Abstract
Most chlamydial strains have a pyruvoyl-dependent decarboxylase protein that converts l-arginine to agmatine. However, chlamydiae do not produce arginine, so they must import it from their host. Chlamydophila pneumoniae has a gene cluster encoding a putative outer membrane porin (CPn1033 or aaxA), an arginine decarboxylase (CPn1032 or aaxB), and a putative cytoplasmic membrane transporter (CPn1031 or aaxC). The aaxC gene was expressed in Escherichia coli producing an integral cytoplasmic membrane protein that catalyzed the exchange of l-arginine for agmatine. Expression of the aaxA gene produced an outer membrane protein that enhanced the arginine uptake and decarboxylation activity of cells coexpressing aaxB and aaxC. This chlamydial arginine/agmatine exchange system complemented an E. coli mutant missing the native arginine-dependent acid resistance system. These cells survived extreme acid shock in the presence of l-arginine. Biochemical and evolutionary analysis showed the aaxABC genes evolved convergently with the enteric arginine degradation system, and they could have a different physiological role in chlamydial cells. The chlamydial system uniquely includes an outer membrane porin, and it is most active at a higher pH from 3 to 5. The chlamydial AaxC transporter was resistant to cadaverine, l-lysine and l-ornithine, which inhibit the E. coli AdiC antiporter.
Chlamydial bacteria persist as nonreplicative elementary bodies that infect eukaryotic epithelial cells. Inside inclusion vacuoles in the host cell, the elementary bodies differentiate into replicative reticulate bodies (7). These chlamydial inclusions do not appear to fuse with lysosomes. No lysosomal or endocytic markers have been detected in the inclusions, and they appear to maintain a neutral pH (18, 47, 48). Chlamydial inclusions must draw nutrients from the host cell, because chlamydia require glucose-6-phosphate, amino acids, cofactors, and nucleotides (33, 35). Understanding the metabolic relationship between intracellular chlamydia and their host cells will be crucial for long-term efforts to develop axenic chlamydial cultures and to resolve persistent infections.
All sequenced chlamydial genomes share a cluster of three genes with putative arginine uptake and decarboxylation functions. The Chlamydophila pneumoniae CPn1032 gene encodes a pyruvoyl-dependent arginine decarboxylase (ArgDC) (12). This enzyme functions optimally in acidic conditions (pH 3.4), catalyzing the decarboxylation of l-arginine and producing agmatine. The CPn1032 gene is flanked by the upstream CPn1033 gene, encoding a putative outer membrane protein, and by the downstream CPn1031 gene, encoding a putative cytoplasmic membrane transporter protein (23). The CPn1033 protein belongs to the OprB family of outer membrane porins, but it was originally annotated as a hypothetical protein due to its low sequence similarity to any characterized member of that porin family (1). A peptide from the Chlamydia muridarum homolog of CPn1031 was presented by major histocompatibility complex class I molecules from murine dendritic cells (24). The CPn1031 protein belongs to the basic amino acid/polyamine antiporter family of transporters (APA; TC 2.A.3.2) (21), and it was originally annotated as an arginine/ornithine antiporter based on its similarity to the Pseudomonas aeruginosa ArcD transporter (51). In Chlamydia trachomatis L2 the three orthologous genes are cotranscribed (Derek Fisher, Uniformed Services University of the Health Sciences, unpublished data), although a nonsense mutation in the L2 homolog of CPn1032 suggests this system may not be functional in the invasive lymphogranuloma venerum strains (50). This gene order is conserved in all chlamydial genomes, but no homologs are found in the anciently diverged Candidatus Protochlamydia amoebophila UWE25 (an endosymbiont of Acanthamoeba sp.).
Many bacteria have arginine uptake and degradation systems, used for a variety of metabolic or defensive purposes. P. aeruginosa and diverse members of the Firmicutes phylum use an arginine deiminase pathway to couple arginine fermentation to ATP formation (5, 51). These cells import l-arginine, hydrolyze it to l-ornithine and ammonium bicarbonate, and export ornithine. Chlamydiae have no homologs of the arginine deiminase gene, and they do not appear to ferment arginine. Numerous bacteria and archaea use biosynthetic arginine decarboxylases to produce agmatine (13, 17, 34). The agmatine ureohydrolase enzyme converts agmatine into putrescine, the core polyamine for spermidine and spermine synthesis. However, chlamydiae also lack agmatine ureohydrolase.
Alternatively, many enteric bacteria, including Escherichia coli and Salmonella enterica, express an arginine-dependent acid resistance system, comprising a pyridoxal 5′-phosphate dependent ArgDC (AdiA) and an arginine/agmatine antiporter (AdiC) (15, 20, 26, 30). This system helps to raise the cytoplasmic pH and invert the cell's membrane potential (43). Chlamydiae could use their arginine decarboxylase with an arginine-agmatine antiporter in an analogous system. Yet there is no evidence for acidification during chlamydial infections (11). Chlamydia could also use this system to deplete the arginine pool in macrophages, reducing the substrate for inducible nitric oxide synthase (iNOS or NOS2). Inhibiting this aspect of the innate immune response could help chlamydial cells resist destruction by macrophages. The extracellular pathogen Helicobacter pylori uses a similar strategy, expressing an arginine deiminase enzyme to deplete arginine, thereby reducing iNOS abundance and activity (4, 14, 29).
We expressed the complete arginine uptake and utilization operon from C. pneumoniae in E. coli from a multicopy plasmid. The CPn1033 gene encoded a porin, localized to the E. coli outer membrane. The CPn1032 gene encoded the previously described pyruvoyl-dependent ArgDC (12). Finally, the CPn1031 gene encoded an integral cytoplasmic membrane protein, functioning as an arginine/agmatine antiporter. Coexpression of the CPn1032 and CPn1031 genes complemented a deletion of the full E. coli adi operon, restoring arginine-dependent acid resistance. E. coli cells coexpressing these proteins showed significant whole-cell arginine uptake and decarboxylase activity at pH 4 to 5, higher than the pH optimum of the arginine decarboxylase alone. Coexpression of all three chlamydial proteins significantly enhanced arginine uptake and decarboxylase activity compared to cells expressing only the CPn1032 and CPn1031 proteins, although it did not increase arginine-dependent acid resistance. We designate these genes as aaxA (CPn1033), aaxB (CPn1032), and aaxC (CPn1031) based on their identified functions in an arginine-agmatine exchange system.
MATERIALS AND METHODS
Strains and DNA.
C. pneumoniae Kajaani 6 chromosomal DNA was a gift from Claudio Cortes Miranda and Benjamin Wizel (University of Texas Health Center at Tyler) (Table 1) (9). E. coli EF1021 was a gift from John Foster (University of South Alabama) (15). E. coli MG1655 (CGSC 7740) was obtained from the E. coli Genetic Stock Center (Yale). E. coli DH5α (Invitrogen) was used as a general cloning host. Bacteriophage P1vir was a gift from Ian Molineux (University of Texas at Austin).
TABLE 1.
Microorganisms and plasmids in this study
| Strain or plasmid | Description and partial genotype | Source or reference |
|---|---|---|
| Strains | ||
| Chlamydophila pneumoniae | Kajaani 6 | 9 |
| Escherichia coli | ||
| BL21(DE3) | Expression strain with T7 RNA polymerase gene | Novagen |
| BW25113 | ΔaraBADAH33 | 6 |
| MG1655 | Sequenced strain (CGSC 7740) | 3 |
| DH5α | General cloning host | Invitrogen |
| DEG0100 | MG1655 ΔadiC1::kan | This study |
| DEG0121 | MG1655 ΔadiA2::kan | 12 |
| DEG0124 | MG1655 ΔadiC1 | This study |
| DEG0147 | MG1655 ΔadiAYC::kan | This study |
| EF1021 | EK227 ΔadiC1::kan | 15 |
| Plasmids | ||
| pBAD/HisA | Expression vector with PBAD promoter | Invitrogen |
| pCDFDuet-1 | Expression vector with CDF replicon | Novagen |
| pCOLADuet-1 | Expression vector with COLA replicon | Novagen |
| pCP20 | Expression vector for FLP recombinase | 6 |
| pDG133 | aaxC in pET-43.1b | This study |
| pDG170 | aaxC in pBAD/HisA | This study |
| pDG183 | aaxC in pTrcHisA | This study |
| pDG193 | aaxC-HSV in pTrcHisA | This study |
| pDG194 | aaxC-HSV in pBAD/HisA | This study |
| pDG219 | pBAD/HisA with modified MCS | This study |
| pDG339 | aaxB in pBAD/HisA | 12 |
| pDG360 | aaxBC in pET-43.1b | This study |
| pDG366 | aaxBC-HSV in pBAD/HisA | This study |
| pDG379 | aaxBC in pBAD/HisA | This study |
| pDG484 | aaxABC in pBAD/HisA | This study |
| pDG512 | aaxA-HSV in pBAD/HisA | This study |
| pDG552 | ompX in pCOLADuet | This study |
| pDG561 | lepA in pCDFDuet-1 | This study |
| pET-43.1b | Expression vector with HSV tag | Novagen |
| pKD13 | Vector with FLP-kan cassette | 6 |
| pKD46 | Expression vector for red recombinase | 6 |
| pTrcHisA | Expression vector with Ptrc promoter | Invitrogen |
Cloning.
The multiple cloning site of vector pBAD/HisA was amplified by PCR using primers pBADMCS2 and pBAD-Rev (all primer sequences are listed in Table 2). The purified product was digested with NcoI and HindIII restriction enzymes and then ligated into the same sites of pBAD/HisA to produce vector pDG219 (Table 1). Primers 5CPn1031N and 3CPn1031H were used to amplify the CPn1031 gene from C. pneumoniae K6 chromosomal DNA. The PCR product was ligated between NdeI and HindIII sites in vector pET-43.1b to produce vector pDG133. Primers 5CPn1031Nc and 3CPn1031H2 were used to amplify CPn1031 from pDG133. The product was ligated between NcoI and HindIII sites of pBAD/HisA to produce pDG170 and between the same sites of pTrcHisA to produce pDG183. Primers 5CPn1031Nc and COLIDOWN-RI were used to amplify the CPn1031-HSV cassette from pDG133. The cassette encodes the CPn1031 protein fused to a carboxy-terminal herpes simplex virus (HSV) glycoprotein D epitope sequence (Novagen). This cassette was ligated between the NcoI and EcoRI sites of pTrcHisA to produce vector pDG193 and between the same sites of pBAD/HisA to produce vector pDG194. Primers 5CPn1032Nc and 3CPn1031H were used to amplify the CPn1032 and CPn1031 gene pair. This PCR product was digested with NcoI and HindIII and ligated into the same sites of plasmid pET-43.1b to produce plasmid pDG360 and in plasmid pBAD/HisA to produce pDG379. The CPn1032-CPn1031-HSV cassette was amplified from pDG360 using the primers 5CPn1032Nc and COLIDOWN. This product was digested with NcoI, phosphorylated with polynucleotide kinase, and ligated between the NcoI and SmaI sites of pDG219 to produce vector pDG366. The CPn1033-CPn1032-CPn1031 cluster was amplified by PCR using the primers 5CPn1033Nc and 3CPn1031X. The product was ligated between the NcoI and XhoI sites of pBAD/HisA to produce pDG484. The CPn1033 gene was amplified using the primers 5CPn1033Nc and 3CPn1033Not and ligated between the NcoI and NotI sites of pDG366 to produce pDG512. Recombinant DNA was sequenced at the Institute for Cellular and Molecular Biology Core Labs DNA Sequencing facility (University of Texas at Austin). Both the CPn1031 and the CPn1033 gene sequences from C. pneumoniae K6 were identical to their orthologous sequences reported for C. pneumoniae CWL029 (GenBank accession numbers AE001363.1 and AE002161.1) (41).
TABLE 2.
Oligodeoxyribonucleotide primers
| Primer | Sequencea |
|---|---|
| 3CPn1031H | GCAAGCTTAAATTTTGATTCTTCC |
| 3CPn1031H | GCAAGCTTAATTTTGATTCTTCC |
| 3CPn1031H2 | GCAAGCTTAAATTTTGATTCTTCC |
| 3CPn1031X | GCGCTCGAGTTAAATTTTGATTCTTCCTGTTAAG |
| 3CPn1033Not | GACGCGGCCGCTAAGGAAAGATTTGCACGC |
| 5CPn1031N | GGTCATATGACCTCAAGGACTAAATCC |
| 5CPn1031Nc | GCCCATGGCCTCAAGGACTAAATCC |
| 5CPn1032Nc | CGGCCATGGCTTACGGAACTCG |
| 5CPn1033Nc | GCGCCATGGTATCCTTTCGTTTTCTTTTACTTTC |
| COLIDOWN | TTCACTTCTGAGTTCGGCATG |
| COLIDOWN-RI | CCGAATTCTTCACTTCTGAGTTCGGCATG |
| EcadiAFwd1 | GGAAGATACTTGCCCGCAACGAAGATTCCTTCATAACCGTGTAGGCTGGAGCTGCTTC |
| EcadiAFwd2 | CGAAAAGGCCGGAAGATACT |
| EcadiARev1 | CGTAATGTTATTTAAACAATTACGCCTTCAGCGGAAATTCCGGGGATCCGTCGACC |
| EcadiCDOWN | CAGCGAATCCAGCGGCAG |
| EcadiCUP | CAGTCCTGCGGCTACAAC |
| 5lepAH | GCCAAGCTTCACTGGCTTTGTTGCATTTTGC |
| 3lepAX | GCACTCGAGTTTGTTGTCTTTGCCGACGTG |
| 5ompXH | GCCAAGCTTCGAAGAGTTTCCCATTA |
| 3ompXX | GCACTCGAGGAAGCGGTAACCAACACC |
| pBADMCS2 | GACCATGGATCGATAACCCGGGCTCGAGATCTGCAGCTGG |
| pBAD-Rev | GATTTAATCTGTATCAGGCTG |
| T7-Promoter | GTAATACGACTCACTATAGGG |
| T7-Term | GCTAGTTATTGCTCAGCGG |
Restriction sites are underlined, and the initiator codon is shown in italics.
Two plasmids containing E. coli membrane proteins were constructed to provide markers for chlamydial membrane protein localization. LepA (EF4) is a 67-kDa cytoplasmic membrane-associated ribosomal back translocase (32, 40). The lepA gene with its upstream sequence was amplified from E. coli DNA by using the primers 5lepAH and 3lepAX. The restriction-digested PCR product was ligated to the HindIII and XhoI sites of vector pCDFDuet-1 to produce plasmid pDG561. This multicopy plasmid encodes the LepA sequence fused to a carboxy-terminal S-Tag sequence (Novagen), under the control of the native lepA promoter. OmpX is an 18-kDa outer membrane protein that is overexpressed during chemical stress conditions (8). The ompX gene with its upstream sequence was amplified from E. coli DNA using the primers 5ompXH and 3ompXX. The product was ligated to the HindIII and XhoI sites of the multicopy vector pCOLADuet-1 to produce plasmid pDG552. The pDG552 plasmid encodes the native OmpX fused to a carboxy-terminal S-Tag sequence. Both the pDG561 and the pDG552 plasmids were transformed into E. coli BW25113 to create a marker strain. In addition, the pDG552 plasmid was transformed into E. coli BL21(DE3) to express ompX from a T7 RNA polymerase promoter.
Deletion of E. coli adiAYC genes.
An adiAYC deletion mutation was generated by the gene disruption method of Datsenko and Wanner (6). The ΔadiC1::kan allele from E. coli strain EF1021 was transduced into strain MG1655 using bacteriophage P1vir, producing strain DEG0100. DEG0100 cells were transformed with vector pCP20, and grown under nonpermissive conditions for plasmid replication (37°C). The kanamycin-sensitive (Kans) recombinant strain DEG0124 was screened by PCR using the primers EcadiCUP and EcadiCDOWN, confirming the excision of the FLP-kan cassette. Primers EcAdiAFwd1 and EcAdiARev1 were used to amplify the kanamycin resistance cassette from pKD13 (6). E. coli DEG0124(pKD46) was transformed with the resulting adiA1::kan PCR product. Recombinant strains were selected by growth on LB agar containing kanamycin (25 mg ml−1). The ΔadiAYC::kan allele in a Kanr recombinant (DEG0147) was confirmed by PCR using the primers EcadiAFwd2 and EcadiCDOWN. The purified 1.6-kbp PCR product was sequenced, confirming the deletion.
Protein expression.
Expression vectors were transformed into E. coli strains by electroporation. E. coli strains carrying pBAD/HisA or derivative plasmids were grown aerobically at 37°C for 22 h in LB Miller medium supplemented with ampicillin (100 μg ml−1) and l-arabinose (0.15% [wt/vol]). For protein expression analysis by Western blotting, cells were grown with l-arabinose for 6 h to reduce proteolytic cleavage. E. coli strains carrying pET-43.1a or pTrcHisA derivative plasmids were grown with ampicillin and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside).
Arginine uptake and decarboxylation assays using whole cells.
Approximately 3 × 109 cells were collected by centrifugation at 18,000 × g for 10 min. These cells were suspended in E medium containing 73 mM K2HPO4, 17 mM Na2HPO4, 0.8 mM MgSO4, and 10 mM sodium citrate and adjusted to the indicated pH (52). Suspensions were prewarmed at 37°C, and the reactions were initiated by the addition of 1 mM l-arginine and 20 nCi of l-[U-14C]arginine (305 mCi mmol−1; GE Healthcare) at 37°C for 15 min. Reactions were terminated by the addition of 100 μl of 4 M HCl. These solutions were heated at 70°C for 15 min, releasing 14CO2 that was trapped, and measured by liquid scintillation counting as described previously (12).
Potential inhibitors of arginine transport or decarboxylation were added to whole-cell assays containing 0.5 mM l-arginine and 20 nCi of l-[U-14C]arginine. The arginine analogs screened at 2 mM concentrations were agmatine, l-arginine O-methyl ester, Nα-acetyl l-arginine, NG-nitro-l-arginine methyl ester, d-arginine, l-argininamide, cadaverine, l-citrulline, l-homoarginine, l-histidine, l- lysine, and l-ornithine.
Cell-free arginine decarboxylase assays.
Cells were suspended in 20 mM Tris-HCl buffer (pH 7.5), and lysed by sonication on ice. Centrifugation at 18,000 × g for 15 min removed insoluble particles. Total protein concentrations of these cell extracts were determined using the Bradford assay (Pierce) with bovine serum albumin as a standard. ArgDC activity in cell lysates was determined by using the 14CO2 capture assay described previously (12).
Reaction product analysis.
The unlabeled agmatine produced by transport and arginine decarboxylation reactions was identified by liquid chromatography-electrospray ionization mass spectrometry (LC-MS) as the trifluoroacetyl derivative. A reaction mixture (300 μl) containing 3 × 109 DEG0147(pDG484) cells with 25 mM l-arginine and ammonium acetate buffer (pH 4) was incubated for 2 h at 37°C. Cells were removed by centrifugation, and the solution was evaporated to dryness under nitrogen. Trifluoroacetyl derivatives were prepared using trifluoroacetic anhydride and analyzed by LC-MS in the positive ion mode, as described previously (16). Tandem mass spectra (MS/MS) were acquired by using collision-induced dissociation of the [MH]+ ions. Peaks corresponding to the molecular ions ([MH]+) are shown first, followed by characteristic ion fragments listed in decreasing order of intensity. The acyl derivative of l-arginine eluted at 2.91 min producing peaks at 271 and 251 m/z; MS/MS of the ion at 271 m/z produced peaks at 213, 255, and 230 m/z. The acyl derivative of agmatine eluted between 2.9 and 3.9 min, producing peaks at 227, 169, and 185 m/z; MS/MS of the ion at 227 m/z produced peaks at 169, 211, and 186 m/z.
For radiolabeled product analysis, reaction mixtures (2 ml) contained 109 cells, suspended in E medium at pH 2.5 or 5.0. Mixtures were preincubated at 37°C for 10 min before reactions were initiated by the addition of 1 mM l-arginine and 4 μCi of l-[2,3,4,5-3H]arginine (50 Ci mmol−1; American Radiolabeled Chemicals). Samples were removed immediately after initiation and again after 2 h of incubation: these were centrifuged at 17,000 × g for 5 min to remove cells. Unlabeled agmatine was added to samples as a carrier. Supernatants were applied to a Luna strong cation-exchange high-pressure liquid chromatography (HPLC) column (150 by 4.6 mm, 5 μm; Phenomenex) with a guard column (4 by 3 mm); equilibrated at 35°C in mobile phase buffer containing 50 mM KH2PO4, 100 mM K2SO4, 10% CH3CN, and water; and adjusted to pH 7.2 with phosphoric acid. Isocratic elution with this buffer was used to separate compounds at a flow rate of 1 ml min−1. Fractions (1 ml) were collected, and the radioactivity was determined by liquid scintillation counting. Using this method, arginine eluted at 1.8 min and agmatine eluted at 6.1 min.
Arginine-dependent acid resistance assay.
E. coli DEG0147 cells transformed with the indicated plasmids were grown in Luria-Bertani broth with 0.15% l-arabinose at 37°C. Approximately 3 × 107 E. coli cells in 20 μl of medium were added to 2 ml of E medium (pH 2.5) supplemented with 1.5 mM l-arginine at 37°C. Survival after 1 h of acid shock treatment was determined as described previously (12). The survival rate is the percentage of viable cells detected after 1 h, relative to the number of viable cells detected immediately after the introduction of cells to acid shock medium.
Western blotting.
Protein expression was determined by immunoblotting. Samples were mixed with sodium dodecyl sulfate (SDS) in loading dye but were not boiled. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membranes (0.2 μm, Pall) by using a MiniVE semiwet blotter (GE Healthcare) at 100 mA for 2 h. Prestained protein marker (New England Biolabs) was used to confirm transfer and measure the apparent molecular masses of proteins detected by immunoblotting. The membrane was blocked with 3% bovine serum albumin in Tris-buffered saline containing 0.05% Tween 20. For the analysis of HSV epitope-tagged proteins, blots were incubated with HSV-tag monoclonal antibody (1:2,000 dilution; Novagen), followed by goat anti-mouse immunoglobulin G (IgG) secondary antibody conjugated to horseradish peroxidase (1:2,500 dilution; Pierce) for 1 h at room temperature. The affinity-purified NusA-HSV-His6 protein from E. coli BL21(DE3) (pET-43.1b) cells was used as a positive control. For the analysis of E. coli LepA-S-Tag or OmpX-S-Tag proteins, blots were incubated with a mouse monoclonal antibody raised against the S-Peptide (1:1,000 dilution; Affinity BioReagents), followed by detection with goat anti-mouse IgG as described above. Blots were developed by using a Super Signal West Pico mouse IgG detection kit (Pierce). Chemiluminescence was detected by using an Image Station 4000R instrument (Carestream Health) with Molecular Imaging software (version 4.0).
Membrane fractionation by sucrose-density gradient centrifugation.
E. coli cells were lysed by sonication, and the lysates were cleared of debris by low-speed centrifugation. Membranes were prepared by ultracentrifugation of these samples at 100,000 × g for 1 h at 4°C in a Beckman TLA-100.3 rotor (53). For total membrane protein analysis, the pellets were resuspended in 2% SDS with 20 mM Tris-HCl (pH 7.5). Suspensions were centrifuged at 100,000 × g for 1 h at 4°C. The supernatant was concentrated by using a centrifugal ultrafiltration device (10,000 molecular weight cutoff; Pall Life Sciences). For fractionation by sucrose-density gradient centrifugation, the membrane pellets were resuspended in a solution containing 25% sucrose, 20 mM Tris-HCl, and 0.5 mM EDTA (pH 7.5). This suspension was layered on top of 30, 35, 40, 45, 50, and 55% sucrose layers and centrifuged at 100,000 × g for 6 h at 4°C (53). The visible cytoplasmic membrane layer and the outer membrane layer (inner membrane-depleted layer) were extracted with a syringe, concentrated by centrifugation, and washed three times with 20 mM Tris-HCl (pH 7.5) to remove sucrose and EDTA. Outer membrane fractions were washed with 5 M urea to remove peripheral or aggregated proteins (31).
Cellular transport assays.
A suspension of 3 × 109 E. coli cells was prepared in 0.1 ml of E medium at 37°C. Transport was initiated by adding 1 mM l-arginine-HCl with 2 μCi of l-[2,3,4,5-3H]arginine. After 10 min of incubation, 1 ml of 0.1 M LiCl was added to stop the reaction, and cells were collected by vacuum filtration on a polyethersulfonate membrane filter (0.2 μm, 25-mm diameter; Pall). The filters were washed with 2 ml of 0.1 M LiCl and then removed for liquid scintillation counting.
Phylogenetic analysis.
Amino acid sequences from 24 members of the APA transporter family were aligned by using the T-Coffee program (version 5.57) (38). The full alignment containing 568 positions was analyzed by using the proml program from the Phylip package (J. Felsenstein, University of Washington, version 3.67) to infer a maximum-likelihood phylogeny from this alignment, with the Jones-Taylor-Thornton model of amino acid changes and a γ-distribution of rates (α = 2.4) approximated by three states. Bootstrap analysis was performed with 100 replicates. Complete organism names and sequence accession numbers are listed in the supplemental material.
RESULTS
Expression of chlamydial proteins in E. coli confers arginine uptake and decarboxylase activities.
To characterize the putative chlamydial arginine uptake and decarboxylation system, we constructed E. coli strain DEG0147 by deleting the adi operon, encoding an inducible arginine-dependent acid resistance system. Transformation of E. coli DEG0124(pKD46) with a ΔadiA1::kan PCR product resulted in recombination with the homologous chromosomal adiA sequence and the FLP site of the scar sequence in the ΔadiC1 allele. This strain (DEG0147) had the genotype ΔadiAYC::kan, and it grew normally during aerobic growth on LB medium. Control experiments with whole DEG0147(pBAD/HisA) cells incubated with l-[U-14C]arginine did not release significant amounts of 14CO2 (Fig. 1). Negligible ArgDC activity was detected in lysates of this control strain (<2.1 × 10−5 U mg−1).
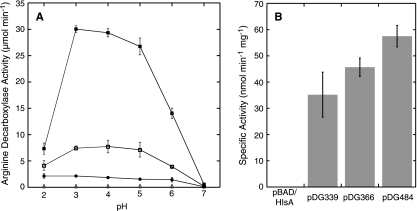
FIG. 1.
E. coli DEG0147 (ΔadiAYC::kan) cells expressing the aaxABC genes transport l-arginine and catalyze its decarboxylation. (A) Reactions containing 3 × 109 cells were incubated with 0.5 mM [U-14C]-l-arginine for 15 min at 37°C in 100 μl of E medium with buffer at the indicated pH. 14CO2 was collected and analyzed as described in Materials and Methods. The DEG0147 strains contained empty vector pBAD/HisA (○), aaxB in pDG339 (•), aaxBC in pDG366 (□), and aaxABC in pDG484 (▪). (B) Cell-free lysates from DEG0147 cells carrying the indicated plasmids were assayed for arginine decarboxylase activity at pH 4. No significant decarboxylase activity was detected in lysates of DEG0147(pBAD/HisA) cells. Error bars in both charts show the standard deviations from the mean of triplicate experiments.
Coexpression of the CPn1032 (aaxB) and CPn1031-HSV (aaxC) genes from the PBAD promoter of plasmid pDG366 in strain DEG0147 increased whole-cell arginine uptake and decarboxylase activity fourfold compared to expression of the aaxB decarboxylase alone (Fig. 1A). This system had maximal activity at pH 4. Similar activities were observed using plasmid pDG379 to produce the untagged proteins. However, expression of all three genes in the C. pneumoniae operon, aaxA (CPn1033), aaxB, and aaxC, in plasmid pDG484 caused a 15-fold increase in the arginine uptake and decarboxylation rate (Fig. 1A). This substantial increase in activity could be due to the transport activities of the AaxA and AaxC proteins, or it could be due to enhanced expression of the AaxB decarboxylase. To test the latter hypothesis, ArgDC activity was determined in cell extracts of each strain (Fig. 1B). Arginine decarboxylase specific activity was 63% higher in the cells expressing all three proteins from pDG484 than in cells expressing only AaxB from pDG339. Therefore, increased AaxB expression does contribute to the increase in whole-cell arginine decarboxylase activity. Yet this increased AaxB activity in cells expressing aaxABC is too low to account for the 1500% increase in whole-cell ArgDC activity, so the AaxA and AaxC transporters must enhance this activity. Whole-cell ArgDC activity did not depend on any components of the E medium buffer. Omitting potassium or sodium ions had no effect on activity, nor did replacing citrate with tartrate buffer. Chloride ions could be replaced by acetate with no loss of activity.
Agmatine production.
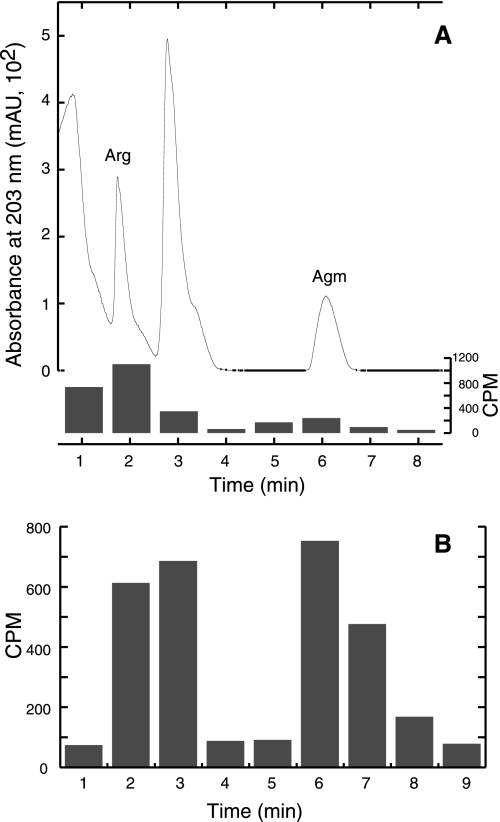
Whole-cell assays showed that E. coli cells expressing the aaxB and aaxC genes from pDG379 released the carboxylate group of radiolabeled arginine as CO2. LC-MS analysis of the extracellular reaction products as the trifluoroacetyl derivatives identified significant amounts of agmatine, as well as residual arginine substrate (data not shown). To quantitatively demonstrate agmatine was exported from these cells, radiolabeled compounds in the supernatant were separated by strong cation exchange HPLC and analyzed by liquid scintillation counting. E. coli DEG0100(pDG379) cells incubated with l-[3H]arginine at pH 2.5 for 2 h released [3H]agmatine, confirming the CPn1032 protein acts an arginine/agmatine antiporter (Fig. 2A). At pH 2.5, DEG0147(pDG379) cells converted 20% ± 3% of the labeled arginine to agmatine. At pH 5.0, the same cells converted 50% of the arginine to agmatine (Fig. 2B). No agmatine was detected in control reactions with DEG0147(pBAD/HisA) cells (data not shown).
FIG. 2.
E. coli cells expressing aaxBC converted tritium-labeled l-arginine to agmatine. (A) Separation of radiolabeled compounds by cation-exchange HPLC from reactions with E. coli DEG0100(pDG379) cells at pH 2.5. Unlabeled agmatine was added as a carrier for product analysis. The chromatogram shows the UV absorbance of arginine (Arg) and agmatine (Agm) correlates with radioactivity (CPM) determined by liquid scintillation counting of 1-ml fractions. Cells expressing the aaxBC genes converted 20% of the l-arginine to agmatine. (B) E. coli DEG0147(pDG379) cells expressing the aaxBC genes converted 50% of the l-arginine to agmatine at pH 5.
Inhibitors of arginine transport and decarboxylation.
We previously showed l-argininamide inhibits the AaxB decarboxylase, and l-canavanine is an alternative substrate for that enzyme (12). A series of arginine analogs was tested to identify inhibitors of the combined arginine uptake and decarboxylation system using the whole-cell decarboxylase assay (Table 3). Compared to reactions containing E. coli DEG0147(pDG366) cells and l-[U-14C]arginine alone at pH 6, the addition of 2 mM l-argininamide had no affect on activity. However, l-argininamide reduced ArgDC activity in lyaste from DEG0147(pDG366) cells. Therefore, l-argininamide does not appear to be transported by the AaxC protein. On the other hand, 2 mM l-canavanine or d-arginine significantly reduced activity, indicating these compounds were imported. Previous studies showed d-arginine had no effect on the purified ArgDC (12), so E. coli crude extract may contain arginine racemase activity, converting d-arginine into l-arginine. An arginine racemase enzyme was previously reported from Pseudomonas graveolens but not sequenced (49). No significant inhibition was observed when 2 mM concentrations of the following analogs were added to the reactions (data not shown): agmatine, l-arginine O-methyl ester, Nα-acetyl l-arginine, cadaverine, l-citrulline, l-homoarginine, NG-nitro-l-arginine methyl ester, l-ornithine, l-histidine, or l-lysine.
TABLE 3.
Inhibitors of arginine uptake and decarboxylation
| Assaya | Relative activity (%) ± SD with added analog
|
|||
|---|---|---|---|---|
| None | l-Argininamide | l-Canavanine | d-Arginine | |
| Whole-cell ArgDC activity (AaxBC) | 100 ± 3 | 92 ± 4 | 30 ± 5 | 81 ± 8 |
| Arginine uptake (AaxC) | 100 ± 1 | 113 ± 15 | 18 ± 2 | 43 ± 0.6 |
| Lysate ArgDC activity (AaxBC) | 100 ± 2 | 74 ± 7 | 6 ± 0.3 | 38 ± 8 |
| Purified AaxB ArgDC activity | 100 | 33 | NDb | 100 |
Whole-cell ArgDC assays included radiolabeled 2 mM l-arginine and E. coli DEG0147(pDG366) cells expressing the aaxBC genes. The mean relative activity values are shown with the associated standard deviations (n = 3). The arginine uptake assays included radiolabeled 1 mM l-arginine and E. coli DEG0147(pDG170) cells expressing the aaxC gene. The mean relative activity values are shown with the associated standard deviations (n = 3). The relative ArgDC activities were measured using cell-free lysates of E. coli DEG0147(pDG366) cells, preincubated with analogs for 5 min before arginine was added to initiate the reactions. The mean relative activity values are shown with the associated standard deviations (n = 3). The relative ArgDC activities using purified C. pneumoniae AaxB protein with 5 mM l-arginine were reported previously (12).
ND, not determined. l-Canavanine was shown to be a substrate for the AaxB protein.
Complementation of E. coli adiAYC with aax genes.
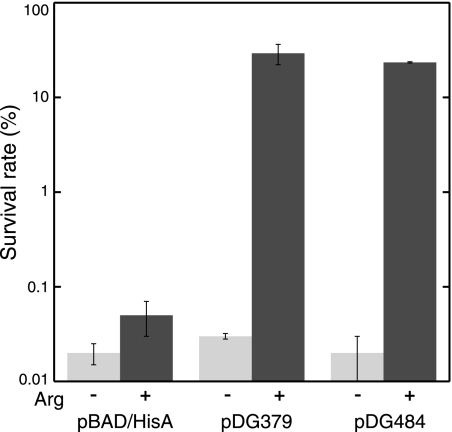
Wild-type E. coli cells resist extreme acid shock by using several pairs of amino acid antiporters and decarboxylases to increase their cytoplasmic pH and invert their membrane potential. Acid shock at pH 2.5 for 1 h reduced E. coli DEG0147(pBAD/HisA) viability by more than 3 orders of magnitude (Fig. 3). When these cells expressed the aaxB and aaxC genes in the presence of 1.5 mM l-arginine, almost 30% of the cells survived treatment. These cells survived acid shock only in the presence of l-arginine. Although coexpression of aaxA substantially increased arginine decarboxylation rates in whole-cell assays, it had no significant effect on survival in these complementation assays.
FIG. 3.
Expression of the C. pneumoniae aaxBC genes restored arginine-dependent acid resistance in E. coli DEG0147. Cells containing pBAD/HisA (vector control), pDG379 (aaxBC), or pDG484 (aaxABC) were incubated in E medium at pH 2.5 for 1 h in the absence (light gray bars) or presence (dark gray bars) of 1.5 mM l-arginine. The mean survival rates and standard deviations are shown for each assay repeated in triplicate.
Heterologous expression of AaxC in the cytoplasmic membrane of E. coli.
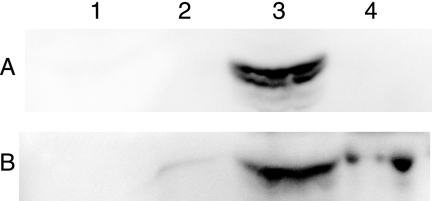
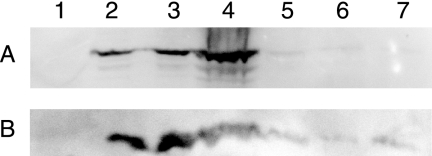
The AaxC protein was predicted to be an integral membrane protein with 12 membrane-spanning α-helices, by the TMHMM program (version 2.0) (27). To monitor heterologous expression, aaxC was fused to a carboxy-terminal HSV epitope tag sequence in vector pDG133. This construct was subcloned behind a Ptrc promoter (in pDG193) and behind a PBAD promoter (in pDG194). Proteins from cell-free lysates or concentrated membrane fractions were separated by SDS-PAGE and analyzed by Western blotting with a monoclonal HSV antibody. No expression was observed in E. coli BL21(DE3) (pDG133) cells or in E. coli DEG0100(pDG183) cells. However, Western blots of E. coli DEG0147(pDG366) and DEG0147(pDG194) extracts identified a new protein with an apparent molecular mass of 41 kDa for AaxC-HSV (56.1 kDa calculated). Many integral membrane proteins migrate anomalously fast during SDS-PAGE, including the E. coli AdiC protein (observed 34 kDa, calculated 46.8 kDa) (10). Fractionation of the cells' total membrane component by sucrose-density gradient centrifugation showed the AaxC-HSV protein was specifically localized to the cytoplasmic membrane (Fig. 4). The AaxC-HSV protein colocalized with the peripheral cytoplasmic membrane marker protein LepA (32) in E. coli BW25113(pDG194, pDG552, pDG561). HSV-tagged proteolytic degradation products with apparent masses of 38, 12, and 5 kDa were also identified in the cytoplasmic membrane fractions (data not shown). No OmpX-S-Tag protein was expressed in this strain (discussed below).
FIG. 4.
Western blots show the AaxC-HSV protein was expressed in the cytoplasmic membrane of E. coli. (A) A 41-kDa band corresponding to AaxC-HSV detected using an anti-HSV monoclonal antibody. (B) A 69-kDa band corresponding to the E. coli LepA-S-Tag protein, detected using an S-peptide monoclonal antibody. The same protein samples were used for both immunoblots. Lane 1 contains the total membrane fraction from E. coli BW25113(pBAD/HisA) cells. Sucrose density-gradient centrifugation was used to separate the total membrane fraction from BW25113(pDG194, pDG552, pDG561) cells into an outer membrane fraction (lane 2), a cytoplasmic membrane fraction (lane 3), and a high-density pellet (lane 4). The LepA protein identified in the high-density pellet may be associated with ribosomes (40). The immunoblot with S-peptide monoclonal antibody also identified a 46-kDa band in the cytoplasmic membrane fraction that may represent a degradation product of LepA (data not shown).
Arginine transport by AaxC.
The activity of the AaxC arginine transporter was analyzed in E. coli DEG0147(pDG170) cells, independent of decarboxylase activity. In the absence of ArgDC activity, this putative arginine/agmatine antiporter was expected to catalyze the exchange of radiolabeled arginine for intracellular arginine as reported for E. coli AdiC (10). Washed cells were mixed with 1 mM l-[3H]arginine for 10 min and then collected by filtration. Maximal transport activity occurred at pH 5 to 6, and no uptake was observed below pH 5 (Fig. 5). As observed in whole-cell ArgDC assays, l-argininamide had no effect on arginine uptake, while 2 mM concentrations of d-arginine and l-canavanine significantly reduced uptake (Table 3). Together with the ArgDC results described above, these transport assays indicate the AaxC protein specifically binds l-arginine, l-canavanine, and d-arginine but discriminates against analogs with less polar substituents.
FIG. 5.
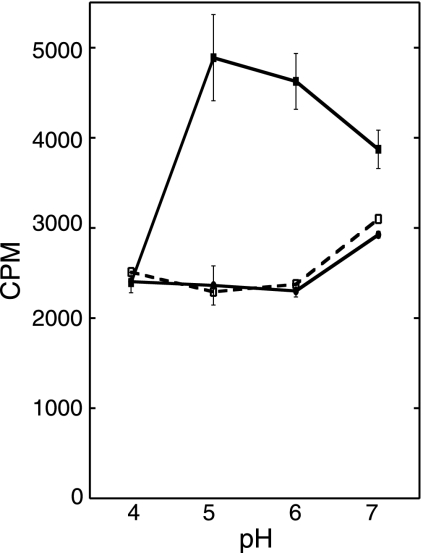
Expression of aaxC promoted arginine uptake. E. coli DEG0147(pDG170) cells expressing aaxC alone optimally transported l-[U-14C]arginine from pH 5 to 6 (▪). Arginine transport by E. coli DEG0147(pBAD/HisA) cells (•) was not significantly different from background levels due to membrane binding of arginine in control reactions without cells (□). The net radioactivity measured in reactions containing DEG0147(pDG170) cells at pH 5 corresponds to 0.4% arginine uptake. No net arginine transport was detected below pH 5 in these cells.
Heterologous expression of AaxA protein in the outer membrane of E. coli.
The AaxA protein belongs to the OprB family of bacterial outer membrane porins. The SignalP program (version 3.0, gram-negative bacterial model) identified an amino-terminal secretory signal peptide in this sequence, predicted to be cleaved between the Ala21 and Glu22 residues (2). An HSV monoclonal antibody specifically bound the AaxA-HSV protein expressed in E. coli DEG0147(pDG512) or E. coli BW25113(pDG512, pDG552, pDG561) cells (data not shown). E. coli BL21(DE3)(pDG512, pDG552) cells coexpressed AaxA-HSV and the E. coli OmpX-S-Tag marker protein in their outer membrane fractions (Fig. 6). The cytoplasmic and outer membranes were separated by sucrose density-gradient centrifugation, and the 44-kDa AaxA-HSV band (49-kDa calculated) was only identified in the outer membrane fraction. Washing the outer membrane fraction with 5 M urea released little epitope-tagged protein, demonstrating the AaxA protein is an integral outer membrane protein, and it is not expressed in an inclusion body of similar density (Fig. 6A) (31). Although no OmpX-S-Tag protein was identified in salicylate-induced BW25113 cells carrying the pDG552 vector, BL21(DE3)(pDG552) cells expressed ompX from a strong T7 RNA polymerase promoter to produce high levels of this protein (Fig. 6B). Most of the heterologously expressed OmpX-S-Tag protein formed insoluble aggregates, as reported previously (39). However, a significant portion of the protein was incorporated into the outer membrane fraction, partially resistant to urea treatment (31). Only trace amounts of OmpX were identified in the cytoplasmic membrane fraction. The copurification of C. pneumoniae AaxA and E. coli OmpX confirmed that AaxA is expressed in the outer membrane.
FIG. 6.
Western blots show the AaxA-HSV protein was expressed in the outer membrane of E. coli. Lane 1 contains total membrane extract from E. coli BW25113(pBAD/HisA) control cells. Lane 2 contains the soluble portion of lysate from BL21(DE3)(pDG512, pDG552) cells expressing C. pneumoniae AaxA-HSV and E. coli OmpX-S-Tag proteins, after centrifugation for 15 min at 18,000 × g. Lane 3 contains the insoluble portion of the lysate from BL21(DE3)(pDG512, pDG552) cells. Lane 4 contains the outer membrane fraction of the same cells, purified by sucrose density-gradient centrifugation and washed with 5 M urea. Lane 5 contains urea-soluble material from washing the outer membrane fraction shown in lane 4. Lane 6 contains the cytoplasmic membrane fraction from sucrose density-gradient centrifugation. Lane 7 contains the pellet from the sucrose density-gradient centrifugation. (A) The AaxA-HSV protein was detected using an anti-HSV monoclonal antibody. (B) The OmpX-S-Tag protein was detected using an anti-S-Tag monoclonal antibody.
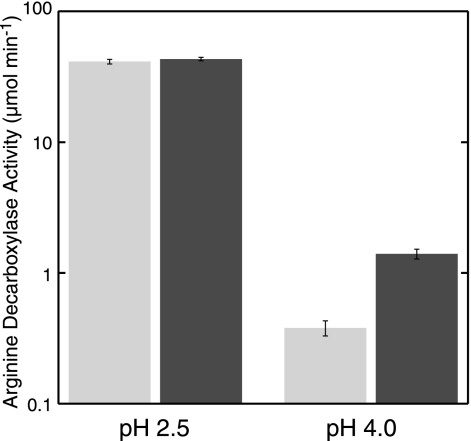
The AaxA protein facilitates arginine uptake with the AaxB and AaxC proteins, so we would expect it to enhance whole-cell arginine decarboxylase activity in wild-type cells expressing the native AdiAYC proteins as well. The E. coli arginine-dependent acid resistance system (AR2) is most active at pH 2.5, but expression of the AaxA protein did not affect ArgDC activity at that pH (Fig. 7). However, at pH 4.0, AaxA expression caused ArgDC activity to increase almost fourfold compared to a control strain. Therefore, AaxA expression enhances arginine uptake in moderately acidic conditions, independent of the AaxB and AaxC proteins.
FIG. 7.
Expression of AaxA stimulates whole-cell arginine decarboxylase activity in wild-type E. coli cells at pH 4. E. coli MG1655 cells (░⃞) or MG1655(pDG512) cells (▪) were incubated with [1-14CO2]-l-arginine at pH 2.5 or pH 4.0, as described in the legend to Fig. 1. The mean values for decarboxylase activity are shown with the associated standard deviations (n = 3).
Evolution of the AaxC arginine/agmatine antiporter.
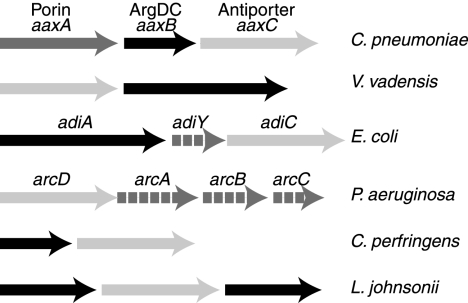
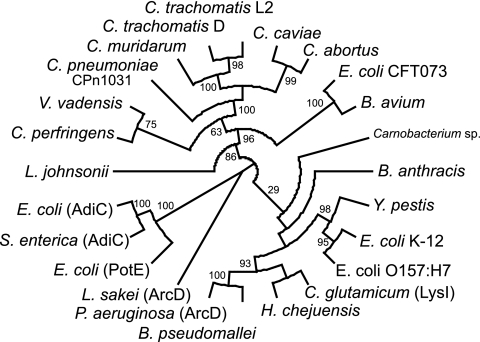
All Chlamydia spp. have operons containing orthologs of the aaxA outer membrane porin, the aaxB arginine decarboxylase, and the aaxC antiporter genes, as shown for C. pneumoniae in Fig. 8. This gene cluster is unique to the Chlamydiales, although most members of the APA family, including AaxC, are associated with amino acid decarboxylases. Phylogenetic analysis of AaxC homologs shows this protein is not specifically related to the functionally equivalent E. coli AdiC arginine/agmatine antiporter (Fig. 9). The most similar homologs of AaxC are found in the anaerobic bacteria Victivallis vadensis and Clostridium perfringens. V. vadensis, a member of the Verrucomicrobia sister group of the Chlamydiales, encodes a transporter homolog adjacent to a PLP-dependent amino acid decarboxylase that is 60% identical to E. coli adiA. The C. perfringens gene is associated with a pyruvoyl-dependent histidine decarboxylase gene for histidine uptake and histamine release (42). In Lactobacillus johnsonii a transporter gene is flanked by two pyruvoyl-dependent phosphatidylserine decarboxylase homologs: the function of this system is unknown. Finally, in P. aeruginosa the arcD gene, encoding a well-characterized arginine/ornithine antiporter, lies in an operon with other genes in the arginine deiminase pathway (51). Members of this family of basic amino acid transporters are intimately associated with decarboxylases or deiminases, but are evolutionarily modular. A variety of gene combinations has evolved through horizontal gene transfer to produce coordinately regulated systems. However, we do not know the physiological functions for most of these systems.
FIG. 8.
Bacterial gene clusters containing aaxC homologs usually include either PLP- or pyruvoyl-dependent amino acid decarboxylase genes (black arrows) and amino acid antiporter genes (light gray arrows). The C. pneumoniae aax operon encodes an arginine-agmatine exchange system consisting of an outer membrane porin (gray arrow, aaxA), a pyruvoyl-dependent arginine decarboxylase (ArgDC, aaxB), and an arginine/agmatine antiporter (aaxC). The Victivallis vadensis cluster encodes putative arginine transporter and PLP-dependent arginine decarboxylase genes. The analogous E. coli adi system is encoded by a PLP-dependent arginine decarboxylase (adiA), a putative transcriptional regulator (dashed line, adiY), and an arginine/agmatine antiporter (adiC) genes. The P. aeruginosa arc operon encodes an arginine fermentative deiminase pathway consisting of an arginine/ornithine antiporter (arcD), arginine deiminase (dashed line, arcA), ornithine carbamoyltransferase (dashed line, arcB), and carbamate kinase (dashed line, arcC) genes. The Clostridium perfringens system encodes a histidine-histamine exchange system composed of a pyruvoyl-dependent histidine decarboxylase and an antiporter. Lactobacillus johnsonii has a gene cluster of unknown function, with two pyruvoyl-dependent phosphatidylserine decarboxylase homologs flanking an antiporter gene.
FIG. 9.
The phylogeny of selected aaxC homologs was inferred by the protein maximum-likelihood method. The C. pneumoniae AaxC protein is more similar to transporter proteins from V. vadensis, C. perfringens, and Bordetella avium than the analogous E. coli AdiC protein. Bootstrap values are indicated for branches supported by a plurality of replicates. The tree construction methods are listed in Materials and Methods, and sequence details and accession numbers are listed in the supplemental material.
DISCUSSION
None of the chlamydial genomes encodes arginine biosynthetic proteins, so the cells must import arginine from their host. In C. pneumoniae, the ArgR repressor regulates transcription of an operon encoding a putative ABC-type arginine transporter (46). Therefore, the arginine utilization system described here should not be required for protein synthesis. Instead, it converts extracellular arginine to agmatine, similar to the E. coli arginine-dependent acid resistance system. However, several aspects of the chlamydial system differ fundamentally from the enteric extreme acid resistance system.
E. coli cells have maximal arginine-agmatine exchange activity near pH 2.5 (15, 20). In E. coli and S. enterica, adiA and adiC expression is enhanced by anaerobic and acidic conditions (26). The C. pneumoniae system reconstituted in E. coli functions over a broad pH range from 3 to 5. Arginine uptake assays suggest this pH optimum reflects the activity of AaxC transporter, rather than the AaxB decarboxylase that functions optimally at pH 3.4 (12). Although the factors determining expression of the chlamydial aax operon are unknown, transcriptional microarray analysis indicated the C. trachomatis L2 homologs CT373 (aaxB) and CT372 (aaxC) were expressed 18 to 36 h postinfection (36). Most metabolic genes were expressed during this period of reticulate body replication. Although aaxB in the L2 strain has a nonsense codon replacing tryptophan at amino acid position 128, this mutation apparently has not stopped transcription of the operon. Future studies will compare the arginine utilization systems in chlamydial cultures to determine the effects of gene inactivation.
l-Lysine, l-ornithine, and cadaverine inhibited arginine transport by the E. coli AdiC protein (10). None of these amino acids interfered with C. pneumoniae AaxC activity, but d-arginine and l-canavanine did inhibit l-arginine transport, indicating different modes of substrate recognition by the transporters. It is unclear whether these inhibitors are productively transported.
The AdiC protein's specificity reflects its evolutionary relationship to a putrescine/ornithine antiporter (Fig. 9). The two systems evolved convergently from different ancestral protein scaffolds. The chlamydial ancestor recruited an archaeal-type pyruvoyl-dependent ArgDC (12) and a transporter from the APA family of antiporters. The enteric system, shared by E. coli and Salmonella enterica, is specifically related to a paralogous acid-inducible system that uses a PLP-dependent ornithine decarboxylase (SpeF) and a putrescine/ornithine antiporter (PotE) (25). Both acid resistance systems probably evolved within the gammaproteobacterial lineage (20).
The aaxA gene is highly conserved among chlamydia, and yet it has low sequence similarity to other putative bacterial porins. The C. pneumoniae AaxA protein shares only 24% sequence identity with its most similar nonchlamydial homolog, a hypothetical protein from Methylobacterium nodulans. This homology extends only over the carboxy-terminal 75% of the AaxA protein. While the AaxA porin is highly diverged from most bacterial outer membrane proteins, its structure probably resembles those of carbohydrate-specific OprB family members (19). Chlamydial cells have at least two other porins, including the highly expressed, broad-specificity major outer membrane protein and the dicarboxylate-specific porin PorB (28).
Stimulation of arginine uptake by the C. pneumoniae AaxA outer membrane protein was unexpected, since E. coli expresses two general porins, OmpC and OmpF, which should permit arginine diffusion into the periplasm. In similar studies of E. coli lysine decarboxylation and cadaverine export, OmpC- and OmpF-mediated permeability was significantly reduced by cadaverine and acidic pH (44). Cadaverine accumulation in the periplasm was proposed to close the OmpF pore, decreasing the outer membrane permeability (37, 45). This inhibition may be advantageous for acid resistance. If agmatine has similar effects on chlamydial porin permeability, then expression of the AaxA porin could help maintain arginine flux into the cell. While expression of AaxA significantly enhanced whole-cell arginine decarboxylase activity in our assay, it had no effect on E. coli's ability to survive acid shock using the chlamydial system. This discrepancy shows that AaxA functions optimally at a neutral or moderately acidic pH, rather than the highly acidic pH of the acid shock assays.
Microbes evolved arginine utilization pathways many times for disparate purposes, suggesting free arginine is commonly available in their environments. Although the chlamydial arginine-agmatine exchange system complements adi mutations in E. coli restoring arginine-dependent acid resistance, the chlamydial system may not have the same function in vivo. The system's relatively high pH optimum, expression profile, and outer membrane protein suggest it could have a more specific effect on the host cell's arginine metabolism. A growing number of pathogens have been shown to consume arginine as a virulence factor against the innate immune response. Arginine uptake regulates macrophage translation of iNOS mRNA, so arginine depletion has the combined effect of reducing iNOS protein abundance and preventing iNOS from converting l-arginine to NO· and l-citrulline (4, 29). The gastric pathogen Helicobacter pylori produces an argininase enzyme, reducing macrophage production of NO· by hydrolyzing l-arginine to urea and l-ornithine (14). P. aeruginosa, a severe respiratory pathogen and a denitrifier, may use nitric oxide reductase to directly counter NO·-mediated toxicity (22). At the same time, these cells encode an active arginine deiminase system, consuming the iNOS substrate. In vivo studies of the chlamydial arginine utilization system will be required to identify the physiological function.
Supplementary Material
Acknowledgments
This study was supported in part by Public Health Service grant AI064444 from the National Institute of Allergy and Infectious Diseases.
We thank Teresa Giles and Gisela Kramer for helpful discussions, and Derek Fisher for sharing unpublished data.
Footnotes
Published ahead of print on 12 September 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adewoye, L. O., L. Tschetter, J. O'Neil, and E. A. Worobec. 1998. Channel specificity and secondary structure of the glucose-inducible porins of Pseudomonas spp. J. Bioenerg. Biomembr. 30257-267. [DOI] [PubMed] [Google Scholar]
- 2.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1462. [DOI] [PubMed] [Google Scholar]
- 4.Chaturvedi, R., M. Asim, N. D. Lewis, H. M. S. Algood, T. L. Cover, P. Y. Kim, and K. T. Wilson. 2007. l-Arginine availability regulates inducible nitric oxide synthase-dependent host defense against Helicobacter pylori. Infect. Immun. 754305-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunin, R., N. Glansdorff, A. Pierard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Mol. Biol. Rev. 50314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dautry-Varsat, A., A. Subtil, and T. Hackstadt. 2005. Recent insights into the mechanisms of Chlamydia entry. Cell. Microbiol. 71714-1722. [DOI] [PubMed] [Google Scholar]
- 8.Dupont, M., C. E. James, J. Chevalier, and J.-M. Pagès. 2007. An early response to environmental stress involves regulation of OmpX and OmpF, two enterobacterial outer membrane pore-forming proteins. Antimicrob. Agents Chemother. 513190-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ekman, M. R., J. T. Grayston, R. Visakorpi, M. Kleemola, C. C. Kuo, and P. Saikku. 1993. An epidemic of infections due to Chlamydia pneumoniae in military conscripts. Clin. Infect. Dis. 17420-425. [DOI] [PubMed] [Google Scholar]
- 10.Fang, Y., L. Kolmakova-Partensky, and C. Miller. 2007. A bacterial arginine-agmatine exchange transporter involved in extreme acid resistance. J. Biol. Chem. 282176-182. [DOI] [PubMed] [Google Scholar]
- 11.Fields, K. A., and T. Hackstadt. 2002. The chlamydial inclusion: escape from the endocytic pathway. Annu. Rev. Cell Dev. Biol. 18221-245. [DOI] [PubMed] [Google Scholar]
- 12.Giles, T. N., and D. E. Graham. 2007. Characterization of an acid-dependent arginine decarboxylase enzyme from Chlamydophila pneumoniae. J. Bacteriol. 1897376-7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giles, T. N., and D. E. Graham. 2008. Crenarchaeal arginine decarboxylase evolved from an S-adenosylmethionine decarboxylase enzyme. J. Biol. Chem. 28325829-25838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gobert, A. P., D. J. McGee, M. Akhtar, G. L. Mendz, J. C. Newton, Y. Cheng, H. L. T. Mobley, and K. T. Wilson. 2001. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc. Natl. Acad. Sci. USA 9813844-13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong, S., H. Richard, and J. W. Foster. 2003. YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J. Bacteriol. 1854402-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham, D. E., and H. K. Huse. 2008. Methanogens with pseudomurein use diaminopimelate aminotransferase in lysine biosynthesis. FEBS Lett. 5821369-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham, D. E., H. Xu, and R. H. White. 2002. Methanococcus jannaschii uses a pyruvoyl-dependent arginine decarboxylase in polyamine biosynthesis. J. Biol. Chem. 27723500-23507. [DOI] [PubMed] [Google Scholar]
- 18.Grieshaber, S., J. A. Swanson, and T. Hackstadt. 2002. Determination of the physical environment within the Chlamydia trachomatis inclusion using ion-selective ratiometric probes. Cell. Microbiol. 4273-283. [DOI] [PubMed] [Google Scholar]
- 19.Hancock, R. E. W., and F. S. L. Brinkman. 2002. Function of Pseudomonas porins in uptake and efflux. Annu. Rev. Microbiol. 5617-38. [DOI] [PubMed] [Google Scholar]
- 20.Iyer, R. K., C. Williams, and C. Miller. 2003. Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J. Bacteriol. 1856556-6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jack, D. L., I. T. Paulsen, and M. H. Saier. 2000. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 1461797-1814. [DOI] [PubMed] [Google Scholar]
- 22.Kakishima, K., A. Shiratsuchi, A. Taoka, Y. Nakanishi, and Y. Fukumori. 2007. Participation of nitric oxide reductase in survival of Pseudomonas aeruginosa in LPS-activated macrophages. Biochem. Biophys. Res. Commun. 355587-591. [DOI] [PubMed] [Google Scholar]
- 23.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21385-389. [DOI] [PubMed] [Google Scholar]
- 24.Karunakaran, K. P., J. Rey-Ladino, N. Stoynov, K. Berg, C. Shen, X. Jiang, B. R. Gabel, H. Yu, L. J. Foster, and R. C. Brunham. 2008. Immunoproteomic discovery of novel T-cell antigens from the obligate intracellular pathogen Chlamydia. J. Immunol. 1802459-2465. [DOI] [PubMed] [Google Scholar]
- 25.Kashiwagi, K., S. Shibuya, H. Tomitori, A. Kuraishi, and K. Igarashi. 1997. Excretion and uptake of putrescine by the PotE protein in Escherichia coli. J. Biol. Chem. 2726318-6323. [DOI] [PubMed] [Google Scholar]
- 26.Kieboom, J., and T. Abee. 2006. Arginine-dependent acid resistance in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1885650-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305567-580. [DOI] [PubMed] [Google Scholar]
- 28.Kubo, A., and R. S. Stephens. 2001. Substrate-specific diffusion of select dicarboxylates through Chlamydia trachomatis PorB. Microbiology 1473135-3140. [DOI] [PubMed] [Google Scholar]
- 29.Lee, J., H. Ryu, R. J. Ferrante, S. M. Morris, Jr., and R. R. Ratan. 2003. Translational control of inducible nitric oxide synthase expression by arginine can explain the arginine paradox. Proc. Natl. Acad. Sci. USA 1004843-4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, J., I. S. Lee, J. Frey, J. L. Slonczewski, and J. W. Foster. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 1774097-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marani, P., S. Wagner, L. Baars, P. Genevaux, J.-W. De Gier, I. Nilsson, R. Casadio, and G. Von Heijne. 2006. New Escherichia coli outer membrane proteins identified through prediction and experimental verification. Protein Sci. 15884-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.March, P. E., and M. Inouye. 1985. GTP-binding membrane protein of Escherichia coli with sequence homology to initiation factor 2 and elongation factors Tu and G. Proc. Natl. Acad. Sci. USA 827500-7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClarty, G. 1999. Chlamydial metabolism as inferred from the complete genome sequence, p. 69-100. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, DC.
- 34.Morris, D. R., and A. B. Pardee. 1966. Multiple pathways of putrescine biosynthesis in Escherichia coli. J. Biol. Chem. 2413129-3135. [PubMed] [Google Scholar]
- 35.Moulder, J. W. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Mol. Biol. Rev. 55143-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicholson, T. L., L. Olinger, K. Chong, G. Schoolnik, and R. S. Stephens. 2003. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J. Bacteriol. 1853179-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302205-217. [DOI] [PubMed] [Google Scholar]
- 39.Pautsch, A., J. Vogt, K. Model, C. Siebold, and G. E. Schulz. 1999. Strategy for membrane protein crystallization exemplified with OmpA and OmpX. Proteins Struct. Funct. Genet. 34167-172. [PubMed] [Google Scholar]
- 40.Qin, Y., N. Polacek, O. Vesper, E. Staub, E. Einfeldt, D. N. Wilson, and K. H. Nierhaus. 2006. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell 127721-733. [DOI] [PubMed] [Google Scholar]
- 41.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 281397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Recsei, P. A., W. M. Moore, and E. E. Snell. 1983. Pyruvoyl-dependent histidine decarboxylases from Clostridium perfringens and Lactobacillus buchneri. Comparative structures and properties. J. Biol. Chem. 258439-444. [PubMed] [Google Scholar]
- 43.Richard, H., and J. W. Foster. 2004. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J. Bacteriol. 1866032-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samartzidou, H., and A. H. Delcour. 1999. Excretion of endogenous cadaverine leads to a decrease in porin-mediated outer membrane permeability. J. Bacteriol. 181791-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samartzidou, H., M. Mehrazin, Z. Xu, M. J. Benedik, and A. H. Delcour. 2003. Cadaverine inhibition of porin plays a role in cell survival at acidic pH. J. Bacteriol. 18513-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaumburg, C. S., and M. Tan. 2006. Arginine-dependent gene regulation via the ArgR repressor is species specific in Chlamydia. J. Bacteriol. 188919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schramm, N., C. R. Bagnell, and P. B. Wyrick. 1996. Vesicles containing Chlamydia trachomatis serovar L2 remain above pH 6 within HEC-1B cells. Infect. Immun. 641208-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scidmore, M. A. 2006. Chlamydial exploitation of host signaling, cytoskeletal, and membrane trafficking pathways, p. 255-295. In P. M. Bavoil and P. B. Wyrick (ed.), Chlamydia: genomics and pathogenesis. Horizon Bioscience, Norfolk, United Kingdom.
- 49.Soda, K., T. Yorifuji, and K. Ogata. 1967. Occurrence of arginine racemase in bacterial extract. Biochim. Biophys. Acta 146606-608. [DOI] [PubMed] [Google Scholar]
- 50.Thomson, N. R., M. T. G. Holden, C. Carder, N. Lennard, S. J. Lockey, P. Marsh, P. Skipp, C. D. O'Connor, I. Goodhead, H. Norbertzcak, B. Harris, D. Ormond, R. Rance, M. A. Quail, J. Parkhill, R. S. Stephens, and I. N. Clarke. 2008. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 18161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhoogt, H. J., H. Smit, T. Abee, M. Gamper, A. J. Driessen, D. Haas, and W. N. Konings. 1992. arcD, the first gene of the arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa, encodes an arginine-ornithine exchanger. J. Bacteriol. 1741568-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 21897-106. [PubMed] [Google Scholar]
- 53.Ward, A., N. M. Sanderson, J. O'Reilly, N. G. Rutherford, B. Poolman, and P. J. G. Henderston. 2000. The amplified expression, identification, purification, assay, and properties of hexahistidine-tagged bacterial membrane transport proteins, p. 141-166. In S. A. Baldwin (ed.), Membrane transport: a practical approach. Oxford University Press, New York, NY.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.