Abstract
Upon prolonged exposure to cholate and other toxic compounds, Lactococcus lactis develops a multidrug resistance phenotype that has been attributed to an elevated expression of the heterodimeric ABC-type multidrug transporter LmrCD. To investigate the molecular basis of bile acid resistance in L. lactis and to evaluate the contribution of efflux-based mechanisms in this process, the drug-sensitive L. lactis NZ9000 ΔlmrCD strain was challenged with cholate. A resistant strain was obtained that, compared to the parental strain, showed (i) significantly improved resistance toward several bile acids but not to drugs, (ii) morphological changes, and (iii) an altered susceptibility to antimicrobial peptides. Transcriptome and transport analyses suggest that the acquired resistance is unrelated to elevated transport activity but, instead, results from a multitude of stress responses, changes to the cell envelope, and metabolic changes. In contrast, wild-type cells induce the expression of lmrCD upon exposure to cholate, whereupon the cholate is actively extruded from the cells. Together, these data suggest a central role for an efflux-based mechanism in bile acid resistance and implicate LmrCD as the main system responsible in L. lactis.
Multidrug transporters are responsible for active efflux of structurally and functionally unrelated drugs and are ubiquitous in nature (21, 32). The majority of the described multidrug transporters in bacteria depend on ion motive forces for extrusion activity, while only few systems use ATP hydrolysis to drive efflux. However, recent reports and functional genomic predictions suggest that the role of these bacterial ABC-type transporters in multidrug resistance (MDR) might be underestimated (32).
Lactococcus lactis is a nonpathogenic lactic acid bacterium that serves as a model organism to study bacterial MDR (31). The genome of L. lactis contains 40 putative drug transporter genes of which several have been characterized in detail. These are LmrP (8), a member of the major facilitator superfamily (MFS) transporter, and the ABC-type transporters LmrA (48) and LmrCD (31). L. lactis MG1363 readily acquires a stable MDR phenotype when challenged with increasing concentrations of cholate (51) or other drugs (7). An ATP-dependent transporter has been implicated in cholate resistance, but its identity has remained obscure (51). Recent transcriptome analysis revealed that a common response of the selected MDR strains is the elevated expression of lmrCD compared to the wild-type strain (31). Furthermore, the deletion of the lmrCD genes makes L. lactis more susceptible to a wide range of toxic compounds, while the drug resistance phenotype can be restored by the overexpression of lmrCD from a plasmid (33).
Although previous studies implicated LmrCD as a major determinant of drug resistance in L. lactis, it has remained unclear whether other putative MDR transporters contribute to resistance in the selected strains as the observed phenotype covers both anionic and cationic drugs such as daunomycin, rhodamine, and ethidium bromide. Moreover, apart from the lmrCD upregulation, the cholate-resistant L. lactis strain (51) showed a different gene expression profile from the strains selected for resistance against cationic drugs (31). The cholate-resistant strain also showed strong upregulation of the arc operon genes involved in the arginine deiminase pathway and of a gene encoding a putative MDR transporter of the MFS, i.e., llmg2513 (formerly named yxbD).
Cholate resistance is of particular interest as lactic acid bacteria like Lactobacillus are important constituents of the human intestinal microflora and are widely used as probiotics in food supplements (22, 42, 50). Probiotic survival depends on resistance against inhibitory compounds present in the intestine such as bile acids (29). These compounds are synthesized from cholesterol in the liver and have an important role in the intestine, where they are involved in the absorption of dietary fats and lipid soluble vitamins (43). Bile acids are released as conjugates of glycine or taurine in the intestine, where indigenous microbes such as Bifidobacteria employ bile acid hydrolases that enzymatically liberate the free bile acids (FBA) from their conjugated forms (45). FBA are toxic, weakly acidic, and hydrophobic molecules that strongly inhibit the growth of various intestinal bacteria (6). Cholate, which is one of the most frequently encountered FBA in the gastrointestinal tract, is produced by the gut microflora from conjugated bile salts, such as taurocholate and glycocholate (29). The exact mechanism of inhibition is still not clear but may involve permeabilization of the cytoplasmic membrane (28). The autochthonous microbiota as well as many gram-negative pathogens have evolved to survive the bactericidal effects of FBA (49). The mechanisms underlying bile acid resistance have been attributed to diverse physiological and cellular responses such as changes in cell envelope (11) and increased activity of bile acid modifying enzymes in many gram-positive bacteria (4) while MDR efflux pumps have been implicated in gram-negative bacteria (35). In contrast, little is known about the role of MDR efflux pumps in bile acid extrusion in gram-positive organisms.
Here, we have investigated the adaptive response of L. lactis cells that lack the major MDR transporter, LmrCD, when challenged with cholate. The data suggest that in the absence of LmrCD, cholate resistance relies on a multitude of transport-unrelated cellular responses. This investigation also establishes for the first time that LmrCD-mediated extrusion of cholate is the primary mechanism of bile acid resistance in L. lactis.
MATERIALS AND METHODS
Bacterial strains, medium, and growth conditions.
L. lactis NZ9000 is a derivative of the plasmid-free L. lactis MG1363 strain containing pepN::nisRK (13, 14) and is referred to as the wild-type strain. The derivative L. lactis NZ9000 ΔlmrCD strain lacks the MDR transporter LmrCD (31). In all cases, cells were grown at 30°C in M17 medium (Difco) containing 0.5% (wt/vol) glucose (GM17).
Cholate adaptation assay.
Cholate-resistant cells were selected by growth of the L. lactis NZ9000 ΔlmrCD strain in GM17 medium containing increasing concentrations of cholate. Exponentially growing cells were diluted 1:100 in 5 ml of fresh GM17 containing cholate and grown overnight. This procedure was repeated several times with a concomitant stepwise increase of the cholate concentration until a significant increase in MIC had occurred. The final concentration of cholate used was 4 mM. To obtain single colonies, the adapted culture was spread-plated on GM17 medium with 1.8% (wt/vol) agar containing 3 mM cholate. Thirty colonies were selected and subcultured in fresh GM17 broth without cholate. Cultures were supplemented with glycerol at a final concentration of 10% (vol/vol) and stored at −80°C. Growth analysis of the individual colonies indicated essentially identical 50% inhibitory concentrations (IC50s) and MICs for cholate. One of the obtained cholate-adapted NZ9000 ΔlmrCD colonies was selected and used for further characterization and is referred to as the ΔLmrCDr strain.
Growth studies.
Overnight cultures of L. lactis wild-type, ΔlmrCD and ΔLmrCDr strains were diluted in fresh GM17 medium and grown to an optical density at 600 nm (OD660) of 0.6. In the case of strains bearing a plasmid, the GM17 medium was supplemented with chloramphenicol at a final concentration of 5 μg/ml. Cells were then diluted to an OD660 of 0.05, and aliquots of 150 μl were transferred to 96-well microtiter plates that contained 50 μl of GM17 medium containing a range of drugs at various concentrations. Sterile silicon oil (50 μl) was pipetted on top of the samples to prevent evaporation. Growth was monitored at 30°C every 6 min for 8 to 12 h at 660 nm using a multiscan photometer (spectraMax 340; Molecular Devices). The maximum specific growth rate μ was calculated from the exponential growth phase (52) and plotted against the concentration of the different drugs. Concentrations that inhibited growth by 50% (IC50s) and 100% (MIC) were determined. The experiments were carried out in triplicate, and the data shown are averaged to obtain the standard error of the mean.
Cholate transport assay.
Exponentially growing cells were harvested at an OD660 of ∼1, washed once with 50 mM potassium phosphate (KPi), pH 7.0, containing 1 mM MgSO4, and resuspended in this buffer to an OD660 of ∼20. The cells were de-energized by incubation with 10 mM 2-deoxyglucose for 30 min at 30°C, washed three times with KPi buffer, and finally resuspended in this buffer to an OD660 of ∼8. Aliquots (3 ml) of the cell suspension were dispensed in glass tubes and preincubated for 5 min at 30°C with gentle stirring. Next, 8 μl of 1.82 mM [14C]cholate (55 mCi/mmol) was mixed with 4 μl of 500 mM nonradioactive cholate in 1 ml of MilliQ water, and 150 μl of the mix was added to the cells (final cholate concentration is ∼100 μM), followed by incubation for 14 min after which glucose (230 mM) was added as a source of metabolic energy. At the time points indicated in the figures, the amount of [14C]cholate associated with cells was determined by a filtration method. Herein, aliquots of 200 μl were passed over 0.2-μm-pore-size cellulose-acetate filters that were prewetted in 100 mM KPi, pH 7.0. Retained cells were then washed two times with 2 ml of 100 mM LiCl. Finally, the radioactivity associated with the cells on the filter was measured by liquid scintillation counting. Values were corrected for the background level of radioactivity obtained for control incubations without cells. The amount of accumulated cholate was related to the quantity of cells. Due to morphological differences (see results) the cellular dry weight rather than the OD660 was used. A similar experiment was performed using an L. lactis NZ9000 ΔlmrCD strain carrying the pILlmrCD plasmid (lmrCD expression under the control of native promoter) and control plasmid pIL252 (31).
Scanning electron microscopy.
A drop of a bacterial culture was placed on a freshly cleaved mica surface pretreated with a 0.5% poly-l-lysine solution. Next, cells were washed with 0.1 M cacodylate buffer, fixed with 2% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for 30 min, and washed with the cacodylate buffer. The specimen was dehydrated with ethanol in a sequence of 30, 50, 70, and 100% (10 min each), followed by two times in 100% for 30 min. Finally, the specimen was dried in a Bal-Tec Critical Point Dryer with CO2 and sputter-coated with 2 to 3 nm of Au/Pd (Bal-Tec sputter coater). Observation was done with a Jeol FE-SEM 6301F (cold-field emission scanning electron microscope).
Expression analysis by RT-PCR.
Overnight cultures of L. lactis NZ9000 were diluted in GM17 medium to an OD660 of 0.05 and grown at 30°C until they reached an OD660 of ∼0.6. Cholate (1 mM) was added to the cultures, and at various time points, aliquots were removed, and cells were harvested by centrifugation (3,500 rpm at 4°C for 5 min). Total RNA was isolated using TRIzol reagent (Invitrogen). The RNA concentration was determined, and equal amounts of RNA were transferred into Illustra Ready To-Go reverse transcription-PCR (RT-PCR) tubes to generate cDNA from the RNA templates. The first synthesis of a cDNA strand was performed at 42°C for 40 min, followed by standard PCR conditions. RT-PCR products of the lmrC, lmrD, and secY genes were obtained with gene-specific primer pairs (Table 1), and samples were analyzed on a 2% (wt/vol) agarose gel.
TABLE 1.
Oligonucleotide primers used for RT-PCR analysis
| Primer namea | Primer sequence (5′→ 3′) |
|---|---|
| lmrC RT-PCR FW | GTTGAAGAACGTGGGAATAATTTCTCAGGTGG |
| lmrC RT-PCR RV | CCTCCTGTGCTTTCTGTGTATCGTAGATTTC |
| lmrD RT-PCR FW | CGTTTCTGATGATGAATCAGTCTTCTCAGTTGG |
| lmrD RT-PCR RV | CAAAAACGAATTGATTATGATAAAGTTCAGAG |
| secY RT-PCR FW | TACAACTGCTCCAGCTACGA |
| secY RT-PCR RV | GTTCCTCCAAGAGCGACAAT |
FW, forward; RV, reverse.
DNA microarray analysis.
DNA microarray experiments were essentially performed as described previously (14, 47). In brief, RNA was isolated from two replicate cultures of both L. lactis ΔlmrCD and cholate-adapted L. lactis ΔLmrCDr. Cultures were grown at 30°C in GM17 medium in the absence of cholate, and cells were harvested at an OD660 of ∼1. Next, single-strand RT (amplification) and indirect labeling of 20 μg of total RNA, with either Cy3 or Cy5 dye, were performed (including samples in which the dyes were swapped to correct for dye-specific effects). Labeled cDNA samples were hybridized to microarray slides containing probes representing 2,496 open reading frames (ORFs) of L. lactis MG1363 spotted in duplicate. After overnight hybridization, slides were washed for 5 min at 37°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.5% sodium dodecyl sulfate followed by 5 min in 1× SSC containing 0.25% sodium dodecyl sulfate to remove nonspecifically hybridized cDNAs. Slides were scanned using a GenePix 4200AL instrument (Westburg). Subsequently, individual spot intensities were determined using ArrayPro, version 4.5 (Media Cybernetics Inc., Silver Spring, MD). Slide data were processed and normalized using MicroPrep, which yielded average gene expression ratios of the mutant to the control strain. Expression of a gene was considered to be significantly altered at a Cyber-T Bayesian P value of ≤0.001. From the set of genes that exhibited a significant change in expression, only genes that exhibited a strong, i.e., ≥1.8-fold, change in expression are discussed.
Microarray data accession number.
The array data reported in this publication have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE10203.
RESULTS
L. lactis can acquire resistance to cholate in the absence of LmrCD.
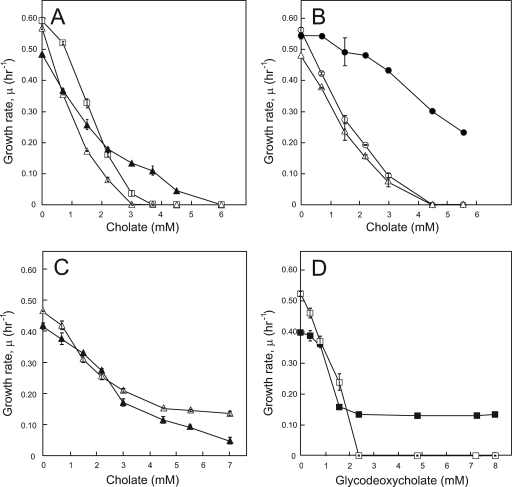
Previously, we have demonstrated that L. lactis cells lacking the genes encoding the heterodimeric ABC-type MDR transporter LmrCD become about twofold more susceptible to cholate than wild-type cells (31). Transcriptome analysis of the MDR strain of L. lactis selected for cholate resistance showed, among other things, a marked induction of lmrCD expression (31) that could be related to inactivation of the transcriptional regulator LmrR (1). These data suggest an important role of LmrCD in resistance to cholate but do not exclude an involvement of other transporters or non-efflux-based mechanisms in cholate resistance. Therefore, the cholate-sensitive L. lactis ΔlmrCD strain was repeatedly exposed to stepwise increasing sublethal concentrations of cholate to induce resistance. In this manner a strain with significantly improved resistance to cholate was obtained (ΔLmrCDr) (Fig. 1A and Table 2). The observed phenotype was found to be stable as the ΔLmrCDr strain remained resistant after growth for 7 days in the absence of cholate (data not shown). Cholate-adapted ΔLmrCDrcells were found to be 1.7-fold more resistant to cholate than the parental ΔlmrCD strain, as judged from the MICs (1.7 and 1 mM, respectively). However, in the absence of cholate, the ΔLmrCDr cells showed a lower growth rate than parental and wild-type L. lactis cells. For reasons that remain unclear, the IC50 of cholate obtained for the ΔlmrCD strain was 2.5-fold lower than that reported previously (1 and 2.5 mM, respectively) (25). However, these IC50s were found consistently in at least six independent experiments, while the relative resistance levels between the wild-type and ΔlmrCD strains obtained in both studies were very similar. Plasmid-based overexpression of lmrCD in the ΔlmrCD strain increased cholate resistance up to approximately fourfold (Fig. 1B), which greatly exceeded the cholate resistance level of both the ΔLmrCDr and the wild-type strain (Fig. 1A and B). Overall, these data demonstrate that LmrCD provides resistance to cholate.
FIG. 1.
Bile acid resistance of L. lactis cells harboring or lacking the lmrCD genes. (A) Cholate resistance of L. lactis wild-type (□), ΔlmrCD (▵), and ΔLmrCDr (▴) strains. (B) Cholate resistance of L. lactis ΔlmrCD (○) and ΔlmrCD containing plasmid pILlmrCD (•) or control plasmid pIL252 (▵). (C) Cholate resistance of cholate-adapted ΔLmrCDr cells containing pILlmrCD (▵) or pIL252 (▴). (D) Glycodeoxycholate resistance of ΔLmrCDr cells containing pILlmrCD (▪) or pIL252 (□). Note that the plasmid-encoded lmrCD genes are under the control of their native promoter. Cells were grown for 8 h in GM17 medium in the absence or presence of various concentrations of bile acid, and the maximum specific growth rate, μ, was determined.
TABLE 2.
Susceptibility of L. lactis wild-type, ΔlmrCD, and ΔLmrCDr strains to various bile acids and drugs
| Drug | IC50 (μM) of the indicated straina
|
Relative resistancef | ||
|---|---|---|---|---|
| Wild type | ΔlmrCD | ΔLmrCDr | ||
| Lithocholatec | 85 | 85 | 80 | 0.9 |
| Deoxycholateb,d | 125 ± 15 | 120 ± 18 | 190 ± 18 | 1.6 |
| Chenodeoxycholateb,d | 115 | 105 | 160 | 1.5 |
| Cholateb,e | 1650 ± 273 | 1000 ± 146 | 1650 ± 261 | 1.7 |
| Glycodeoxycholateb | 6800 ± 566 | 1063 ± 53 | 1625 ± 35 | 1.5 |
| Taurocholateb | NS | NS | NS | |
| Glycocholateb | NS | NS | NS | |
| Hoechst 33342 | 1.55 ± 0.13 | 0.3 ± 0.21 | 0.35 ± 0.17 | 1.2 |
| Daunomycin | 25.5 | 2.8 | 2.8 | 1.0 |
| Rhodamine 6G | 5.7 | 4.9 | 4.5 | 0.9 |
| Ethidium bromide | 4.8 | 3.7 | 3.4 | 0.9 |
| Quinine | 850 | 775 | 350 | 0.5 |
| Tetracycline | 0.26 | 0.2 | 0.18 | 0.9 |
| Erythromycin | 0.065 | 0.067 | 0.061 | 0.9 |
| Kanamycin | 35 ± 9 | 33 ± 9.5 | 41 ± 6.8 | 1.2 |
| Chloramphenicol | 4.0 ± 0.5 | 4.2 ± 0.9 | 4.5 ± 0.6 | 1.1 |
| Puromycin | 27.5 | 26.5 | 27 | 1.0 |
Values are means and standard errors of IC50s obtained from at least three independent experiments. NS, not sensitive (no appreciable growth inhibition at 6 mM).
Sodium salts (anionic detergent).
One hydroxyl group.
Two hydroxyl groups.
Three hydroxyl groups.
Calculated as (IC50 of ΔLmrCDr cells)/(IC50 of ΔlmrCD cells).
Exposure to one specific drug may evoke cross-resistance to other toxic compounds in bacteria (18, 41). Therefore, the resistance to structurally and functionally diverse toxic compounds was determined. To this end, ΔLmrCDr cells were grown in the presence of a variety of drugs, among which a range of bile acids, several commonly used antibiotics, and structurally unrelated fluorescent dyes like rhodamine 6G, ethidium bromide, and Hoechst 33342 (Table 2). Compared to the wild-type cells, L. lactis ΔlmrCD cells proved to be susceptible to glycodeoxycholate in addition to cholate, Hoechst 33342, daunomycin, rhodamine 6G, and ethidium bromide. Compared to the ΔlmrCD strain, the cholate-adapted ΔLmrCDr cells showed a significant increase in resistance to the unconjugated bile acids deoxycholate (1.6-fold) and chenodeoxycholate (1.5-fold) and the glycoconjugate of deoxycholate (1.5-fold). Interestingly, the acquired resistance to several of these cholate derivatives even exceeded that of the wild-type cells. However, this was certainly not the case for glycodeoxycholate. Compared to the wild-type strain, ΔLmrCDr cells were 4.2-fold more susceptible to glycodeoxycholate (Table 2). Note that the deletion of the lmrCD genes from the wild-type strain results in a more than sixfold increase in sensitivity to glycodeoxycholate (Table 2.). These data suggest that glycodeoxycholate is an important substrate for LmrCD.
In contrast to the observed enhanced resistance to bile acids, ΔLmrCDr cells did not show a significant improvement in resistance to any of the antibiotics and fluorescent dyes tested. On the other hand, the ΔLmrCDr strain was twofold more susceptible to quinine than the ΔlmrCD and wild-type strains. Taken together, the results indicate that the acquired resistance of the ΔLmrCDr strain does not arise from MDR but is specific for cholate and related bile acids.
Next, we tested whether the resistance to bile acids of the ΔLmrCDr strain could be enhanced by reintroducing the lmrCD genes. The ΔLmrCDr strain carrying either the control plasmid or the plasmid harboring the lmrCD genes (under the control of their own promoter) was grown in the presence of cholate (Fig. 1C) or glycodeoxycholate (Fig. 1D). Expression of lmrCD in ΔLmrCDr cells resulted in a relatively small increase in cholate resistance, i.e., the IC50 increased from 2.4 to 3.2 mM (Fig. 1C). For glycodeoxycholate (Fig. 1D), the IC50 essentially did not change upon expression of lmrCD (i.e., it remained at ∼1.5 mM). In contrast, the MICs, differed greatly; ΔLmrCDr cells readily die in the presence of 2.4 mM glycodeoxycholate, but when expressing LmrCD they can withstand concentrations up to 8 mM (Fig. 1D), which is consistent with the notion that glycodeoxycholate is an excellent substrate for LmrCD. The ΔLmrCDr strain carrying the lmrCD plasmid was also grown in the presence of ethidium bromide, rhodamine 6G, or Hoechst 33342. As expected, the ΔLmrCDr strain gained the MDR phenotype associated with LmrCD expression (data not shown).
During growth L. lactis produces lactic acid, which results in acidification of the growth medium (40). Since the pH may affect the solubility (and, thus, toxicity) of the bile acids tested, the acidification of the growth medium caused by the wild-type, ΔlmrCD, and ΔLmrCDr strains, either in the absence or presence of cholate (0.7 mM and 2.2 mM), was monitored in time (data not shown). All strains showed similar degrees of acidification with a drop of the pH from pH 7.3 at the early stages of growth to pH 5.4 in the stationary growth phase. This shows that the bile acid resistance of the ΔLmrCDr strain is not due to alteration in primary metabolism.
Cholate-adapted L. lactis ΔlmrCD cells exhibit an altered cell morphology.
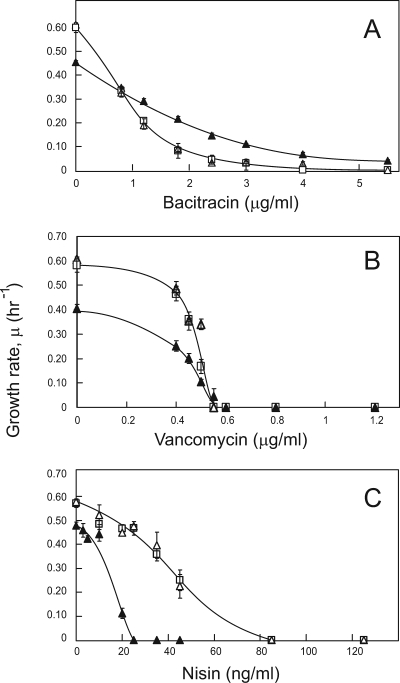
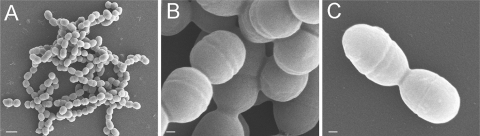
In the absence of drugs, ΔLmrCDr cells had a slower growth rate than the parental ΔlmrCD and wild-type strains (Fig. 1A and 3). Interestingly, ΔLmrCDr cells exhibited an unusual flaky morphology when cultivated in GM17 medium, and they sedimented more readily than ΔlmrCD and wild-type cells. Scanning electron microscopy revealed that ΔLmrCDr cells were similar in size to the ΔlmrCD and wild-type cells but appeared to grow in unusually long strings of cocci clumped together as large aggregates (Fig. 2). Although multiplication of the ΔLmrCDr cells seemed unaffected, the final stage of cell division, i.e., cell separation, was clearly impaired. Such an altered overall cellular morphology likely results from changes in the cell envelope.
FIG. 3.
Sensitivity of L. lactis wild-type (□), ΔlmrCD (▵), and the cholate-adapted ΔLmrCDr (▴) strain to bacteriocins. Cells were grown for 8 h in GM17 medium containing the bacteriocins bacitracin (A), vancomycin (B), or nisin (C) at various concentrations. The maximum specific growth rate, μ, is plotted against the bacteriocin concentration.
FIG. 2.
Morphological changes in L. lactis ΔlmrCD cells following cholate adaptation. Scanning electron micrographs of ΔLmrCDr cells at magnifications of ×6,000 (A) or ×45,000 (B) and of the parental ΔlmrCD strain at a magnification of ×45,000 (C). Bars, 1 μm (A) and 100 nm (B and C).
To investigate the latter possibility, the sensitivity of the cells to various bacteriocins was tested. This included peptides that specifically act on the cell membrane either by a combination of pore formation and inhibition of peptidoglycan synthesis (nisin) or by exclusively inhibiting peptidoglycan synthesis (bacitracin and vancomycin) (Fig. 3). Wild-type and ΔlmrCD cells displayed very similar sensitivities to the tested bacteriocins. In contrast, ΔLmrCDr cells were found to be highly susceptible to the bacteriocin nisin (fourfold lower IC50), while improved resistance to bacitracin (twofold higher IC50) was observed. The sensitivity to vancomycin was similar to that of the control strains. The altered sensitivity of ΔLmrCDr cells to membrane- and peptidoglycan-acting compounds likely arises from an altered cell envelope. Such altered properties of the cell surface may also account for the increased susceptibility to quinine (Table 2).
Transcriptome analysis of cholate-adapted L. lactis ΔlmrCD cells.
To investigate the underlying adaptive mechanism(s) of the cholate-adapted strain, DNA microarray analysis was performed on the global gene expression profiles of exponentially growing ΔlmrCD and ΔLmrCDr cells in the absence of cholate. Compared to the parental ΔlmrCD strain, 124 genes of the ΔLmrCDr strain showed ≥1.8-fold change in expression with a Bayesian P value of ≤0.001 (the full array data set is available under accession number GSE10203 at http://www.ncbi.nlm.nih.gov/geo/accessible). Of these genes, 87 and 37 showed an increased and decreased expression, respectively. The maximum level of gene induction and repression was 5-fold (matR) and 11.6-fold (llmg1356), respectively. The differentially expressed genes were grouped into functional classes (Table 3), taking into account the categories defined earlier for the related L. lactis strain IL1403 (9). Responsive genes were related to cell envelope biogenesis, stress response and chaperones, general metabolism, housekeeping functions, and sex factor. Remarkably, the expression of none of the 40 genes encoding putative MDR transporters, among which are the well-characterized LmrA and LmrP, was significantly altered in the ΔLmrCDr strain. This also includes the llmg2513 (yxbD) gene that was upregulated in the MDR strain of L. lactis selected by cholate (31). Expression of a large number of hypothetical ORFs was also significantly changed including llmg1960, a putative di- and tricarboxylate transporter. However, none of the remaining upregulated hypothetical ORFs could be related to a (drug) transport function (see full array set).
TABLE 3.
Differential expression of genes in cholate-adapted L. lactis ΔLmrCDr cells versus ΔlmrCD cells
| Functional class and gene name | Bayesian P value | Fold change | Proposed name and/or description |
|---|---|---|---|
| Sex factor/conjugation | |||
| matR | 1.38E-10 | 5.0 | Retron-type reverse transcriptase/LtrA |
| cluA | 2.73E-09 | 3.2 | Cell surface antigen I/II precursor |
| llmg1399 | 8.14E-09 | 3.2 | Putative cell surface antigen |
| ltrC | 7.05E-08 | 2.7 | Relaxosome formation |
| ltrB | 1.51E-06 | 2.5 | Group II intron-interrupted relaxase LtrB (mobA) |
| ltrD | 5.47E-07 | 2.4 | Relaxosome formation |
| ltrE | 1.46E-06 | 2.0 | Relaxosome formation |
| traD | 5.81E-06 | 1.8 | Conjugal transfer protein TraD |
| llmg1353 | 6.39E-11 | −11.0 | Putative tellurite resistance protein |
| telB | 1.73E-13 | −10.8 | Putative tellurite resistance protein |
| telC | 5.43E-11 | −7.1 | Putative tellurite resistance protein |
| telA | 1.32E-10 | −6.6 | Putative tellurite resistance protein |
| Stress and chaperones | |||
| groES | 7.77E-09 | 2.9 | GroES/Hsp10 chaperone |
| nah | 3.96E-09 | 2.8 | Na+/H+ antiporter |
| htrA | 4.91E-08 | 2.7 | Housekeeping protease |
| groEL | 2.83E-08 | 2.5 | GroEL/Hsp60 chaperone |
| hslO | 5.55E-08 | 2.4 | Heat shock protein; 33-kDa chaperone |
| llmg2047 | 1.51E-08 | −2.8 | Universal stress protein E |
| uspA2 | 2.67E-06 | −2.1 | Universal stress protein A2 |
| uspA | 2.02E-07 | −2.1 | Universal stress protein A |
| sodA | 1.32E-07 | −2.1 | Superoxide dismutase |
| clpE | 6.57E-07 | −2.0 | ATP-dependent Clp protease ATP-binding subunit E |
| gshR | 5.95E-07 | −2.0 | Glutathione reductase |
| trxA | 3.66E-07 | −1.9 | Thioredoxin |
| Cell envelope | |||
| chiC | 8.22E-09 | 2.8 | Acidic endochitinase precursor |
| llmg1148 | 1.65E-07 | 2.5 | Putative cell surface antigen |
| llmg2420 | 1.24E-07 | 2.3 | Putative glycosyltransferase |
| chb | 2.43E-07 | 2.2 | Chitin binding protein, putative |
| cdsA | 7.95E-07 | 2.1 | Phosphatidate cytidylyltransferase |
| llmg0538 | 4.95E-07 | 2.0 | (3R)-Hydroxymyristoyl-(acyl-carrier-protein) dehydratase |
| lplL | 1.29E-05 | 1.9 | Lipoate-protein ligase A |
| murC | 1.93E-04 | 1.9 | UDP-N-acetyl muramate-alanine ligase |
| llmg2421 | 1.67E-05 | 1.9 | Putative glycosyltransferase |
| llmg0215 | 6.65E-04 | 1.8 | Predicted membrane protein |
| rgpE | 2.80E-04 | 1.8 | Glycosyltransferase RgpE |
| cfa | 1.82E-06 | −2.0 | Cyclopropane-fatty-acyl-phospholipid synthase |
| mvk | 7.80E-07 | −2.0 | Mevalonate kinase |
| apbE | 1.82E-06 | −1.9 | Thiamine biosynthesis lipoprotein ApbE precursor |
| General metabolism | |||
| rrma | 5.78E-09 | 3.1 | Putative rRNA (guanine-N1-)-methyltransferase |
| gltX | 3.69E-06 | 2.6 | Glutamyl-tRNA synthetase |
| llmg1361 | 8.96E-06 | 2.6 | Putative tyrosine recombinase |
| polC | 4.66E-05 | 2.2 | DNA polymerase III alpha subunit |
| ackA1 | 5.52E-07 | 2.1 | AckA1 protein (Acetate kinase) |
| lacX | 5.87E-07 | 2.1 | Galactose mutarotase related enzyme |
| purA | 8.09E-07 | 2.1 | Putative adenylosuccinate synthetase |
| hisS | 4.30E-08 | 2.0 | Histidyl-tRNA synthetase, class IIa |
| llmg1089 | 1.49E-06 | 2.0 | Carbamoyl-phosphate synthase, large subunit |
| ilvB | 6.31E-05 | 2.0 | Acetolactate synthase large subunit |
| butA | 6.56E-08 | 2.0 | Acetoin reductase |
| llmg2209 | 4.46E-07 | 1.9 | tRNA-dihydrouridine synthase B |
| pheS | 2.84E-06 | 1.9 | Phenylalanyl-tRNA synthetase alpha chain |
| pfl | 9.71E-06 | 1.9 | Pyruvate formate lyase |
| recF | 3.12E-06 | 1.9 | DNA replication and repair protein RecF |
| proS | 2.16E-06 | 1.8 | Prolyl-tRNA synthetase |
| llmg2205 | 5.25E-06 | 1.8 | Conserved hypothetical protein |
| polA | 2.84E-06 | 1.8 | DNA polymerase I |
| metA | 1.32E-06 | 1.8 | Homoserine O-succinyltransferase |
| glmS | 7.67E-09 | −2.7 | Glucosamine-fructose-6-phosphate aminotransferase |
| add | 2.59E-06 | −2.6 | Adenosine deaminase |
| ilvE | 5.20E-04 | −2.2 | Branched-chain amino acid aminotransferase |
| rlrG | 8.25E-07 | −2.2 | Transcriptional regulator, LysR family |
| pepC | 4.72E-06 | −2.1 | Aminopeptidase C |
| serB | 2.74E-06 | −2.0 | Phosphoserine phosphatase |
| llmg1086 | 1.91E-06 | −1.9 | MgtA-like cation transporting ATPase |
| nadE | 5.04E-06 | −1.9 | NAD+ synthase |
| cysD | 1.70E-06 | −1.9 | O-acetylhomoserine sulfhydrylase |
| fbaA | 8.76E-07 | −1.8 | Fructose-bisphosphate aldolase |
| dtpT | 2.09E-06 | −1.8 | Di-/tripeptide transporter |
aStatistically significant changes in expression are given as the ratio of the cholate-adapted ΔLmrCDr strain versus the control ΔlmrCD strain (fold change). For clarity, most hypothetical ORFs (56) were omitted.
Genes related to cell envelope.
Various genes associated with cell envelope biosynthesis or morphology, namely, cdsA, murC, rgpE, llmg0215, llmg0538, and llmg1148, showed increased expression levels in the ΔLmrCDr strain. Decreased expression levels were observed for cfa, which is involved in membrane lipid biosynthesis, and mvk, a key gene of the mevalonate biosynthetic pathway needed for isoprenoid synthesis.
Genes related to stress response and chaperones.
A distinct level of overexpression of the molecular chaperone genes groES, groEL, and hslO was observed in the ΔLmrCDr strain, as well the gene encoding a serine protease, htrA, implicated in protein folding stress. Also nah, encoding an Na+/H+ antiporter involved in sodium toxicity and intracellular pH regulation, was expressed at a higher level. In contrast, oxidative stress response genes trxA, sodA, and gshR and the general stress response genes uspA and uspA2 were distinctly repressed.
Genes related to general metabolism and housekeeping functions.
A variety of genes related to various biosynthetic pathways and DNA replication and repair are differentially expressed. Increases in expression of the genes rrma, polC, polA, and recF involved in DNA replication occurred in the ΔLmrCDr strain. Likewise, an increase in expression of genes involved in amino acid biosynthesis (i.e., ilvB, metA, proS, llmg1089, hisS, gltX, and pheS) and sugar metabolism (i.e., lacX, ackA1, pfl, and butA) was observed, whereas several other genes were repressed, such as serB and cysD involved in glycine and serine biosynthesis, respectively, as well as genes involved in nucleotide biosynthesis. Of the extensive proteolytic system of L. lactis, only pepC, an amino acid peptidase, and dtpT, a di- and tripeptide transporter, exhibited significant repression.
Sex factor.
The sex factor is a chromosomally located ∼55-kb element (llmg1411-llmg1348) involved in conjugation. Forty-one out of 57 sex factor genes were significantly differentially expressed. One of the most prominently upregulated genes is cluA. This gene confers a cell aggregation phenotype in L. lactis (34), which would explain the clumping phenotype observed. A similar role for the product of llmg1399 can be envisaged as it lies in close proximity to cluA and shares high homology with it. In addition, traD, involved in conjugal transfer, was distinctly overexpressed. A group of 10 genes located at the distal end of the element is significantly repressed. Among these, telA, telB, and telC have been implicated in tellurite resistance. In line with this observation, the ΔLmrCDr cells were found to be significantly more susceptible to tellurite than cells of the ΔlmrCD and wild-type strains, with MICs of 1.5, 2.5, and 2.5 mM, respectively (data not shown).
LmrCD is responsible for cholate extrusion.
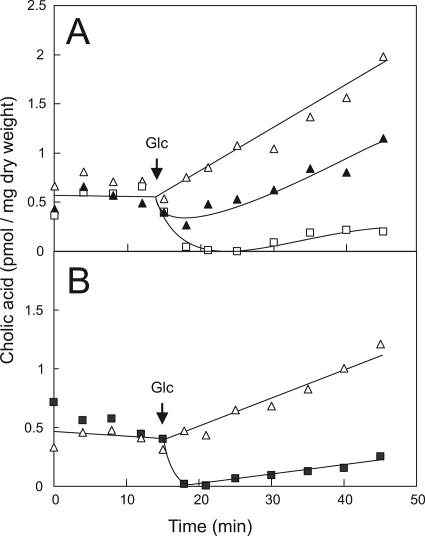
The transcriptome analysis of the cholate-adapted ΔLmrCDr cells showed no evidence of an altered expression of any of the putative MDR transporters. To further analyze the mechanism of cholate resistance, the ability of cholate to enter cells was examined by means of a [14C]cholate transport assay. Wild-type, ΔlmrCD, and ΔLmrCDr cells showed a similar level of [14C]cholate association when tested under energy-deprived conditions (Fig. 4A). Upon the addition of glucose, ΔlmrCD cells accumulated cholate in a time-dependent fashion, while wild-type cells showed a strong extrusion of cholate under identical conditions. For energized ΔLmrCDr cells, initially a minor efflux is observed, followed by an extended cholate accumulation phase similar to that of the ΔlmrCD strain. The ΔLmrCDr cells accumulated cholate to a lesser extent (approximately twofold) than the parental ΔlmrCD strain. The cholate extrusion activity of L. lactis ΔlmrCD cells could be restored by the reintroduction of the lmrCD genes on a plasmid under the control of their native promoter (Fig. 4B). It should be noted that since cholate is a weak acid, the observed energy-dependent uptake of cholate by the ΔlmrCD and ΔLmrCDr cells (Fig. 4A) likely results from passive permeation and a ΔpH-dependent partitioning of the cholate across the membrane. Taken together, these data unequivocally show that in L. lactis, LmrCD efficiently extrudes cholate from the cell in an energy-dependent fashion. Furthermore, the apparent lack of major extrusion activity in the ΔLmrCDr cells suggests that LmrCD is the major contributor in cholate extrusion and lends further support for the notion that the cholate resistance of the ΔLmrCDr strain is unrelated to a transport phenomenon.
FIG. 4.
Accumulation of cholate by L. lactis cells harboring or lacking lmrCD genes. (A and B) De-energized cells were preloaded with [14C]cholate, and after 14 min cells were energized with glucose at a final concentration of 230 mM. The arrow indicates when glucose was added. (A) Cholate accumulation in the wild-type (□), ΔlmrCD (▵), and ΔLmrCDr (▴) strains. (B) Cholate accumulation in the ΔlmrCD strain containing plasmid pILlmrCD (▪) or pIL252 (▵).
The expression of the lmrCD genes in L. lactis is controlled by the transcriptional regulator LmrR (1). Previous studies have demonstrated that binding of drugs like Hoechst 33342 and daunomycin to LmrR relieves the repression of the lmrCD genes, thus leading to the manifestation of the MDR phenotype. Since growth and transport studies suggest that bile acids are natural substrates of the LmrCD transporter, the ability of cholate to induce the expression of the lmrCD genes was investigated. RT-PCR-based detection of mRNA revealed a transient increase of both the lmrC and lmrD transcripts upon a challenge of the cells with cholate (Fig. 5). On the other hand, the mRNA levels of the constitutively expressed secY gene remained unaltered. This indicates that cholate is an inducer of lmrCD expression.
FIG. 5.
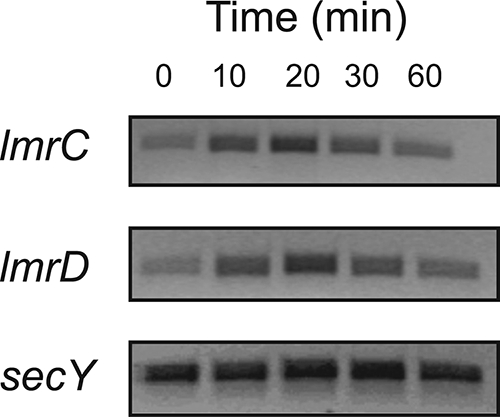
Cell-based RT-PCR analysis of lmrCD expression in L. lactis wild-type cells following cholate induction. PCR products were generated through the use of gene-specific primers for lmrCD. Amplified products were separated on 2.0% agarose gels and were identified by ethidium bromide staining. For each time sample, an RT-PCR with primers specific for the secY gene was run as a control.
DISCUSSION
The most widely used probiotic bacteria are Lactobacillus and Bifidobacteria, which are natural inhabitants of the human gastrointestinal tract. Lactococci are not considered part of the gastrointestinal tract, and only a few studies on their probiotic activity are available. However, lactococci do exhibit probiotic properties, like the ability to (i) survive in the gut, (ii) lower the cholesterol level, and (iii) modulate the immune response of the host (26, 46). Lactococci also tolerate gut secretions such as bile acids (24, 25). The purpose of this study was to examine the role of the ABC-type MDR transporter LmrCD in bile acid resistance. In gram-negative bacteria, drug extrusion-based mechanisms across the outer membrane are major determinants of bile acid resistance. This resistance involves MDR efflux pumps that belong to resistance nodulation division and MFS (30, 38), both of which utilize the proton motive force as an energy source. In gram-positive bacteria such as L. lactis, the exact mechanisms of cholate resistance are less clear, but the involvement of efflux-based mechanisms based on secondary transport have been suggested (20).
Here, we demonstrate that in L. lactis, LmrCD is responsible for an extrusion-based mechanism of resistance against cholate and also provides resistance against the physiologically relevant conjugate glycodeoxycholate. Cells lacking the lmrCD genes are sensitive to these compounds. However, when challenged with increasing concentrations of cholate, these cells regain resistance. Remarkably, this resistance is no longer based on an extrusion mechanism but relates to secondary responses such as cell envelope changes, stress responses, and alterations in metabolism. On the other hand, in wild-type cells the short-term response to a challenge with cholate is the upregulation of the lmrCD genes, resulting in an increased extrusion of cholate. This upregulation involves the transcriptional repressor LmrR that likely interacts directly with cholate, whereupon repression of lmrCD is relieved (1). To our knowledge, this report shows for the first time a central role of an ABC-type transporter in bile acid resistance in prokaryotes. In this respect, ABC-type transporters are generally involved in bile extrusion and transport within the liver (15).
To investigate possible alternative mechanisms of cholate resistance, the cholate-adapted strain, ΔLmrCDr, and parental strain were subjected to transcriptome profiling. About 100 genes involved in cellular metabolism and morphology were found to be differentially expressed in the transcriptome of the cholate-adapted strain. This is in sharp contrast to what was observed in cholate-adapted wild-type L. lactis cells, which showed a prominent defect in the lmrR gene that encodes a transcriptional repressor of lmrCD expression, which resulted in the constitutive expression of the lmrCD genes (1, 32). Interestingly, neither the well-characterized transporters LmrA and LmrP nor any of the other remaining putative MDR transporters were found to be upregulated in the cholate-adapted ΔLmrCDr strain, lending further support to the notion that bile acid extrusion is a key activity of LmrCD.
One of the most strongly induced genes in ΔLmrCDr cells is nah, which encodes the Na+/H+ antiporter that has been implicated in intracellular pH homeostasis and Na+ toxicity. Due to passive permeation of cholic acid, high concentrations of cholate might interfere with intracellular pH regulation, which may explain this cellular response. Alternatively, upregulation of nah is a means to counteract Na+ toxicity as cholate is added as a sodium salt. One of the other remarkable responses is the upregulation of the genes associated with the chromosomally embedded sex factor. This 55-kb region comprises a unique mobile genetic element in L. lactis that can be excised in a closed circular form and is readily lost from cells (17). The genes contained in the proximal region (ltrA-matR, ltrB, ltrC, ltrD, ltrE, and traD) show strongly elevated expression. ltrA-matR codes for a protein with reverse transcriptase, endonuclease, and RNA maturase activity (37) that facilitates retrohoming of ltrB into intronless alleles. The ltrC, ltrD, and ltrE gene products are involved in relaxosome formation during conjugation (12) while traD encodes a coupling protein that links to the DNA transfer intermediate and perhaps leads the DNA through the mating channel (19). It has been shown for L. lactis MG1363 that the sex factor element can mobilize chromosomal genes (16). Possibly during cholate stress, genetic traits that confer resistance to cholate may be transferred to the recipient cells. Another sex factor gene, cluA, associated with cell aggregation phenotype also increased in expression and is involved in the cell-to-cell contact necessary for conjugal transfer (44). CluA is a 136-kDa surface-bound protein covalently linked to the cell wall peptidoglycan. This protein is not only responsible for a constitutive aggregation phenotype in L. lactis MG1363 but also linked to high-frequency conjugation and transfer of the sex factor (34, 42). The upregulation of the sex factor genes might thus be responsible for a major morphological change such as the clumping of the cells that may provide a certain level of protection to the inner cells in the aggregate to cholate. Interestingly, Lactobacillus plantarum also showed morphological changes in response to bile acid stress (10). Challenged cells clumped together (but did not form long strings) and showed elevated expression levels of several genes involved in membrane- and cell wall-associated functions. Thus, altering the properties of the bacterial cell surface may be a common response to bile acid stress.
In the ΔLmrCDr strain several upregulated genes appear to be associated with cell envelope biogenesis such as murC, which is involved in the biosynthesis of the peptidoglycan murein, which catalyzes the addition of l-alanine to the nucleotide precursor UDP-N-acetylmuramoyl, and genes such as rgpE, llmg2420, and llmg2421, encoding putative glycosyltransferases that catalyze the formation of linear glycan chains. The upregulation of these genes may result in an alteration of the cell envelope composition and thus indirectly affect cholate permeation and susceptibility. This is further supported by the altered responses of the cholate-adapted cells to the activity of three peptide antibiotics, i.e., nisin, bacitracin, and vancomycin. These antimicrobials affect the cell envelope by different mechanisms of action (2, 3, 23). The increased nisin sensitivity in the cholate-adapted strain suggests that either the levels of lipid II, the binding site for nisin, have increased (2, 36) or that lipid II is more accessible (27, 36). Nisin sensitivity frequently links to bacitracin resistance in bacteria (36), and this also appears to be the case in the cholate-adapted strain. On the other hand, the vancomycin sensitivity did not change, demonstrating that vancomycin binding sites have not changed in the mutant, thus suggesting that the different responses are due to antimicrobial-dependent differences in cell envelope permeation.
Although the cholate-adapted ΔLmrCDr cells show a greater resistance to bile acids than wild-type cells, this is not the case for all derivatives tested (Table 2). The cholate-adapted cells remained very susceptible to the hydrophobic bile acid lithocholate and are substantially more sensitive to glycodeoxycholate than the wild-type. Our data show that the unconjugated bile acids (cholate, deoxycholate, chenodeoxycholate, and lithocholate) are more toxic to L. lactis than the conjugated forms (glycodeoxycholate, glycocholate, and taurocholate) as is the case with bacteria in general (39). The lack of toxicity of taurocholate and glycocholate may be due to the low pKa values (∼1.4 and ∼2.4, respectively) which render these compounds fully ionized at neutral pH. Thus, these molecules are likely highly membrane impermeable. Overall, the susceptibility of L. lactis for bile acids seems to be directly related to their hydrophobicity. Indeed, membrane permeability decreases with the number of the hydroxyl group additions (28). However, this assessment does not take into account the extrusion-based resistance mechanism that prevails in wild-type cells. Notably, the toxicity of glycodeoxycholate is strongly dependent on the activity of LmrCD. This is intriguing since glyco-forms of bile acid conjugates are more toxic than tauro-conjugates while the former also represent the predominant form of bile salt conjugate in human bile (5). Therefore, the observation that LmrCD renders cells highly resistant to glycodeoxycholate suggests that bile acids are natural substrates of this transporter. As homologs of LmrCD are widely distributed among gut bacteria, ABC-type MDR transporters may be important factors in colonization and survival in the intestine.
Acknowledgments
We gratefully acknowledge the contribution of I. Stokroos of the Department of Cell Biology, Molecular Imaging and Electron Microscopy Section (UMCG), University of Groningen, for the scanning electron microscopy work.
A.Z. was supported by a scholarship from the Higher Education Commission, Government of Pakistan.
Footnotes
Published ahead of print on 12 September 2008.
REFERENCES
- 1.Agustiandari, H., J. Lubelski, H. B. van den Berg van Saparoea, O. P. Kuipers, and A. J. Driessen. 2008. LmrR is a transcriptional repressor of expression of the multidrug ABC transporter LmrCD in Lactococcus lactis. J. Bacteriol. 190759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur, M., and P. Courvalin. 1993. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 371563-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barna, J. C., and D. H. Williams. 1984. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu. Rev. Microbiol. 38339-357. [DOI] [PubMed] [Google Scholar]
- 4.Begley, M., C. Hill, and C. G. Gahan. 2003. Identification and disruption of btlA, a locus involved in bile tolerance and general stress resistance in Listeria monocytogenes. FEMS Microbiol. Lett. 21831-38. [DOI] [PubMed] [Google Scholar]
- 5.Begley, M., C. Hill, and C. G. Gahan. 2006. Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 721729-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binder, H. J., B. Filburn, and M. Floch. 1975. Bile acid inhibition of intestinal anaerobic organisms. Am. J. Clin. Nutr. 28119-125. [DOI] [PubMed] [Google Scholar]
- 7.Bolhuis, H., D. Molenaar, G. Poelarends, H. W. Van Veen, B. Poolman, A. J. Driessen, and W. N. Konings. 1994. Proton motive force-driven and ATP-dependent drug extrusion systems in multidrug-resistant Lactococcus lactis. J. Bacteriol. 1766957-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolhuis, H., G. Poelarends, H. W. Van Veen, B. Poolman, A. J. Driessen, and W. N. Konings. 1995. The lactococcal lmrP gene encodes a proton motive force-dependent drug transporter. J. Biol. Chem. 27026092-26098. [DOI] [PubMed] [Google Scholar]
- 9.Bolotin, A., B. Quinquis, A. Sorokin, and D. S. Ehrlich. 2004. Recent genetic transfer between Lactococcus lactis and enterobacteria. J. Bacteriol. 1866671-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bron, P. A., M. Marco, S. M. Hoffer, M. E. Van, W. M. de Vos, and M. Kleerebezem. 2004. Genetic characterization of the bile salt response in Lactobacillus plantarum and analysis of responsive promoters in vitro and in situ in the gastrointestinal tract. J. Bacteriol. 1867829-7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bron, P. A., D. Molenaar, W. M. de Vos, and M. Kleerebezem. 2006. DNA micro-array-based identification of bile-responsive genes in Lactobacillus plantarum. J. Appl. Microbiol. 100728-738. [DOI] [PubMed] [Google Scholar]
- 12.Chen, I., P. J. Christie, and D. Dubnau. 2005. The ins and outs of DNA transfer in bacteria. Science 3101456-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.den Hengst, C. D., S. A. van Hijum, J. M. Geurts, A. Nauta, J. Kok, and O. P. Kuipers. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 28034332-34342. [DOI] [PubMed] [Google Scholar]
- 14.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 623662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faber, K. N., M. Muller, and P. L. Jansen. 2003. Drug transport proteins in the liver. Adv. Drug Deliv. Rev. 55107-124. [DOI] [PubMed] [Google Scholar]
- 16.Gasson, M. J., J. J. Godon, C. J. Pillidge, T. J. Eaton, K. Jury, and C. A. Shearman. 1995. Characterization and exploitation of conjugation in Lactococcus lactis. Int. Dairy J. 5757-762. [Google Scholar]
- 17.Gasson, M. J., S. Swindell, S. Maeda, and H. M. Dodd. 1992. Molecular rearrangement of lactose plasmid DNA associated with high-frequency transfer and cell aggregation in Lactococcus lactis 712. Mol. Microbiol. 63213-3223. [DOI] [PubMed] [Google Scholar]
- 18.George, A. M. 1996. Multidrug resistance in enteric and other gram-negative bacteria. FEMS Microbiol. Lett. 1391-10. [DOI] [PubMed] [Google Scholar]
- 19.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67277-301, table. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunn, J. S. 2000. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2907-913. [DOI] [PubMed] [Google Scholar]
- 21.Higgins, C. F. 2007. Multiple molecular mechanisms for multidrug resistance transporters. Nature 446749-757. [DOI] [PubMed] [Google Scholar]
- 22.Hord, N. G. 2008. Eukaryotic-microbiota crosstalk: potential mechanisms for health benefits of prebiotics and probiotics. Annu. Rev. Nutr. 28215-231. [DOI] [PubMed] [Google Scholar]
- 23.Hyde, A. J., J. Parisot, A. McNichol, and B. B. Bonev. 2006. Nisin-induced changes in Bacillus morphology suggest a paradigm of antibiotic action. Proc. Natl. Acad. Sci. USA 10319896-19901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimoto, H., J. Kurisaki, N. M. Tsuji, S. Ohmomo, and T. Okamoto. 1999. Lactococci as probiotic strains: adhesion to human enterocyte-like Caco-2 cells and tolerance to low pH and bile. Lett. Appl. Microbiol. 29313-316. [DOI] [PubMed] [Google Scholar]
- 25.Kimoto, H., M. Nomura, M. Kobayashi, K. Mizumachi, and T. Okamoto. 2003. Survival of lactococci during passage through mouse digestive tract. Can. J. Microbiol. 49707-711. [DOI] [PubMed] [Google Scholar]
- 26.Kimoto-Nira, H., K. Mizumachi, M. Nomura, M. Kobayashi, Y. Fujita, T. Okamoto, I. Suzuki, N. M. Tsuji, J. Kurisaki, and S. Ohmomo. 2006. Lactococcus sp. as potential probiotic lactic acid bacteria. Jpn. Agric. Res. Q. 41181-189. [Google Scholar]
- 27.Kramer, N. E., E. J. Smid, J. Kok, K. B. de, O. P. Kuipers, and E. Breukink. 2004. Resistance of gram-positive bacteria to nisin is not determined by lipid II levels. FEMS Microbiol. Lett. 239157-161. [DOI] [PubMed] [Google Scholar]
- 28.Kurdi, P., K. Kawanishi, K. Mizutani, and A. Yokota. 2006. Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J. Bacteriol. 1881979-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurdi, P., H. Tanaka, H. W. Van Veen, K. Asano, F. Tomita, and A. Yokota. 2003. Cholic acid accumulation and its diminution by short-chain fatty acids in bifidobacteria. Microbiology 1492031-2037. [DOI] [PubMed] [Google Scholar]
- 30.Li, X. Z., and H. Nikaido. 2004. Efflux-mediated drug resistance in bacteria. Drugs 64159-204. [DOI] [PubMed] [Google Scholar]
- 31.Lubelski, J., J. A. de, M. R. van, H. Agustiandari, O. P. Kuipers, J. Kok, and A. J. Driessen. 2006. LmrCD is a major multidrug resistance transporter in Lactococcus lactis. Mol. Microbiol. 61771-781. [DOI] [PubMed] [Google Scholar]
- 32.Lubelski, J., W. N. Konings, and A. J. Driessen. 2007. Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol. Mol. Biol. Rev. 71463-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lubelski, J., P. Mazurkiewicz, M. R. van, W. N. Konings, and A. J. Driessen. 2004. ydaG and ydbA of Lactococcus lactis encode a heterodimeric ATP-binding cassette-type multidrug transporter. J. Biol. Chem. 27934449-34455. [DOI] [PubMed] [Google Scholar]
- 34.Luo, H., K. Wan, and H. H. Wang. 2005. High-frequency conjugation system facilitates biofilm formation and pAMβ1 transmission by Lactococcus lactis. Appl. Environ. Microbiol. 712970-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma, D., D. N. Cook, J. E. Hearst, and H. Nikaido. 1994. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 2489-493. [DOI] [PubMed] [Google Scholar]
- 36.Mantovani, H. C., and J. B. Russell. 2001. Nisin resistance of Streptococcus bovis. Appl. Environ. Microbiol. 67808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuura, M., R. Saldanha, H. Ma, H. Wank, J. Yang, G. Mohr, S. Cavanagh, G. M. Dunny, M. Belfort, and A. M. Lambowitz. 1997. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 112910-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 1835803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okoli, A. S., T. Wadstrom, and G. L. Mendz. 2007. Minireview: bioinformatic study of bile responses in Campylobacterales. FEMS Immunol. Med. Microbiol. 49101-123. [DOI] [PubMed] [Google Scholar]
- 40.Rallu, F., A. Gruss, and E. Maguin. 1996. Lactococcus lactis and stress. Antonie van Leeuwenhoek 70243-251. [DOI] [PubMed] [Google Scholar]
- 41.Russell, A. D., U. Tattawasart, J. Y. Maillard, and J. R. Furr. 1998. Possible link between bacterial resistance and use of antibiotics and biocides. Antimicrob. Agents Chemother. 422151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon, G. L., and S. L. Gorbach. 1984. Intestinal flora in health and disease. Gastroenterology 86174-193. [PubMed] [Google Scholar]
- 43.Staggers, J. E., S. C. Frost, and M. A. Wells. 1982. Studies on fat digestion, absorption, and transport in the suckling rat. III. Composition of bile and evidence for enterohepatic circulation of bile salts. J. Lipid Res. 231143-1151. [PubMed] [Google Scholar]
- 44.Stentz, R., K. Jury, T. Eaton, M. Parker, A. Narbad, M. Gasson, and C. Shearman. 2004. Controlled expression of CluA in Lactococcus lactis and its role in conjugation. Microbiology 1502503-2512. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka, H., K. Doesburg, T. Iwasaki, and I. Mierau. 1999. Screening of lactic acid bacteria for bile salt hydrolase activity. J. Dairy Sci. 822530-2535. [DOI] [PubMed] [Google Scholar]
- 46.Timmerman, H. M., L. E. Niers, B. U. Ridwan, C. J. Koning, L. Mulder, L. M. Akkermans, F. M. Rombouts, and G. T. Rijkers. 2007. Design of a multispecies probiotic mixture to prevent infectious complications in critically ill patients. Clin. Nutr. 26450-459. [DOI] [PubMed] [Google Scholar]
- 47.van Hijum, S. A., J. A. de, R. J. Baerends, H. A. Karsens, N. E. Kramer, R. Larsen, C. D. den Hengst, C. J. Albers, J. Kok, and O. P. Kuipers. 2005. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Veen, H. W., K. Venema, H. Bolhuis, I. Oussenko, J. Kok, B. Poolman, A. J. Driessen, and W. N. Konings. 1996. Multidrug resistance mediated by a bacterial homolog of the human multidrug transporter MDR1. Proc. Natl. Acad. Sci. USA 9310668-10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Velkinburgh, J. C., and J. S. Gunn. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 671614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaughan, E. E., H. G. Heilig, K. Ben-Amor, and W. M. de Vos. 2005. Diversity, vitality and activities of intestinal lactic acid bacteria and bifidobacteria assessed by molecular approaches. FEMS Microbiol. Rev. 29477-490. [DOI] [PubMed] [Google Scholar]
- 51.Yokota, A., M. Veenstra, P. Kurdi, H. W. Van Veen, and W. N. Konings. 2000. Cholate resistance in Lactococcus lactis is mediated by an ATP-dependent multispecific organic anion transporter. J. Bacteriol. 1825196-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zwietering, M. H., I. Jongenburger, F. M. Rombouts, and R. K. van't. 1990. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 561875-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]