium) were visualized in glass tubes by crystal violet staining (upper panel) and quantified using the ethanol-solubilization procedure, relative to the WT EAEC strain (lower graph).
Among the T6SS core component genes, one encodes a putative lipoprotein (COG3521 family) (8, 10). In the EAEC sci-1 gene cluster, this gene, called sciN, is the last open reading frame of the locus (Fig. 1A). To test whether this conserved gene is required for the T6SS, we constructed an sciN deletion mutant and a plasmid allowing the IPTG-inducible production of a C-terminally HA-tagged SciN protein. Fractionation experiments further showed that SciD-FLAG was not secreted in the sciN strain carrying the empty vector, but SciD-FLAG secretion was restored when sciN-HA was supplied in trans (Fig. 1B). This result demonstrates that SciN is required for T6S.
No role for the sci-1 T6S gene cluster had been assigned for EAEC virulence. Using the worm C. elegans, previously shown to be a model for testing virulence of pathogenic E. coli (3), we showed that WT EAEC cells killed the worms at a 50% lethal dose of 7 days (data not shown). In contrast, the E. coli K-12 strain DH5α had no effect on C. elegans during the 12 days of the experiments. The EAEC sciN strain displayed a 50% lethal dose similar to that of the WT EAEC on C. elegans (data not shown). Because some T6S gene clusters have been shown to be necessary for biofilm formation or to be expressed during biofilm development (29, 65), we tested bacteria aggregation on polyethylene plastic wells and glass tubes. Crystal violet staining showed that the sciN mutant forms reproducibly lower amounts of biofilm than the WT cells or sciN cells complemented by the HA-tagged version of SciN (Fig. 1C).
SciN is a lipoprotein.
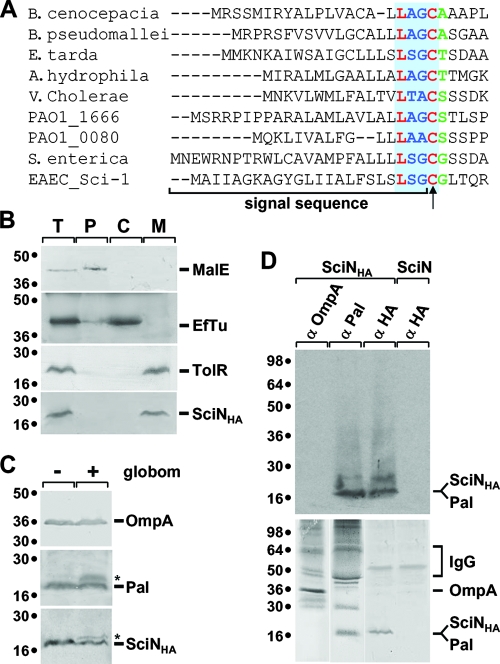
The N terminus of SciN shares characteristic features of classical lipoproteins, which include a predicted lipoprotein signal sequence consensus (lipobox: L-A/S-G/A) followed by a cysteine residue (69) (Fig. 2A). This lipobox is shared by all SciN homologues recovered in T6S gene clusters (Fig. 2A). Lipoprotein signal sequences are cleaved by the signal peptidase II (SPII), contrary to other membrane and periplasmic proteins, which are processed by SPI. After removal of the signal sequence, the α-amino group of the N-terminal cysteine residue is fatty acylated to yield the mature lipoprotein. Thus, the bacterial lipoproteins are membrane proteins processed by SPII and acylated. We first tested whether SciN is membrane associated. Upon cell fractionation, SciN-HA associated with the membrane fraction in EAEC (Fig. 2B). As controls, the TolR protein fractionated with the membrane fraction, and MalE localized in the periplasm whereas EfTu was found in the cytoplasmic fraction. To test whether SciN is a lipoprotein, we used (i) inhibition of SPII with the specific inhibitor globomycin (41) and (ii) [3H]palmitate labeling. The unprocessed forms of SciN-HA and of the Pal lipoprotein were detected when globomycin was added to growing EAEC cells expressing sciN-HA, whereas the OM OmpA protein was correctly processed (Fig. 2C). These results suggest that SciN is processed by SPII. Similarly, both SciN-HA and Pal were labeled in vivo with [3H]palmitate, whereas OmpA was not (Fig. 2D), demonstrating that SciN is fatty acylated. Overall, these results demonstrate that SciN is a lipoprotein.
FIG. 2.
SciN is a lipoprotein. (A) Sequence alignment using Clustal W of the N terminus of the SciN protein from the EAEC Sci-1 T6SS and homologous genes (COG3521 family) from T6SS of Burkholderia pseudomallei strain K96248 (YP_112106), E. tarda (ABW69084), Aeromonas hydrophila strain ATCC7966 (YP_856372), V. cholerae strain V52 (ZP_01680158), P. aeruginosa strain PAO1 HSI-1 (PA0080; NP_248770) and HSI-2 (PA1666; NP_250357), S. enterica serovar Typhi strain CT18 (NP_454883), and B. cenocepacia strain HI2424 (YP_834112). The putative lipobox motif, L-G/A/SG/A/S-C, is indicated by the blue box with the invariable residues in red. The positions of the signal sequence and the acylated N-terminal cysteine residue (arrow) are indicated. The residue in green corresponds to the +2 residue, which is responsible for the final localization of the lipoprotein in the cell envelope. (B) SciN cofractionates with membranes. A fractionation procedure was applied to EAEC cells producing SciN-HA (lane T), allowing separation between the periplasmic (lane P), cytoplasmic (lane C), and membrane fractions (lane M). Samples were loaded on a 15% acrylamide SDS-PAGE gel and subjected to immunodetection with antibodies directed against the periplasmic MalE, the cytoplasmic EfTu, the membrane TolR proteins, and the HA epitope (from top to bottom panel). Molecular weight markers are indicated on the left. (C) SciN is not processed in the presence of globomycin. A total of 2 × 108 EAEC cells producing the HA-tagged SciN protein treated (+) or not (−) by the SPII inhibitor antibiotic globomycin (globom) were loaded on a 15% acrylamide SDS-PAGE gel and subjected to immunodetection by anti-OmpA, anti-Pal, or anti-HA antibody (from top to bottom panel). The unprocessed species of the Pal lipoprotein and SciN-HA are indicated by asterisks. Molecular weight markers are shown on the left. (D) SciN is labeled by [3H]palmitate. A total of 3 × 109 cells of EAEC producing SciN or SciN-HA were labeled in vivo by [3H]palmitic acid, SDS-Triton X-100 solubilized, and immunoprecipitated using anti-OmpA, anti-Pal, and anti-HA antibodies, as indicated. Samples were loaded on 15% acrylamide SDS-PAGE gels and subjected to autoradiography (upper panel) or immunodetection by corresponding antibodies (lower panels). Molecular weight markers are indicated on the left. α, anti; IgG, immunoglobulin G.
SciN fractionates with the OM.
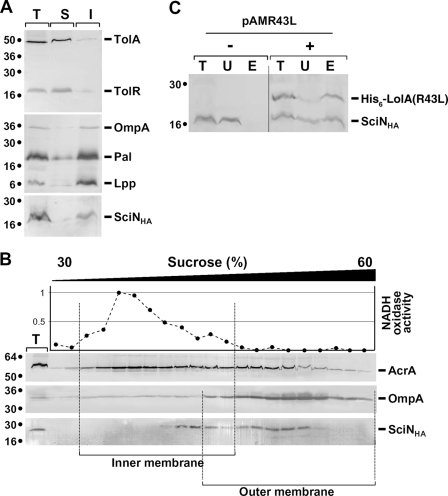
To test whether SciN associates with the IM or OM, we performed (i) selective detergent solubilization using sodium lauroyl sarcosinate (SLS) and (ii) isopycnic sucrose sedimentation gradients. SLS has been previously shown to selectively disrupt the IM (32). Under the conditions used in this study (2% SLS, no Mg2+, and low-ionic-strength buffer), the OM is resistant to solubilization. As shown in Fig. 3A, SciN, as well as the OM OmpA protein and the OM-associated Pal and Lpp lipoproteins, was found in the SLS-insoluble fraction whereas the IM TolR and TolA proteins were extracted with SLS. The OM localization of SciN was then confirmed using sedimentation density gradient centrifugation of membrane fractions, using the OM OmpA and the IM AcrA proteins as controls (Fig. 3B). Because AcrA interacts with the OM TolC proteins and a fraction of AcrA remains associated with the OM (40), we confirmed IM fractions of the gradient using the NADH oxidase enzymatic test (Fig. 3B). Overall, our results clearly demonstrate that SciN associates with the OM in E. coli. However, a portion of SciN is retained in the IM fractions or in fractions with intermediate densities (see Discussion).
FIG. 3.
SciN is an OM protein. (A) SciN is not solubilized by SLS. Total (T) membranes from EAEC cells producing SciN-HA were treated with SLS, and solubilized (S) and insolubilized (I) membranes were separated. Samples from 5 × 108 cells were loaded on 15% acrylamide SDS-PAGE gels and subjected to immunodetection with antibodies directed against TolA, TolR (upper panel), and Pal (which recognizes OmpA and Lpp; middle panel) (11) and against the HA epitope (lower panel). Molecular weight markers are indicated on the left. (B) SciN cofractionates with the OM. Total (T) membranes from EAEC cells producing SciN-HA were separated on a discontinuous sedimentation sucrose gradient. Collected fractions were analyzed for contents using the anti-AcrA, anti-OmpA, and anti-HA antibodies (from top to bottom panel) and with an NADH oxidase activity test (upper graph). NADH oxidase activity is represented relative to the fraction having the highest activity. The positions of the IM- and OM-containing fractions are indicated. (C) SciN coprecipitates with LolA(R43L). Periplasm fractions (lanes T) of EAEC cells producing pSciN-HA and carrying (+) or not (−) plasmid pAMR43L, encoding the His6-LolA(R43L) mutant protein, were subjected to pull-down on cobalt beads. His6-LolA(R43L) was eluted with imidazole. Unbound (U) and eluted (E) materials were loaded on 15% acrylamide SDS-PAGE gels and subjected to immunodetection using the anti-His5 and anti-HA antibodies. Molecular weight markers are indicated on the left.
In contrast to IM lipoproteins, OM lipoproteins are conveyed through the periplasm by the lipoprotein-specific periplasmic carrier LolA protein and are released to the OM LolB protein for assembly (69). The LolA(R43L) mutant has been shown to interact with OM lipoproteins but is unable to release them to LolB (51). As a consequence, OM lipoproteins are sequestered in the periplasm, in complex with LolA(R43L) (51). The periplasm fraction of the lpp strain JE5505 (a strain devoid of the most abundant OM lipoprotein) (68) coproducing His6-LolA(R43L) and SciN-HA was subjected to pull-down experiments using affinity chromatography. Figure 3C shows that SciN-HA was efficiently coprecipitated with His6-LolA(R43L), further adding evidence for SciN OM localization.
SciN is exposed in the periplasm.
To test whether the OM SciN lipoprotein is exposed at the cell surface or protrudes in the periplasm, we performed proteinase K accessibility experiments. Similar to the Pal lipoprotein, SciN-HA was resistant to proteinase K degradation in whole cells but was degraded upon cell permeabilization (Fig. 4). In contrast, the OmpA protein, which possesses surface-exposed loops, was partly degraded to a discrete fragment upon treatment of whole or permeabilized cells with proteinase K. This result suggests that SciN is exposed at the periplasmic side of the OM.
FIG. 4.
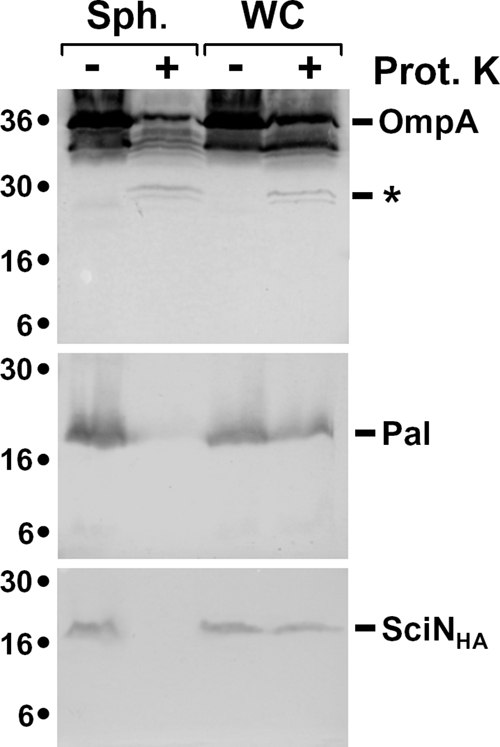
SciN is exposed in the periplasm. Whole cells (WC) or spheroplasts (Sph) of EAEC cells producing SciN-HA were treated (+) or not (−) with proteinase K (Prot K). Samples from 2 × 108 cells were loaded on 15% acrylamide SDS-PAGE gels and subjected to immunodetection with anti-OmpA, anti-Pal, and anti-HA antibodies (from top to bottom panel). The degradation product of OmpA, corresponding of the cleavage of the external loop, is indicated by an asterisk. Molecular weight markers are indicated on the left.
SciN proper localization is required for function.
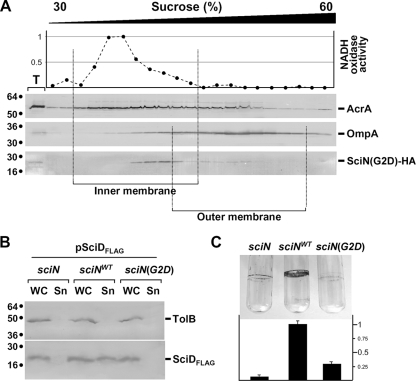
SciN is an OM lipoprotein exposed in the periplasm. To test whether the OM localization of SciN is required for function, we constructed a point mutation targeting the +2 glycine residue, which is responsible for OM localization. E. coli lipoproteins that possess an aspartate residue at position +2 are usually retained in the IM (34, 63, 73). The plasmid producing the Gly2Asp SciN mutant protein, SciN(G2D), was constructed by site-directed mutagenesis using pSciN-HA as a template (see Materials and Methods). Pilot studies showed that SciN(G2D) was solubilized upon SLS treatment (data not shown) and cofractionated with IM fractions in a sedimentation sucrose gradient (Fig. 5A). The SciN(G2D) lipoprotein thus mislocalizes at the IM. We then tested whether this mislocalization alters SciN function. Figure 5B shows that SciD-FLAG was not secreted in sciN cells producing SciN(G2D). Furthermore, sciN cells producing SciN(G2D) formed small amounts of biofilm (Fig. 5C). Overall, the results displayed in Fig. 5 demonstrate that mislocalization of the SciN OM lipoprotein in the IM prevents correct functioning of the EAEC Sci-1 T6SS.
FIG. 5.
OM localization of SciN is required for T6SS function. (A) The SciN(G2D) mutant protein cofractionates with the IM. Total (T) membranes from EAEC cells producing SciN-HA were separated on a discontinuous sedimentation sucrose gradient. Collected fractions were analyzed for contents using anti-AcrA, anti-OmpA, and anti-HA antibodies (from top to bottom panel) and with an NADH oxidase activity test (upper graph). NADH oxidase activity is represented relative to the fraction having the highest activity. The positions of the IM- and OM-containing fractions are indicated. (B) SciD-FLAG secretion was assessed by separating cells (WC) and supernatant (Sn) fractions from WT, and sciN cells complemented with the HA-tagged SciN (sciNWT) or the G2D mutant, sciN(G2D). A total of 2 × 108 cells and the TCA-precipitated material of the supernatant from 5 × 108 cells were loaded on a 15% acrylamide SDS-PAGE gel and subjected to immunodetection using the anti-Flag monoclonal antibody (lower panel) and the anti-TolB polyclonal antibodies (upper panel). (C) Biofilms formed in static cultures of the WT and sciN cells complemented with the HA-tagged SciN (sciNWT) or the G2D mutant, sciN(G2D), were visualized in glass tubes by crystal violet staining (upper panel) and quantified using an ethanol-solubilization procedure, relative to the WT EAEC strain (lower graph).
DISCUSSION
Two gene clusters of the pheU pathogenicity island of EAEC strain 17-2, sci-1 and sci-2, encode subunits of two distinct T6SSs (28). Herein, we showed that the Sci-1 T6S machine is responsible for the specific secretion of the SciD protein, an Hcp-like protein. SciD is released from EAEC cells but not from E. coli K-12 DH5α cells. Further experiments demonstrated that at least two genes of the sci-1 gene cluster are required for secretion of SciD: sciG, which encodes a putative AAA+ ATPase of the ClpV family, and sciN. The homologues of these genes have been found to be necessary for the secretion of the Hcp-like EvpC and the VgrG-like EvpI proteins in E. tarda in a systematic deletion approach (75). As expected, the sci-1 gene cluster is not required for the secretion of the Hcp-like AaiC protein encoded within the second T6SS gene cluster, sci-2. Further experiments showed that in the absence of the sci-2 gene cluster, the sci-1 cannot substitute for secretion of AaiC (M. S. Aschtgen and E. Cascales, unpublished observations), suggesting a tightly regulated mechanism of substrate specificity between the two T6S machines encoded within the EAEC pheU pathogenicity island. It is noteworthy that, under our experimental conditions, only a portion of the produced SciD-FLAG was found in the culture supernatant of WT EAEC cells, suggesting either that the expression level of the plasmid-borne sciD is far more than the T6S machine can secrete or that the C-terminal FLAG epitope somehow alters the secretion mechanism.
Our results also showed that the sci-1 gene cluster is involved in biofilm formation. T6SSs have been shown to be involved in numerous processes linked to the virulence of bacterial pathogens, including cytotoxicity, invasion, intracellular growth, survival, and persistence within the host (10). Our results demonstrating a probable role of the Sci-1 T6S machine for adherence on abiotic surfaces, although preliminary, do not constitute a novel phenotype of T6SS mutations. Mutations in the T6S gene cluster have been found in a screen to identify genes of V. parahaemolyticus involved in biofilm formation (29). Furthermore, proteome studies showed that the Pseudomonas aeruginosa Hcp-like proteins PA0085 and PA0263 and the HSI-1 ClpV-type ATPase PA0090 are produced more abundantly during biofilm formation (58, 65). However, whether the SciN lipoprotein only, the whole T6SS apparatus, or secreted effectors are necessary for biofilm formation remains to be answered.
One of the genes present on all T6S gene clusters encodes a probable lipoprotein. These conserved proteins share a characteristic N-terminal sequence, called the lipobox (69). This signal sequence is specifically processed by LspA, the SPII, releasing an N-terminal cysteine residue which is then acylated to give the mature lipoprotein. In this study, using the EAEC sci-1 gene cluster as a model, we showed experimentally that sciN encodes an acylated protein processed by SPII. We concluded that SciN is a lipoprotein. Because all SciN homologues share the characteristic features of lipoproteins, one may suggest that these proteins are all lipoproteins. After processing, E. coli lipoproteins are distributed to the IMs or OMs, depending of the nature of the amino acid immediately following the acylated cysteine residue (73). Lipoproteins with an aspartate at position +2 are sequestered in the IM, whereas lipoproteins carrying any other amino acid at this position are conveyed to the OM by the LolA protein and inserted through the action of the OM LolB lipoprotein (69). Using sedimentation sucrose gradients and selective solubilization procedures, we demonstrated that SciN is a lipoprotein anchored to the OM. Further indirect evidence has been provided by the coprecipitation of SciN with a His6-tagged LolA protein carrying the R43L mutation, which prevents release of OM lipoproteins from LolA to LolB. Sucrose density gradient analysis also showed that a portion of SciN colocalized with IM fractions or fractions with intermediate densities. The aberrant localization in intermediate density fractions was also observed for the SciN(G2D) protein and has been previously shown for proteins involved in formation of trans-envelope complexes, such as the Tol-Pal system (47). One may suggest that SciN is retained in these fractions through contacts with T6SS components from both membranes. As an argument for this hypothesis, SciN is found only in OM fractions upon production in the E. coli K-12 DH5α strain (data not shown), suggesting that SciN may interact with an IM T6SS component in EAEC. Using a yeast two-hybrid assay, Zheng and Leung showed that the SciN homologue of the E. tarda Evp T6SS, EvpL, interacts with the IcmF-like IM component EvpO (75). Further experiments demonstrated that mislocalization of the SciN lipoprotein at the IM through modification of its glycine +2 residue to an aspartate led to a defect in SciD secretion and the capacity of the strain to produce WT levels of biofilm.
Because homologues of sciN exist in all T6S gene clusters and because of the critical role of this gene in T6SS function, one may suggest that T6SS-linked lipoproteins play an important role in machine assembly or substrate secretion. Lipoproteins have been identified in all secretion systems having a large number of subunits, including T2SS, T3SS, and T4SS (1, 20, 24, 30, 61, 64). In all these secretion systems, lipoproteins are essential components and have been shown to be involved in machine assembly or stabilization of the secretion apparatus. In the prototypic Klebsiella oxytoca Pul T2SS, the OM PulS lipoprotein has been shown to be necessary for proper localization of the homomultimeric OM secretin PulD (37, 38). Further experiments suggested that PulS may convey PulD through the periplasm to the OM, and it has therefore been called “pilotin” (36). In the S. enterica serovar Typhimurium and Yersinia enterocolitica T3SS, the OM lipoproteins InvH and YscW are required for proper localization and stabilization of the OM secretin InvG and YscC, respectively (9, 19, 20). In the Agrobacterium tumefaciens T4SS, the OM VirB7 lipoprotein forms an intermolecular disulfide bridge with the C-terminal domain of the VirB9 protein (2, 6, 66). Whether VirB9 is an OM component has yet to be determined, but this has been suggest by the observation of numerous β-strand transmembrane segments in its central domain (17, 42) and by immunolocalization studies (7). Whether VirB7 is necessary for VirB9 localization is not known, but it has been shown that VirB7 stabilizes most VirB proteins and may thus participate in the early stages of T4S machine morphogenesis, perhaps as a nucleation factor (13, 31). Other examples of the stabilization or assembly of transport apparatus have been demonstrated for several OM lipoproteins including the PilP lipoprotein of the gonococcal type IV pilus (5, 26), the CsgG lipoprotein of the curli apparatus (48), the FlgH lipoprotein of the OM ring of the flagellar basal body (60), or the Rhizobium leguminosarum PssN and P. aeruginosa PelC lipoproteins in the exopolysaccharide export systems (50, 71). The function of the T6SS-associated lipoproteins in the morphogenesis or stabilization of the apparatus has yet to be determined. One additional common feature of secretion system-associated lipoproteins is the interaction with OM components. Bioinformatics analysis of the proteins encoded within the sci-1 gene cluster does not give any clue about putative OM proteins. However, an OM component has been identified recently in the Francisella tularensis T6S gene cluster (49). One alternative may be that the SciN lipoprotein is anchored to the OM through a lipid moiety and is also an acylated OM integral protein, as is the Wza lipoprotein required for group 1 capsular polysaccharide translocation (25, 27). Our results from proteinase K accessibility cannot discriminate between these possibilities since we detected the SciN lipoprotein through a C-terminal epitope tag, which may be degraded by the protease, leaving the integral part of the protein integrated in the OM. Studies aiming at defining whether SciN is a periplasmic OM anchored lipoprotein and whether it associates with any OM partner are currently under way in our laboratory. Future studies of the T6S protein characteristics should provide important information to understand how T6SSs assemble and function.
Acknowledgments
We thank Arlette Darfeuille-Michaud for providing the EAEC strain 17-2, Danièle Cavard for the gift of globomycin, Hajime Tokuda for plasmid pAMR43L, Laure Journet and Denis Duché for critical reading of the manuscript, Aurélie Barnéoud-Arnoulet and members of the Lloubès, Bouveret, and Sturgis groups for fruitful discussions and encouragement, Steve Garvis for testing virulence on C. elegans, and Véronique Viarre and Romé Voulhoux for assistance with sucrose density gradients and discussions. We thank the Sanger Institute for releasing the sequence of strain EAEC 042 prior to publication.
This work is supported by the Centre National de la Recherche Scientifique.
Footnotes
Published ahead of print on 19 September 2008.
REFERENCES
- 1.Allaoui, A., P. J. Sansonetti, and C. Parsot. 1992. MxiJ, a lipoprotein involved in secretion of Shigella Ipa invasins, is homologous to YscJ, a secretion factor of the Yersinia Yop proteins. J. Bacteriol. 1747661-7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, L. B., A. V. Hertzel, and A. Das. 1996. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc. Natl. Acad. Sci. USA 938889-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anyanful, A., J. M. Dolan-Livengood, T. Lewis, S. Sheth, M. N. Dezalia, M. A. Sherman, L. V. Kalman, G. M. Benian, and D. Kalman. 2005. Paralysis and killing of Caenorhabditis elegans by enteropathogenic Escherichia coli requires the bacterial tryptophanase gene. Mol. Microbiol. 57988-1007. [DOI] [PubMed] [Google Scholar]
- 4.Aubert, D., R. Flannagan, and M. A. Valvano. 2008. A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia. Infect. Immun. 761979-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balasingham, S. V., R. F. Collins, R. Assalkhou, H. Homberset, S. A. Frye, J. P. Derrick, and T. Tønjum. 2007. Interactions between the lipoprotein PilP and the secretin PilQ in Neisseria meningitidis. J. Bacteriol. 1895716-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baron, C., Y. R. Thorstenson, and P. C. Zambryski.1997. The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J. Bacteriol. 1791211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayliss, R., R. Harris, L. Coutte, A. Monier, R. Fronzes, P. J. Christie, P. C. Driscoll, and G. Waksman. 2007. NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system. Proc. Natl. Acad. Sci. USA 1041673-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bingle, L., C. Bailey, and M. J. Pallen. 2008. Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 113-8. [DOI] [PubMed] [Google Scholar]
- 9.Burghout, P., F. Beckers, E. de Wit, R. van Boxtel, G. R. Cornelis, J. Tommassen, and M. Koster. 2004. Role of the pilot protein YscW in the biogenesis of the YscC secretin in Yersinia enterocolitica. J. Bacteriol. 1865366-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cascales, E. 2008. The Type VI secretion tool kit. EMBO Rep. 9735-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascales, E., A. Bernadac, M. Gavioli, J. C. Lazzaroni, and R. Lloubès. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cascales, E., and P. J. Christie. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 3041170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cascales, E., and P. J. Christie. 2004. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc. Natl. Acad. Sci. USA 10117228-17233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cascales, E., M. Gavioli, J. N. Sturgis, and R. Lloubès. 2000. Proton motive force drives the interaction of the inner membrane TolA and outer membrane Pal proteins in Escherichia coli. Mol. Microbiol. 3804-15. [DOI] [PubMed] [Google Scholar]
- 16.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Claret, L., S. Miquel, N. Vieille, D. A. Ryjenkov, M. Gomelsky, and A. Darfeuille-Michaud. 2007. The flagellar sigma factor FliA regulates adhesion and invasion of Crohn disease-associated Escherichia coli via a cyclic dimeric GMP-dependent pathway. J. Biol. Chem. 28233275-33283. [DOI] [PubMed] [Google Scholar]
- 19.Crago, A. M., and V. Koronakis. 1998. Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol. Microbiol. 3047-56. [DOI] [PubMed] [Google Scholar]
- 20.Daefler, S., and M. Russel. 1998. The Salmonella typhimurium InvH protein is an outer membrane lipoprotein required for the proper localization of InvG. Mol. Microbiol. 281367-1380. [DOI] [PubMed] [Google Scholar]
- 21.Das, S., and K. Chaudhuri. 2003. Identification of a unique IAHP (IcmF associated homologous proteins) cluster in V. cholerae and other proteobacteria through in silico analysis. In Silico Biol. 3287-300. [PubMed] [Google Scholar]
- 22.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delepelaire, P. 2004. Type I secretion in gram-negative bacteria. Biochim. Biophys. Acta 1694149-161. [DOI] [PubMed] [Google Scholar]
- 24.D'Enfert, C., and A. P. Pugsley. 1989. Klebsiella pneumoniae pulS gene encodes an outer membrane lipoprotein required for pullulanase secretion. J. Bacteriol. 1713673-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong, C., K. Beis, J. Nesper, A. L. Brunkan-Lamontagne, B. R. Clarke, C. Whitfield, and J. H. Naismith. 2006. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature 444226-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drake, S. L., S. A. Sandstedt, and M. Koomey. 1997. PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular-mass multimer. Mol. Microbiol. 23657-668. [DOI] [PubMed] [Google Scholar]
- 27.Drummelsmith, J., and C. Whitfield. 2000. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO J. 1957-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dudley, E. G., N. Thomson, J. Parkhill, N. Morin, and J. P. Nataro. 2006. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol. Microbiol. 611267-1282. [DOI] [PubMed] [Google Scholar]
- 29.Enos-Berlage, J., Z. Guvener, C. Keenan, and L. L. McCarter. 2005. Genetic determinants of biofilm development of opaque and translucent V. parahaemolyticus. Mol. Microbiol. 551160-1182. [DOI] [PubMed] [Google Scholar]
- 30.Fernandez, D., T. A. T. Dang, G. M. Spudich, X. R. Zhou, B. B. Berger, and P. J. Christie. 1996. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J. Bacteriol. 1783156-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez, D., G. M. Spudich, X. R. Zhou, and P. J. Christie. 1996. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J. Bacteriol. 1783168-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Filip, C., G. Fletcher, J. L. Wulff, and C. F. Earhart. 1973. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl-sarcosinate. J. Bacteriol. 115717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filloux, A., A. Hachani, and S. Bleves. 2008. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 1541570-1583. [DOI] [PubMed] [Google Scholar]
- 34.Gennity, J. M., and M. Inouye. 1991. The protein sequence responsible for lipoprotein membrane localization in Escherichia coli exhibits remarkable specificity. J. Biol. Chem. 26616458-16464. [PubMed] [Google Scholar]
- 35.Gerlach, R. G., and M. Hensel. 2007. Protein secretion systems and adhesins: the molecular armory of gram-negative pathogens. Int. J. Med. Microbiol. 297401-415. [DOI] [PubMed] [Google Scholar]
- 36.Guilvout, I., M. Chami, A. Engel, A. P. Pugsley, and N. Bayan. 2006. Bacterial outer membrane secretin PulD assembles and inserts into the inner membrane in the absence of its pilotin. EMBO J. 255241-5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardie, K. R., S. Lory, and A. P. Pugsley. 1996. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15978-988. [PMC free article] [PubMed] [Google Scholar]
- 38.Hardie, K. R., A. Seydel, I. Guilvout, and A. P. Pugsley. 1996. The secretin-specific, chaperone-like protein of the general secretory pathway: separation of proteolytic protection and piloting functions. Mol. Microbiol. 22967-976. [DOI] [PubMed] [Google Scholar]
- 39.Harrington, S. M., E. G. Dudley, and J. P. Nataro. 2006. Pathogenesis of enteroaggregative Escherichia coli infection. FEMS Microbiol. Lett. 25412-18. [DOI] [PubMed] [Google Scholar]
- 40.Husain, F., M. Humbard, and R. Misra. 2004. Interaction between the TolC and AcrA proteins of a multidrug efflux system of Escherichia coli. J. Bacteriol. 1868533-8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inukai, M., M. Takeuchi, K. Shimizu, and M. Arai. 1978. Mechanism of action of globomycin. J. Antibiot. (Tokyo) 311203-1205. [DOI] [PubMed] [Google Scholar]
- 42.Jakubowski, S. J., E. Cascales, V. Krishnamoorthy, and P. J. Christie. 2005. Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J. Bacteriol. 1873486-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson, T. L., J. Abendroth, W. G. Hol, and M. Sandkvist. 2006. Type II secretion: from structure to function. FEMS Microbiol. Lett. 255175-186. [DOI] [PubMed] [Google Scholar]
- 44.Journet, L., K. T. Hughes, and G. R. Cornelis. 2005. Type III secretion: a secretory pathway serving both motility and virulence. Mol. Membr. Biol. 2241-50. [DOI] [PubMed] [Google Scholar]
- 45.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2123-140. [DOI] [PubMed] [Google Scholar]
- 46.Liu, H., S. J. Coulthurst, L. Pritchard, P. E. Hedley, M. Ravensdale, S. Humphris, T. Burr, G. Takle, M. B. Brurberg, P. R. Birch, G. P. Salmond, and I. K. Toth. 2008. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 4e1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lloubès, R., E. Cascales, A. Walburger, E. Bouveret, C. Lazdunski, A. Bernadac, and L. Journet. 2001. The Tol-Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol. 152523-529. [DOI] [PubMed] [Google Scholar]
- 48.Loferer, H., M. Hammar, and S. Normark. 1997. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol. Microbiol. 2611-23. [DOI] [PubMed] [Google Scholar]
- 49.Ludu, J. S., O. M. de Bruin, B. N. Duplantis, C. L. Schmerk, A. Y. Chou, K. L. Elkins, and F. E. Nano. 2008. The Francisella pathogenicity island protein PdpD is required for full virulence and associates with homologues of the type VI secretion system. J. Bacteriol. 1904584-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marczak, M., A. Mazur, J. E. Król, W. I. Gruszecki, and A. Skorupska. 2006. Lipoprotein PssN of Rhizobium leguminosarum bv. trifolii: subcellular localization and possible involvement in exopolysaccharide export. J. Bacteriol. 1886943-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyamoto, A., S. I. Matsuyama, and H. Tokuda. 2001. Mutant of LolA, a lipoprotein-specific molecular chaperone of Escherichia coli, defective in the transfer of lipoproteins to LolB. Biochem. Biophys. Res. Commun. 2871125-1128. [DOI] [PubMed] [Google Scholar]
- 52.Mougous, J., M. Cuff, S. Raunser, A. Shen, M. Zhou, C. Gifford, A. Goodman, G. Joachimiak, C. Ordoñez, S. Lory, T. Walz, A. Joachimiak, and J. J. Mekalanos. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 3121526-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nataro, J. P. 2005. Enteroaggregative Escherichia coli pathogenesis. Curr. Opin. Gastroenterol. 214-8. [PubMed] [Google Scholar]
- 54.Parsons, D., and F. Heffron. 2005. sciS, an icmF homolog in Salmonella enterica serovar Typhimurium, limits intracellular replication and decreases virulence. Infect. Immun. 734338-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pukatzki, S., A. Ma, A. Revel, D. Sturtevant, and J. J. Mekalanos. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. USA 10415508-15513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pukatzki, S., A. Ma, D. Sturtevant, B. Krastins, D. Sarracino, W. Nelson, J. Heidelberg, and J. J. Mekalanos. 2006. Identification of a conserved bacterial protein secretion system in V. cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 1031528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rao, P., Y. Yamada, Y. Tan, and K. Y. Leung. 2004. Use of proteomics to identify novel virulence determinants that are required for E. tarda pathogenesis. Mol. Microbiol. 53573-586. [DOI] [PubMed] [Google Scholar]
- 58.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 1841140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schlieker, C., H. Zentgraf, P. Dersch, and A. Mogk. 2005. ClpV, a unique Hsp100/Clp member of pathogenic proteobacteria. Biol. Chem. 3861115-1127. [DOI] [PubMed] [Google Scholar]
- 60.Schoenhals, G. J., and R. M. Macnab. 1996. Physiological and biochemical analyses of FlgH, a lipoprotein forming the outer membrane L ring of the flagellar basal body of Salmonella typhimurium. J. Bacteriol. 1784200-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schuch, R., and A. T. Maurelli. 2001. MxiM and MxiJ, base elements of the Mxi-Spa type III secretion system of Shigella, interact with and stabilize the MxiD secretin in the cell envelope. J. Bacteriol. 1836991-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sexton, J. A., J. L. Miller, A. Yoneda, T. E. Kehl-Fie, and J. P. Vogel. 2004. Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect. Immun. 725983-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seydel, A., P. Gounon, and A. P. Pugsley. 1999. Testing the “+2 rule” for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol. Microbiol. 34810-821. [DOI] [PubMed] [Google Scholar]
- 64.Shevchik, V. E., and G. Condemine. 1998. Functional characterization of the Erwinia chrysanthemi OutS protein, an element of a type II secretion system. Microbiology 1443219-3228. [DOI] [PubMed] [Google Scholar]
- 65.Southey-Pillig, C., D. Davies, and K. Sauer. 2005. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 1878114-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spudich, G. M., D. Fernandez, X. R. Zhou, and P. J. Christie. 1996. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc. Natl. Acad. Sci. USA 937512-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suarez, G., J. Sierra, J. Sha, S. Wang, T. Erova, A. Fadl, S. Foltz, A. Horneman, and A. Chopra. 2008. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44344-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki, H., Y. Nishimura, S. Yasuda, A. Nishimura, M. Yamada, and Y. Hirota. 1978. Murein-lipoprotein of Escherichia coli: a protein involved in the stabilization of bacterial cell envelope. Mol. Gen. Genet. 1671-9. [DOI] [PubMed] [Google Scholar]
- 69.Tokuda, H., and S. Matsuyama. 2004. Sorting of lipoproteins to the outer membrane in E. coli. Biochim. Biophys. Acta 16935-13. [DOI] [PubMed] [Google Scholar]
- 70.van den Ent, F., and J. Löwe. 2006. RF cloning: a restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods 6767-74. [DOI] [PubMed] [Google Scholar]
- 71.Vasseur, P., C. Soscia, R. Voulhoux, and A. Filloux. 2007. PelC is a Pseudomonas aeruginosa outer membrane lipoprotein of the OMA family of proteins involved in exopolysaccharide transport. Biochimie 89903-915. [DOI] [PubMed] [Google Scholar]
- 72.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100189-194. [DOI] [PubMed] [Google Scholar]
- 73.Yamaguchi, K., F. Yu, and M. Inouye. 1988. A single amino acid determinant of the membrane localization of lipoproteins in Escherichia coli. Cell 53423-432. [DOI] [PubMed] [Google Scholar]
- 74.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 75.Zheng, J., and K. Y. Leung. 2007. Dissection of a type VI secretion system in E. tarda. Mol. Microbiol. 661192-1206. [DOI] [PubMed] [Google Scholar]