Abstract
The consumption of lactate and amino acids is very important for microbial development and/or aroma production during cheese ripening. A strain of Yarrowia lipolytica isolated from cheese was grown in a liquid medium containing lactate in the presence of a low (0.1×) or high (2×) concentration of amino acids. Our results show that there was a dramatic increase in the growth of Y. lipolytica in the medium containing a high amino acid concentration, but there was limited lactate consumption. Conversely, lactate was efficiently consumed in the medium containing a low concentration of amino acids after amino acid depletion was complete. These data suggest that the amino acids are used by Y. lipolytica as a main energy source, whereas lactate is consumed following amino acid depletion. Amino acid degradation was accompanied by ammonia production corresponding to a dramatic increase in the pH. The effect of adding amino acids to a Y. lipolytica culture grown on lactate was also investigated. Real-time quantitative PCR analyses were performed with specific primers for five genes involved in amino acid transport and catabolism, including an amino acid transporter gene (GAP1) and four aminotransferase genes (ARO8, ARO9, BAT1, and BAT2). The expression of three genes involved in lactate transport and catabolism was also studied. These genes included a lactate transporter gene (JEN1) and two lactate dehydrogenase genes (CYB2-1 and CYB2-2). Our data showed that GAP1, BAT2, BAT1, and ARO8 were maximally expressed after 15 to 30 min following addition of amino acids (BAT2 was the most highly expressed gene), while the maximum expression of JEN1, CYB2-1, and CYB2-2 was delayed (≥60 min).
Substrate supply and consumption are very important for microbial growth during cheese ripening. Cheese curd is a complex matrix containing carbon and/or nitrogen sources whose nature and availability play significant roles in the ripening process (15). Lactate, resulting from the conversion of lactose by lactic acid bacteria (LAB), and amino acids resulting from casein proteolysis are the main carbon and/or nitrogen sources necessary for microbial growth and aroma production (10, 21, 22).
Yeasts have an essential role in cheese curd deacidification, a prerequisite for the development of acid-sensitive bacteria on the cheese surface (1, 6, 14). The phenomena leading to deacidification still need to be investigated but are probably related to amino acid and/or lactate catabolism (1, 8). Microorganisms such as Yarrowia lipolytica that develop on the cheese surface constitute an adventitious microflora from the cheese environment (brine, ripening shelves, and personnel), and they rapidly outnumber the commercial cultures (16). Y. lipolytica is a ubiquitous, naturally developing yeast (6, 7) that comprises part of the surface flora. Its adventitious nature and its enzymatic activities make it a good candidate for ripening. Due to its efficient productivity, this yeast has been used for the preparation of cheese flavor compounds (5). Y. lipolytica produces many more volatile sulfur compounds (VSC) than conventional cheese-ripening yeasts, such as Debaryomyces hansenii and Kluveromyces lactis (4, 5).
Amino acid catabolism has been studied in several cheese-ripening yeasts, including Y. lipolytica, Geotrichum candidum, K. lactis, and D. hansenii, with respect to VSC biosynthesis. The degradation of amino acids is initiated by an aminotransferase in which the amino group of an amino acid is transferred to an α-keto acid (e.g., α-ketoglutarate), resulting in the formation of the corresponding amino acid (e.g., glutamate) and keto acids, which are subsequently degraded to flavor compounds (21, 22). It has been found that in Y. lipolytica l-methionine degradation via transamination, by a branched-chain aminotransferase (BAT1), is involved in VSC formation (4). In Saccharomyces cerevisiae, ARO8 expression is subject to general amino acid biosynthesis control. ARO9 expression is induced when aromatic amino acids are present in the growth medium and also in ARO8 mutants grown on minimal ammonia medium (11).
Lactate catabolism by yeasts surely plays a central role during cheese ripening since it coincides with a dramatic increase in the pH at the cheese surface, as shown for D. hansenii, K. lactis, and Kluveromyces marxianus (1, 14). D. hansenii assimilates lactose and lactate, while G. candidum and Y. lipolytica assimilate lactate but not lactose (2). Enzymes and related degradation pathways involved in lactate catabolism by cheese-ripening yeasts have not been studied much. However, we hypothesized that lactate dehydrogenase-type enzymes are possible lactate catabolic enzymes. Nevertheless, the expression patterns of target genes related to l-methionine and lactate/lactose catabolism have been investigated in several cheese-ripening yeasts. The results revealed that D. hansenii and K. marxianus were implicated mainly in lactose and lactate catabolism, whereas Y. lipolytica preferentially consumed l-methionine (5).
This study focused on the catabolism of lactate and amino acids by Y. lipolytica. The first part of this work was devoted to growth and obtaining biochemical data related to amino acid and/or lactate consumption by this yeast. In the second part, the expression patterns of several target genes involved in amino acid and lactate catabolism in Y. lipolytica were studied following addition of amino acids.
MATERIALS AND METHODS
Yeast strain and storage conditions.
The microorganism used in this work was Y. lipolytica 1E07. This strain, originally isolated from Livarot French cheese, was obtained from the UMR-GMPA laboratory collection and was selected because of its biotechnological potential. The yeast was stored in 5% glycerol-nonfat dry milk at −80°C until it was used.
Culture conditions.
The yeast was cultivated in a 500-ml flask containing 100 ml of medium, and a preculture was grown in potato dextrose broth (Difco Laboratories, Detroit, MI) for 1 day at 25°C with agitation (150 rpm). The potato dextrose broth was inoculated with 1 ml of a thawed stock suspension. The preculture served as the inoculum for the subsequent culture. A defined synthetic medium (SM), adapted from the medium described by Otto et al. (18), was used for all culture conditions. It contained 48 components, including lactate and lactose as principal carbon sources, 19 free amino acids, 14 vitamins, five metallic ions, and four nucleic acid bases (Table 1). The pH of the culture medium was adjusted to 6.7. Two amino acid concentrations were used (0.1× and 2×) in the presence and in the absence of lactate. The total amino acid concentrations found in a Camembert cheese ranged from 4 to 50 g kg−1 (12), depending on the amino acid. These concentrations evolved during ripening. No data describing the evolution of free amino acids are available. We assumed that 0.01% (low amino acid concentration) to 10% (high amino acid concentration) of the total amino acids could be released following the microbial degradation of casein. A culture grown in the presence of lactate but without amino acids was also used. Cultures were incubated at 25°C and 150 rpm for either 42 or 56 h. The SM was selected because it contained substrates that allowed us to study lactate and amino acid catabolism. The concentration of lactate and lactose used was chosen because it corresponded to the maximum amount of substrates present in the curd of a soft cheese like Camembert (14).
TABLE 1.
Composition of complete defined SMa
| Constituent | Concn (g liter−1) |
|---|---|
| Lactose | 20 |
| Sodium lactate (60%) | 18 |
| Sodium acetate | 1 |
| Ammonium citrate | 0.6 |
| KH2PO4 | 9 |
| K2HPO4 | 7.5 |
| MgCl2·6H2O | 0.2 |
| FeCl2·4H2O | 0.011 |
| CaCl2·2H2O | 0.050 |
| ZnCl2 | 0.005 |
| CoCl2·6H2O | 0.0025 |
| p-Aminobenzoic acid | 0.010 |
| Biotin | 0.010 |
| Cyanocobalamin | 0.001 |
| Folic acid | 0.001 |
| Inosine | 0.005 |
| Nicotinic acid | 0.001 |
| Orotic acid | 0.005 |
| Calcium pantothenate | 0.001 |
| Pyridoxamine | 0.005 |
| Pyridoxine | 0.002 |
| Riboflavin | 0.001 |
| Thiamine | 0.001 |
| dl-6,8-Thioctic acid | 0.0025 |
| Thymidine | 0.005 |
| Adenine | 0.010 |
| Guanine | 0.010 |
| Uracil | 0.010 |
| Xanthin | 0.010 |
| l-Alanine | 0.24 |
| l-Arginine | 0.12 |
| l-Asparagine | 0.34 |
| l-Glutamine | 0.51 |
| l-Glycine | 0.17 |
| l-Histidine | 0.11 |
| l-Isoleucine | 0.20 |
| Leucine | 0.47 |
| l-Lysine | 0.35 |
| l-Methionine | 0.12 |
| Proline | 0.68 |
| l-Serine | 0.34 |
| Threonine | 0.23 |
| Tryptophan | 0.05 |
| Valine | 0.33 |
| Glutamate | 0.68 |
| Phenylalanine | 0.28 |
| Tyrosine | 0.29 |
| Cysteine | 0.17 |
See reference 18.
Microbial and substrate analyses.
Viable cell counts, expressed in CFU ml−1, were determined by using a standard aerobic plate count procedure. One milliliter of a yeast culture was mixed with 9 ml of physiological saline (9 g liter−1 NaCl), and then 10-fold serial dilutions were prepared and plated on 120-mm-diameter petri dishes; these dishes contained a glucose chloramphenicol agar (Biokar Diagnostics, Paris, France) to ensure immediate differentiation of the colonies based on their size, appearance, and pigmentation and to detect any microbial contamination. Colonies were counted after incubation for 2 days at 25°C.
The amino acids and ammonia production were analyzed using the ninhydrin method previously described by Grunau and Swiader (9).
Lactate was quantified by performing high-performance liquid chromatography (Waters TCM; Waters, Saint Quentin en Yvelines, France) with a cation-exchange column (diameter, 7.8 mm; length, 300 mm; Aminex HPX-87H; Bio-Rad, Ivry-sur-Seine, France) using a thermostat set at 35°C. The culture supernatants were filtered using a polyethersulfone membrane filter (pore size, 0.22 μm; diameter, 25 mm). The mobile phase was 0.01 N sulfuric acid at a flow rate of 0.6 ml min−1. Compounds of interest were detected with a Waters 486 tunable UV/visible detector regulated at 210 nm. All compounds were quantified by using calibration curves established with pure chemicals.
Extraction and purification of total RNA.
Cultures were centrifuged for 5 min at 8,200 × g and 4°C. Each pellet was resuspended in 1.25 ml of Trizol reagent (Invitrogen, Cergy Pontoise, France) and poured into a 2-ml tube containing 800 mg zirconium beads (diameters, 0.1 and 0.5 mm; Biospec Products, Bartlesville, OK). The tubes were vigorously shaken in a bead beater (Fast-Prep-24; MP Biomedicals, France) by using three 60-s mixing sequences at a speed of 6.5 m s−1. They were cooled on ice for 5 min before each mixing sequence. After centrifugation for 10 min at 12,000 × g and 4°C, each supernatant was collected. It was transferred to a 2-ml tube (Phase Lock Gel Heavy; Eppendorf, Hamburg, Germany), and 230 μl of chloroform was added. The tubes were gently mixed by inversion and centrifuged for 15 min at 12,000 × g and 4°C. Each aqueous phase was transferred to a fresh tube, and an equal volume of phenol-chloroform-isoamyl alcohol (pH 4.7) (Sigma) was added. The tubes were gently mixed by inversion and centrifuged for 10 min at 12,000 × g and 4°C. Then the upper phase in each tube was collected. An equal volume of 100% ethanol was added to the aqueous phase, after which purification with an RNeasy kit (Qiagen, Courtaboeuf, France) was performed according to the manufacturer's instructions. RNA samples were treated with DNase using a DNase Turbo DNA-free kit (Ambion, Austin, TX). RNA quality and quantity were analyzed using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and an Agilent 2100 bioanalyzer (Agilent, Palo Alto, CA).
Real-time PCR conditions.
The RNA extraction and purification procedures used are described above. cDNAs were synthesized using the SuperScript III First-Strand synthesis system (Invitrogen). A mixture containing up to 5 μg of total RNA, oligo(dT)20 (50 μM), and deoxynucleoside triphosphates (10 mM) was prepared and incubated at 65°C for 5 min, and then it was placed on ice for at least 1 min. A cDNA synthesis mixture containing 10× reverse transcription buffer, MgCl2 (25 mM), dithiothreitol (0.1 M), RNaseOUT (40 U μl−1), and SuperScript III reverse transcriptase (200 U μl−1) was added to each RNA-primer mixture and then incubated for 50 min at 50°C. The reaction was stopped by incubation for 5 min at 85°C.
The primers used for real-time PCR were designed so that they were about 20 to 25 bases long, had a G+C content of over 50%, and had a melting temperature of about 60°C. The lengths of the PCR products ranged from 90 to 150 bp. LightCycler software (Roche, Mannheim, Germany) was used to select primer sequences. All of the primers were synthesized by Eurogentec (Seraing, Belgium) (Table 2).
TABLE 2.
Primers used in this study
| Primer | Accession no. | Sequence (5′-3′) | Putative functiona |
|---|---|---|---|
| GAP1-R | YALI0B16522g | CGACCACAGCAATGACTTTAATA | Amino acid transporter |
| GAP1-F | YALI0B16522g | AACTACTGGAATGAAGCTAACG | |
| ARO8-R | YALI0E20977g | GGCTCCGACCCAGTTGT | Aromatic aminotransferase |
| ARO8-F | YALI0E20977g | TTCTCCTCCGCCATCGAGTG | |
| ARO9-R | YALI0C05258g | GGTTGGGAAGAGCTCCAGAGAT | Aromatic aminotransferase |
| ARO9-F | YALI0C05258g | ACGACAAGTTCATTTCTGACCGTT | |
| BAT1-R | YALI0D01265g | GTTGGCTCCCAGCTTCTTGT | Branched-chain amino acid aminotransferase |
| BAT1-F | YALI0D01265g | CTCTCGGCGTCGGAACC | |
| BAT2-R | YALI0F19910g | TCCAACGGCCTTGGAGTTCT | Branched-chain amino acid aminotransferase |
| BAT2-F | YALI0F19910g | CCTCAAGCTCTACTGCTCCGA | |
| JEN1-R | YALI0D20108g | TTAATGTGAGCGTCACAGATATCAC | Organic acid transporter |
| JEN1-F | YALI0D20108g | AGCTCCAGCACAATAAATAGAACAC | |
| CYB2-1-R | YALI0D12661g | TCTTCTTCTTCGGCGGCAATCT | Lactate dehydrogenase |
| CYB2-1-F | YALI0D12661g | GCATCCTGGAGGACAGGCTA | |
| CYB2-2-R | YALI0E21307g | CTCGGCGGCGTTGTCAAT | Lactate dehydrogenase |
| CYB2-2-F | YALI0E21307g | GACACTCCCTACCTACCTCGGAAACCGATA | |
| ACT2-R | YALI0D08272gb | GGCCAGCCATATCGAGTCGCA | Gene encoding actin |
| ACT2-F | YALI0D08272gb | TCCAGGCCGTCCTCTCCC |
The annotations are Génolevures annotations (http://cbi.labri.fr/Genolevures/).
See reference 3.
SYBR green I PCR amplification was performed using a LightCycler (Roche). Amplification was carried out by using a 10-μl (final volume) mixture containing 250 ng of RNA sample, 4 mM MgCl2, 0.5 μM primer, and 1 μl of LightCycler-FastStart DNA Master SYBR green I (Roche). Five dilutions of cDNA were prepared to determine the efficiencies of real-time PCR. A negative control without cDNA was systematically included. The amplification procedure involved incubation at 95°C for 8 min for initial denaturation, followed by 45 cycles consisting of (i) denaturation at 95°C for 10 s, (ii) annealing at a temperature that was 5°C below the melting temperature of the primers for 7 s, (iii) extension at 72°C for 6 s, and (iv) fluorescence acquisition (530 nm) at the end of extension. The temperature transition rate was 20°C/s for each step. After real-time PCR, a melting curve analysis was performed by continuously measuring fluorescence during heating from 65 to 95°C at a transition rate of 0.1°C/s. The cycle threshold values were determined with LightCycler software (version 3.3), using the second-derivative method. Standard curves were generated by plotting the cycle threshold values as a function of the log initial RNA concentration. PCR efficiency (E) was then calculated using the following formula: E = 10−1/slope. The actin gene (3) was used as a suitable internal control gene to normalize the results. The Pffafl method (19) was used to calculate the changes in transcript abundance normalized to the actin gene and relative to the sample collected before the pulse.
RESULTS
Growth and substrate assimilation by Y. lipolytica. (i) Yeast growth and pH over time.
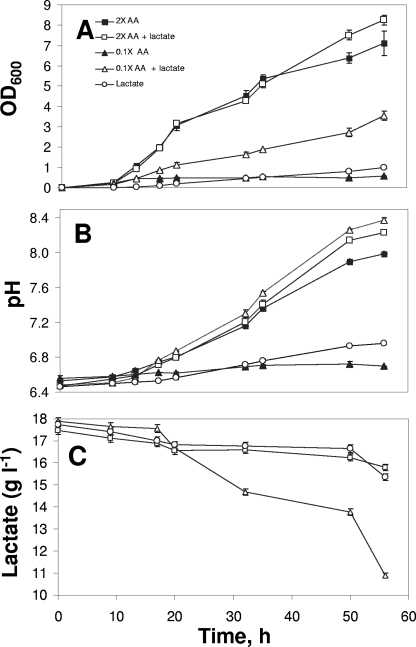
Y. lipolytica 1E07 was cultivated in SM containing amino acids at two concentrations (0.1× and 2×) in the presence and in the absence of lactate. A fifth culture condition was medium containing lactate but no amino acids. The growth of Y. lipolytica under the five conditions is shown in Fig. 1A. Yeast growth was more effective in SM containing the high concentration of amino acids, regardless of the presence of lactate; the concentrations of the yeast at 56 h were 8.25 × 107 CFU ml−1 in the presence of lactate and 7.11 × 107 CFU ml−1 in the absence of lactate. The increase in the yeast concentration was accompanied by a dramatic increase in the pH, from pH 6.5 at the beginning of the culture to pH 8 and 8.2 after 56 h (Fig. 1). In contrast, limited growth and a limited increase in the pH were observed in SM containing 0.1× amino acids or lactate. Limited growth was observed at 56 h in SM containing lactate (107 CFU ml−1) or 0.1× amino acids (6 × 106 CFU ml−1). A dramatic increase in yeast growth (3.3 × 107 CFU ml−1) was observed when 0.1× amino acids and lactate were supplied together. On the basis of these data, it appears that deacidification was highly correlated with lactate and/or amino acid degradation.
FIG. 1.
Growth (A), increase in the pH (B), and lactate consumption (C) of Y. lipolytica 1E07 in SM containing lactate and/or amino acids. AA, amino acids; OD600, optical density at 600 nm.
(ii) Lactate utilization.
Lactate consumption data are shown in Fig. 1C. The yeast grew well in SM containing lactate and 0.1× amino acids and consumed a large amount of lactate (7 g liter−1 of lactate), while growth was significantly reduced in SM containing only lactate, in which this carbon source was poorly consumed (1.5 g liter−1 of lactate). Moreover, only 2 g liter−1 of lactate was utilized in the presence of 2× amino acids. When lactate was provided, some of it was always consumed by Y. lipolytica, but it was not totally depleted.
(iii) Amino acid utilization.
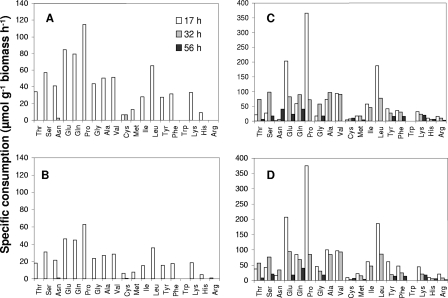
The overall amino acid consumption rates are shown in Fig. 2. Amino acids were used during the growth of Y. lipolytica 1E07, regardless of their initial concentration. The amino acid utilization correlated well with the increase in the biomass. In SM containing 0.1× amino acids with or without lactate, amino acids could be limiting for the growth of Y. lipolytica since all of the amino acids were depleted at 32 h (Fig. 2A and 2B). In medium supplemented with the low level of amino acids, amino acids were more efficiently consumed without lactate (Fig. 2A and 2B), in contrast to the results for medium supplemented with 2× amino acids, in which lactate had no effect on the amino acid consumption rate (Fig. 2C and 2D).
FIG. 2.
Overall rates of consumption of amino acids by Y. lipolytica 1E07 cultivated in SM containing low and high concentrations of amino acids. (A) 0.1× amino acids. (B) 0.1× amino acids plus lactate. (C) 2× amino acids. (D) 2× amino acids plus lactate.
In SM containing 2× amino acids (regardless of the presence of lactate), amino acids were not limiting since they were not exhausted at the end of the exponential phase, although they were efficiently consumed as an energy source for the growth of Y. lipolytica (the amino acids were totally consumed after 56 h). Regardless of the amino acid concentration, proline, glutamate, leucine, and glutamine were the amino acids that were most efficiently consumed by Y. lipolytica.
(iv) Ammonia production.
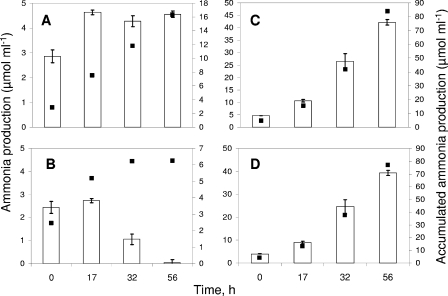
The ammonia production and accumulated production are shown in Fig. 3. Ammonia production was correlated with amino acid degradation. In SM with 0.1× amino acids (Fig. 3A), ammonia production increased at 17 h during culture and was stable up to 56 h. In SM containing 0.1× amino acids and lactate, ammonia production decreased after 32 h, which coincided with total depletion of amino acids and the onset of lactate consumption. In SM containing amino acids at the high concentration, ammonia production increased during yeast growth and there was not a significant difference between growth in the presence of lactate and growth in the absence of lactate. The results for production of ammonia were consistent with extensive amino acid degradation (Fig. 2C, 2D, 3C, and 3D).
FIG. 3.
Ammonia production (bars) by Y. lipolytica 1E07 cultivated in SM containing low and high concentrations of amino acids. (A) 0.1× amino acids. (B) 0.1× amino acids plus lactate. (C) 2× amino acids. (D) 2× amino acids plus lactate. The accumulated ammonia production is indicated by filled squares.
(v) Analyses of expected and actual cell yields.
In SM containing only lactate, the sole nitrogen source available, ammonium citrate, supported a maximum yield of about 0.72 g liter−1 of cells, which essentially corresponded to the measured biomass (Table 3). Assuming that nitrogen is used to make only biomass and that the cell yield on lactate is about 50%, this suggests that there was only enough nitrogen present to support the utilization of 1.5 g liter−1 of lactate. The actual amount of lactate consumed, 2.5 g liter−1, may be explained by nitrogen limitation.
TABLE 3.
Expected and actual cell yields after 56 h for Y. lipolytica grown under various culture conditionsa
| Culture conditions | Substrate consumed (g liter−1)
|
Theoretical biomass provided by the nitrogen sources available (g liter−1)
|
Total theoretical biomass (g liter−1) | Theoretical biomass provided by the carbon sources available (g liter−1)
|
Total theoretical biomass (g liter−1) | Measured biomass (g liter−1) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino acids | Ammonium citrate | Lactate | Amino acids | Ammonium citrate | Amino acids | Ammonium citrate | Lactate | ||||
| Lactate | 0.6 | 2.5 | 0.72 | 0.72 | 0.26 | 1.25 | 1.51 | 0.61b | |||
| 0.1× amino acids | 0.568 | 0.6 | 0.91 | 0.72 | 1.63 | 0.24 | 0.26 | 0.50 | 0.36c | ||
| 0.1× amino acids + lactate | 0.568 | 0.6 | 7 | 0.91 | 0.72 | 1.63 | 0.24 | 0.26 | 3.5 | 4 | 2.19b |
| 2× amino acids | 11.36 | 0.6 | 18.18 | 0.72 | 18.9 | 4.77 | 0.26 | 5.03 | 4.40c | ||
| 2× amino acids + lactate | 11.36 | 0.6 | 2 | 18.18 | 0.72 | 18.9 | 4.77 | 0.26 | 1 | 6.03 | 5.12c |
The yields for theoretical calculation of biomass were 10% for nitrogen and 50% for carbon.
Nitrogen limitation.
Carbon limitation.
The same analysis could be performed for SM supplemented only with 0.1× amino acids. Calculation of theoretical cell yields showed that there should have been sufficient nitrogen to support production of 1.63 g liter−1 of cells (where 0.72 g liter−1 of cells came from ammonium citrate and 0.91 g liter−1 of cells came from 0.1× amino acids) and sufficient carbon to support 0.5 g liter−1 of cells (where 0.24 g liter−1 of cells came from 0.1× amino acids and 0.26 g liter−1 of cells came from citrate), which essentially corresponded to the actual cell mass. This result shows that there was probably carbon limitation.
In SM containing 0.1× amino acids and lactate, the nitrogen available should have supported production of about 1.63 g liter−1 of cells, and the actual yield was 2.19 g liter−1 of cells (Table 3). The theoretical biomass yield on carbon is 4 g liter−1 of cells. The results showed that there was nitrogen limitation and that at least some lactate was converted to something besides cell mass. In addition, the production of ammonia indicated that not all the nitrogen was used to make cell mass.
The media containing 2× amino acids without and with lactate should have contained enough nitrogen to support production of about 18 g liter−1 of cell mass. However, the available carbon was limited and could support theoretical cell mass yields of only 5.03 and 6.03 g liter−1, respectively. Since the actual cell mass yields were 4.4 and 5.12 g liter−1, respectively, it was clear that lactate was poorly consumed to make biomass when a large amount of amino acids was provided. The observed lactate consumption essentially corresponded to the excess biomass produced (1 g liter−1) in the presence of 2× amino acids plus lactate compared to the biomass produced in the presence of only 2× amino acids (Table 3).
Reverse transcription-PCR analysis after an amino acid pulse in a culture of Y. lipolytica 1E07 cultivated on lactate.
We suspect that amino acids are the molecules primarily used by Y. lipolytica, rather than lactate. In order to better understand the complementary catabolism, a pulse experiment in which 0.1× amino acids was added to a batch culture of Y. lipolytica containing lactate as the carbon source was conducted. Total RNA was extracted before the pulse (after 42 h of culture) and then 15, 30, and 60 min later. At 42 h, the lactate consumption was steady and the cell concentration was 5 × 106 CFU ml−1. Reverse transcription-PCR and real-time quantitative PCR were then performed with primers specific for genes involved in amino acid and lactate catabolism. Four aminotransferase genes and one amino acid transporter-encoding gene (ARO8, ARO9, BAT1, BAT2 and GAP1) were selected as markers for amino acid catabolism. Two lactate dehydrogenase genes and one lactate transporter-encoding gene (CYB2-1, CYB2-2, and JEN1) were chosen as markers for lactate catabolism.
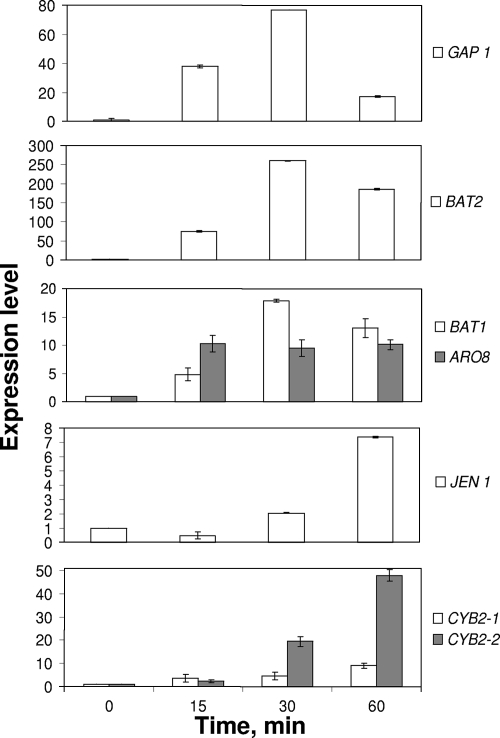
The levels of expression of these genes are shown in Fig. 4. Fifteen minutes after addition of 0.1× amino acids, the levels of GAP1, BAT1, BAT2, and ARO8 expression dramatically increased. Thirty minutes after amino acid addition, the levels of expression were 78-fold greater for GAP1, 18-fold greater for BAT1, and 250-fold greater for BAT2, and 60 min after the pulse the values were 17-, 14-, and 180-fold greater, respectively. Furthermore, the level of expression of ARO8 was 10-fold greater 15 min after the amino acid pulse and remained stable thereafter. ARO9 was not expressed, even following amino acid addition. The levels of expression of JEN1, CYB2-1, and CYB2-2 increased considerably after the amino acid pulse. However, the expression of these genes was delayed compared to the expression of aminotransferase genes and the amino acid transporter GAP1 gene, since expression was induced most after 60 min (7-, 10-, and 50-fold increases, respectively).
FIG. 4.
Levels of expression of the GAP1, BAT2, BAT1, ARO8, JEN1, CYB2-1, and CYB2-2 genes, as measured by real-time PCR after an amino acid pulse (0.1× amino acids), for a Y. lipolytica 1E07 pure culture grown in SM containing lactate.
Five hours after amino acid addition, the levels of expression of all aminotransferase and lactate dehydrogenase genes were significantly lower than the levels of expression 60 min after amino acid addition (data not shown).
DISCUSSION
In this work, we were able to identify complementary roles of lactate and amino acids in the yeast Y. lipolytica. These molecules are major carbon and/or nitrogen sources for microbial growth and aroma production during cheese ripening. At the beginning of the cheese-ripening process, lactose is rapidly transformed into lactate by the LAB. After a few days of ripening, lactate is the most important carbon source available in the cheese curd. In addition, lactose, which may not be converted to lactate by LAB, is not assimilated by Y. lipolytica (2). Therefore, lactate catabolism in this yeast was investigated. Amino acid degradation, including degradation of aromatic amino acids (phenylalanine, tyrosine, and tryptophan), branched amino acids (leucine, isoleucine, and valine), or methionine, has been shown to play an important role in aroma compound production during cheese ripening (11). However, the possible use of amino acids as an energy source for growth has never been fully investigated for any cheese-ripening yeast, although amino acids could play an important role in microbial development during the ripening process. The competitive consumption of lactate and amino acids was investigated using the yeast Y. lipolytica 1E07, which originated from cheese. In this yeast, amino acids are degraded first, and lactate is consumed only after total amino acid depletion. This suggests that amino acids are likely to be the main nitrogen and carbon sources for growth of Y. lipolytica and that a high amino acid concentration probably has a negative effect on lactate catabolism. Since there is redundancy of genes encoding high-affinity amino acid permeases in the Y. lipolytica genome, we suspect that such amino acid transporters may provide a competitive advantage to Y. lipolytica for efficient growth on amino acids compared with other cheese-ripening yeasts, which primarily use lactate and/or lactose, as suggested previously for methionine (5). Furthermore, it has been reported that lactate is not consumed in cheeses inoculated with Y. lipolytica (used as the sole yeast) in association with bacteria (17, 20).
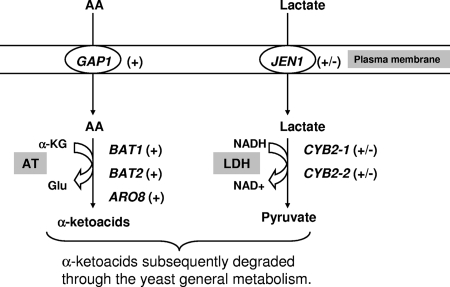
The expression of several genes involved in amino acid catabolism was investigated (Fig. 5). Our data show that the expression of GAP1, BAT2, BAT1, and ARO8 was highly induced 15 to 30 min following addition of amino acids. Amino acids are probably transported through the amino acid transporter encoded by GAP1 into the cell, where the aminotransferase reactions are activated. Among the aminotransferase-encoding genes, BAT2 is by far the most highly expressed, an indication of the major role of BAT2 in amino acid transamination. It has recently been reported that the levels of expression of AGP1 (which encodes an amino acid permease with a low affinity and a broad spectrum) and MUP1 (involved in l-methionine transport) were increased following addition of amino acids (after 4 h) in S. cerevisiae during alcoholic fermentation (13).
FIG. 5.
Possible relationship between amino acid catabolism (AA) and lactate catabolism in Y. lipolytica 1E07. +, early induction of the genes in italics; +/−, delayed induction of the genes following addition of amino acids. AT, amino transferase; LDH, lactate dehydrogenase; α-KG, α-ketoglutarate; Glu, glutamate. Gene functions are shown in Table 2.
The lactate transporter gene JEN1 and two lactate dehydrogenase-encoding genes, CYB2-1 and CYB2-2, which may be involved in lactate catabolism, were also studied (Fig. 5). The maximum levels of expression of JEN1, CYB2-1, and CYB2-2 were delayed (≥60 min) compared to the expression of genes involved in amino acid catabolism.
The pH increased considerably in the medium containing the low concentration of amino acids plus lactate, while the pH increased similarly regardless of the presence of lactate in SM containing the high concentration of amino acids. Furthermore, the pH variations observed with Y. lipolytica in SM (around 2 pH units) are comparable to what was observed by other workers for various cheese environments (for instance, in a cheese agar medium [8] or in a model cheese [17]). It was shown previously that lactate was not degraded in cocultures when Y. lipolytica was the sole yeast, while the pH increased significantly, and that Y. lipolytica produced large amounts of ammonia (17). The increase in the pH was attributed to amino acids that could control ammonia release in yeast colonies (13). The possible role of ammonia in the increase in the pH on the cheese surface has been studied by measuring ammonia production in single colonies of different yeast species, including D. hansenii, G. candidum, and Y. lipolytica (8). On cheese agar, the greatest increase in pH was observed around colonies of Y. lipolytica that efficiently produced and accumulated ammonia. A 10-fold increase in ammonia production was observed with Y. lipolytica compared to the ammonia production with the other yeasts (8). Our data suggest that amino acid and lactate catabolism has a dual effect on the deacidifying capacities of Y. lipolytica, in contrast to what was observed for other yeasts of cheese origin, in which deacidification is believed to coincide with lactate degradation (1). Furthermore, it was shown that ammonia produced by colonies essentially originated from amino acid catabolism (23).
This is the first study of Y. lipolytica in which lactate catabolism and amino acid catabolism were studied at the level of gene expression. The results show for the first time that lactate catabolism and amino acid catabolism complement each other with respect to growth and that amino acids are degraded first. They also show that both amino acid degradation and lactate degradation by Y. lipolytica actively participate in deacidification, which is very important for the development of acid-sensitive bacteria during cheese ripening. Ammonia production by yeasts may therefore be taken into consideration as an additional technological parameter when starter cultures are selected for surface-ripened cheeses.
Acknowledgments
Soulaf Mansour is grateful to the ABIES Doctoral School for providing a Ph.D. scholarship.
We thank J. Bailly and C. Monnet for helpful discussions. We also thank J. Colle and P. Courtin for their excellent technical assistance.
Footnotes
Published ahead of print on 5 September 2008.
REFERENCES
- 1.Arfi, K., M. N. Leclercq-Perlat, H. E. Spinnler, and P. Bonnarme. 2005. Importance of curd-neutralising yeasts on the aromatic potential of Brevibacterium linens during cheese ripening. Int. Dairy J. 15:883-891. [Google Scholar]
- 2.Barnett, J. A., R. W. Payne, and D. Yarrow. 2000. Yeasts: characteristics and identification. Cambridge University Press, Cambridge, United Kingdom.
- 3.Blanchin-Roland, S., G. D. Costa, and C. Gaillardin. 2005. ESCRT-I components of the endocytic machinery are required for Rim101-dependent ambient pH regulation in the yeast Yarrowia lipolytica. Microbiology 151:3627-3637. [DOI] [PubMed] [Google Scholar]
- 4.Bondar, D. C., J.-M. Beckerich, and P. Bonnarme. 2005. Involvement of a branched-chain aminotransferase in production of volatile sulfur compounds in Yarrowia lipolytica. Appl. Environ. Microbiol. 71:4585-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cholet, O., A. Henaut, S. Casaregola, and P. Bonnarme. 2007. Gene expression and biochemical analysis of cheese-ripening yeasts: focus on catabolism of l-methionine, lactate, and lactose. Appl. Environ. Microbiol. 73:2561-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corsetti, A., J. Rossi, and M. Gobbetti. 2001. Interactions between yeasts and bacteria in the smear surface-ripened cheeses. Int. J. Food Microbiol. 69:1-10. [DOI] [PubMed] [Google Scholar]
- 7.Cosentino, S., M. E. Fadda, M. Deplano, A. F. Mulargia, and F. Palmas. 2001. Yeasts associated with Sardinian ewe's dairy products. Int. J. Food Microbiol. 69:53-58. [DOI] [PubMed] [Google Scholar]
- 8.Gori, K., H. D. Mortensen, N. Arneborg, and L. Jespersen. 2007. Ammonia production and its possible role as a mediator of communication for Debaryomyces hansenii and other cheese-relevant yeast species. J. Dairy Sci. 90:5032-5041. [DOI] [PubMed] [Google Scholar]
- 9.Grunau, J. A., and J. M. Swiader. 1992. Chromatography of 99 amino acids and other ninhydrin-reactive compounds in the Pickering lithium gradient system. J. Chromatogr. A 594:165-171. [Google Scholar]
- 10.Iraqui, I., S. Vissers, B. Andre, and A. Urrestarazu. 1999. Transcriptional induction by aromatic amino acids in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:3360-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iraqui, I., S. Vissers, M. Cartiaux, and A. Urrestarazu. 1997. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol. Gen. Genet. 257:239-248. [DOI] [PubMed] [Google Scholar]
- 12.Ireland, J., J.-C. Favier, and M. Feinderg. 2002. Répertoire générale des aliments. Tome 2. Produits laitiers, 2nd ed. INRA-Editions TEC & DOC, Paris, France.
- 13.Jimenez-Marti, E., and M. l. del Olmo. 2008. Addition of ammonia or amino acids to a nitrogen-depleted medium affects gene expression patterns in yeast cells during alcoholic fermentation. FEMS Yeast Res. 8:245-256. [DOI] [PubMed] [Google Scholar]
- 14.Leclercq-Perlat, M. N., A. Oumer, J. L. Bergere, H. E. Spinnler, and G. Corrieu. 2000. Behavior of Brevibacterium linens and Debaryomyces hansenii as ripening flora in controlled production of smear soft cheese from reconstituted milk: growth and substrate consumption dairy foods. J. Dairy Sci. 83:1665-1673. [DOI] [PubMed] [Google Scholar]
- 15.McSweeney, P. L. H., and M. J. Sousa. 2000. Biochemical pathways for the production of flavour compounds in cheeses during ripening: a review. Lait 80:293-324. [Google Scholar]
- 16.Mounier, J., S. Goerges, R. Gelsomino, M. Vancanneyt, K. Vandemeulebroecke, B. Hoste, N. M. Brennan, S. Scherer, J. Swings, G. F. Fitzgerald, and T. M. Cogan. 2006. Sources of the adventitious microflora of a smear-ripened cheese. J. Appl. Microbiol. 101:668-681. [DOI] [PubMed] [Google Scholar]
- 17.Mounier, J., C. Monnet, T. Vallaeys, R. Arditi, A.-S. Sarthou, A. Helias, and F. Irlinger. 2008. Microbial Interactions within a cheese microbial community. Appl. Environ. Microbiol. 74:172-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otto, R., B. ten Brink, H. Veldkamp, and W. N. Konings. 1983. The relationship between growth rate and electrochemical proton gradient of Streptococcus cremoris. FEMS Microbiol. Lett. 16:69-74. [Google Scholar]
- 19.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sinigaglia, M., R. Lanciotti, and M. E. Guerzoni. 1994. Biochemical and physiological characteristics of Yarrowia lipolytica strains in relation to isolation source. Can. J. Microbiol. 40:54-59. [DOI] [PubMed] [Google Scholar]
- 21.Smit, G., B. A. Smit, and W. J. M. Engels. 2005. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 29:591-610. [DOI] [PubMed] [Google Scholar]
- 22.Yvon, M., and L. Rijnen. 2001. Cheese flavour formation by amino acid catabolism. Int. Dairy J. 11:185-201. [Google Scholar]
- 23.Zikanova, B., M. Kuthan, M. Ricicova, J. Forstova, and Z. Palkova. 2002. Amino acids control ammonia pulses in yeast colonies. Biochem. Biophys. Res. Commun. 294:962-967. [DOI] [PubMed] [Google Scholar]